Significance
To navigate large cargo through the viscous cytoplasm, cells use a variety of energy-consuming machines that, akin to ropewalkers, move on intracellular “tracks” in a stepwise “bipedal” fashion. To prevent waste of energy by futile walking, several control mechanisms have evolved. In the case described here for one group of yeast myosins, crystallographic and biophysical analyses revealed that a single myosin molecule associates with an intertwined middle region of two “adaptor” molecules. The adaptor also contains distinct binding sites for cargo. Only when cargo is attached to the myosin-bound adaptor are two of the myosin–adaptor complexes joined into a pair, akin to converting a uniped (unable to walk) into a biped (able to walk).
Keywords: in vitro assembly, cytoplasmic mRNA localization, endoplasmic reticulum inheritance
Abstract
Myosin 4 protein (Myo4p), one of five distinct myosins of yeast, is dedicated to cytoplasmic transport of two types of cargos, zipcoded messenger ribonucleoprotein particles (mRNPs) and tubular endoplasmic reticulum (tER). Neither cargo binds directly to Myo4p. Instead, swi5p-dependent HO expression 3 protein (She3p) serves as an “adaptor” that contains three binding modules, one for Myo4p and one each for zipcoded mRNP and tER. The assembly of a transport-competent motor complex is poorly understood. Here, we report that Myo4p•She3p forms a stable 1:2 heterotrimer in solution. In the Myo4p•She3p crystal structure, Myo4p’s C-terminal domain (CTD) assumes a lobster claw-shaped form, the minor prong of which adheres to a pseudocoiled-coil region of She3p. The extensive Myo4p•She3p interactome buries 3,812 Å2 surface area and is primarily hydrophobic. Because the Myo4p•She3p heterotrimer contains only one myosin molecule, it is not transport-competent. By stepwise reconstitution, we found a single molecule of synthetic oligonucleotide (representing the mRNA zipcode element) bound to a single tetramer of zipcode binding protein She2p to be sufficient for Myo4p•She3p dimerization. Therefore, cargo initiates cross-linking of two Myo4p•She3p heterotrimers to an ensemble that contains two myosin molecules obligatory for movement. An additional crystal structure comprising an overlapping upstream portion of She3p showed continuation of the pseudocoiled-coil structure and revealed another highly conserved surface region. We suggest this region as a candidate binding site for a yet unidentified tER ligand. We propose a model whereby zipcoded mRNP and/or tER ligands couple two Myo4p•She3p heterotrimers and thereby generate a transport-competent motor complex either for separate transport or cotransport of these two cargos.
The mRNA transport to specific regions in the cytoplasm, followed by localized translation, is an important means to establish cellular asymmetry. The mRNAs to be localized are each marked by one (or several) cis-acting elements, referred to as zipcodes. Cognate zipcode-binding proteins mediate assembly of zipcoded mRNA into a motor complex. In general, actin-based motors are used for short-distance mRNA transport whereas microtubule-based motors are used for long-range transport (see reviews in refs. 1 and 2).
Zipcoded mRNA transport in the budding yeast (Saccharomyces cerevisiae) is among the most extensively studied of the myosin-based mRNA transport systems. More than twenty zipcoded mRNAs are removed from the yeast mother-cell cytoplasm and transported through the bud neck to the daughter cell (3). Transport proceeds along the few actin filaments that in mitotic yeast cells stretch for about 2 μm from minus ends in the mother-cell cytoplasm, across the bud neck, to plus ends near the tip of the daughter cell (4, 5). The founding member of this family of zipcoded mRNAs is ASH1 mRNA that codes for a transcriptional repressor in the daughter cell’s mating-type switch (4, 6). ASH1 mRNA happens to contain four modular zipcodes, termed E1, E2a, E2b, and E3 (7–10). These zipcode elements are up to 100 nucleotides long and share predicted features in their structure (9, 10).
The components of the motor complex for the zipcoded mRNA transport system in yeast were identified some time ago (11): (i) Myosin 4 protein (Myo4p) (also known as She1p), one of five distinct myosin heavy chains in yeast (each Myo4p “lever arm” portion is predicted to be associated with five identical light chains); (ii) She2p, a tetramer that specifically binds to mRNA zipcode (12–15); and (iii) swi5p-dependent HO expression 3 protein (She3p), a multimodule “adaptor” protein that serves as linker between She2p and the C-terminal domain (CTD) of Myo4p (16, 17). She4p is another member of the “She family”; it forms a homodimer that interacts with the motor domain of Myo4p (18).
The zipcode region of nascent mRNA associates cotranscriptionally with She2p, as well as other, more generic mRNA binding proteins (19). Some of the cotranscriptionally recruited mRNA-binding proteins remain mRNA-associated even after extensive protein stripping mediated by helicase at the cytoplasmic surface of the nuclear pore (20). Once in the cytoplasm, a zipcoded and She2p-marked messenger ribonucleoprotein particle (mRNP) associates with Myo4p and She3p, an assembly process that yields a transport-competent motor complex and enables “processive” movement along actin filaments from the mother to the daughter cell (21, 22). Although crystal structures of fragments of some components of the yeast motor complex have been reported (12, 17), to date, no structural analyses of any of the hetero oligomeric interactomes of this yeast multiprotein ensemble have been carried out. Consequently, neither the mechanism of assembly, nor its regulation, or even its stoichiometry, are well understood.
As Myo4p belongs to a class of intrinsically monomeric myosins (23, 24), as long as it is not assembled into a complex containing two Myo4p molecules, it is not capable of moving on an actin cable. The proposed mechanisms by which an intrinsically monomeric Myo4p might be assembled to yield such a transport-competent complex containing two Myo4p molecules are controversial. In one model, two Myo4p molecules and an unidentified number of She3p molecules were proposed to interact with each other to yield a complex [e.g., see figure 9 of Muller et al. (25)]. In this model, neither zipcode RNA nor She2p was directly involved in generating a cytoplasmic Myo4p•She3p complex containing two myosin molecules. Another model proposed that a dedicated helical region downstream from the lever arm of Myo4p interacts with a helical region of She3p to form a heterocoiled coil, i.e., a 1:1 Myo4p•She3p heterodimer (13, 16, 26). To obtain a transport-competent motor complex with two Myo4p molecules, two such Myo4p•She3p heterodimers had to be linked together. Initially, it was reported that only She2p tetramer, i.e., without added zipcode RNA, was sufficient to carry out coupling of two Myo4p•She3p heterodimers and thereby to generate a motor complex with the obligatory two Myo4p molecules (13). However, in an in vitro reconstitution experiment (concurrent to our work) carried out at physiological salt concentration, both She2p and zipcoded mRNA were reported to be required for the assembly of a transport-competent motor complex. Although the stoichiometry of the assembled components was not determined, it was assumed to contain two Myo4p•She3p heterodimers (26).
She3p contains at least three binding modules, one for Myo4p and two for cargos, as diverse as zipcoded mRNP and tubular endoplasmic reticulum (tER) (27–29). She3p’s binding site for Myo4p has been localized to the middle region of She3p (30), with a genetically determined upstream binding site for tER and a downstream binding site for zipcoded mRNP (29). Recently, the C-terminal region of She3p has also been reported to contain an RNA-binding site (25, 31). The modular arrangement of two distinct cargo-binding sites, one for mRNP and the other one for tER, would permit simultaneous transport of these two cargos. Indeed, coupled transport of zipcoded mRNPs and tER has been reported although cotransport appears not to be obligatory (29, 32).
To resolve the question of how Myo4p and She3p interact with each other, we carried out biochemical, biophysical, and crystallographic studies. Contrary to the proposal (13, 16, 26) that Myo4p•She3p form a 1:1 heterodimer, our data show that Myo4p and She3p associate instead in a 1:2 heterotrimer. Specifically, we found that a pseudocoiled-coil region of two She3p molecules forms an extensive binding platform for the C-terminal domain (CTD) of a single Myo4p molecule. Notably, Myo4p displays two distinct domains, which are separated in the Myo4p primary structure by 264 residues but, in the quaternary structure, are located adjacent to each other. In addition, we carried out stepwise in vitro reconstitution at physiological salt concentrations and monitored assembly by size-exclusion chromatography and multiangle light scattering (MALS). These experiments used the CTD (residues 918–1,471) of Myo4p, full-length She3p, full-length She2p, and/or synthetic oligonucleotides that represented zipcodes of ASH1 mRNA E2b, E3, or of a shortened E3. We found that Myo4p•She3p heterotrimers, when incubated by themselves, do not form higher-order complexes; this finding ruled out a model according to which a complex comprising two Myo4p and an undetermined number of She3p molecules preexists in the cytoplasm (25). Moreover, we showed that in vitro incubation of the 1:2 Myo4p•She3p heterotrimers with She2p tetramers yielded only unstable higher-order complexes. However, when Myo4p•She3p heterotrimers were incubated with She2p together with synthetic oligonucleotide (representing either E2b or E3 zipcode of ASH1 mRNA), a stable complex of Myo4p•She3p•She2p•zipcode in a 2:4:4:1 stoichiometry was formed. These results suggest that two Myo4p•She3p heterotrimers are cooperatively “cross-linked” by a single She2p tetramer bound to a single synthetic oligonucleotide representing a zipcode. These results contradict a model in which heterodimers of Myo4p•She3p are the cytoplasmic starting material for the assembly of higher-order complexes (26) or that She3p is sufficient for Myo4p dimerization (26). In further support of the crucial role of zipcode in the assembly of stable higher-order complexes, we found that a shortened zipcode (E3-33), replacing “full-length” (E3-51) zipcode, yielded only formation of an unstable higher-order complex.
Results
Myo4p and She3p Form 1:2 Heterotrimer.
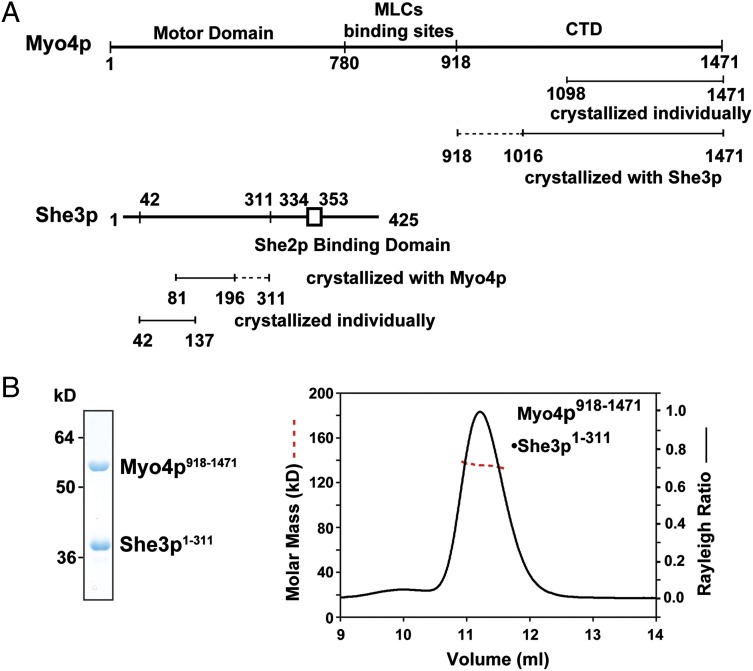
We coexpressed various fragments (“protomers”) of Myo4p (containing a cleavable tag) and She3p in bacteria and characterized pull-downs by SDS/PAGE (Fig. 1 and Fig. S1). In close agreement with previously reported data (17, 33), we found that She3p residues 148–197 and Myo4p residues 1,021–1,471 (Fig. S1B) form likely borders of interacting regions. One of these pull-downs, Myo4p918-1471•She3p1-311, was further characterized by size-exclusion chromatography coupled to MALS. The observed molar mass, 134.9 kDa, of this complex suggested that it formed a 1:2 heterotrimer consisting of one molecule of Myo4p and two molecules of She3p. These data were unexpected as these two molecules were thought to form a 1:1 heterodimer (26).
Fig. 1.
Boundaries and stoichiometry of the Myo4p-She3p interactome. (A) Domain organization of Myo4p and She3p is indicated by lines and was independently assessed by pull-down experiments (Fig. S1). Fragments (“protomers”) of Myo4p and She3p, crystallized either individually or as a complex, are shown as a straight line with dashed line extensions representing N- or C-terminally disordered regions in the crystal (Figs. S2A and S3A); CTD, C-terminal domain. (B) Analyses of a purified Myo4p918-1471•She3p1-311 complex by SDS/PAGE and Coomassie Blue staining (Left) and by size-exclusion chromatography coupled to MALS (Right) (100 µg, 2 mg/mL). The Rayleigh ratio (right ordinate, black line) and the molar mass distribution (left ordinate, red dashed line) indicate a molar mass of 134.9 ± 1.4 kDa, which is close to a theoretical molar mass of 135.5 kDa for a heterotrimeric Myo4p•She3p complex in a 1:2 ratio.
Structure of Myo4p•She3p.
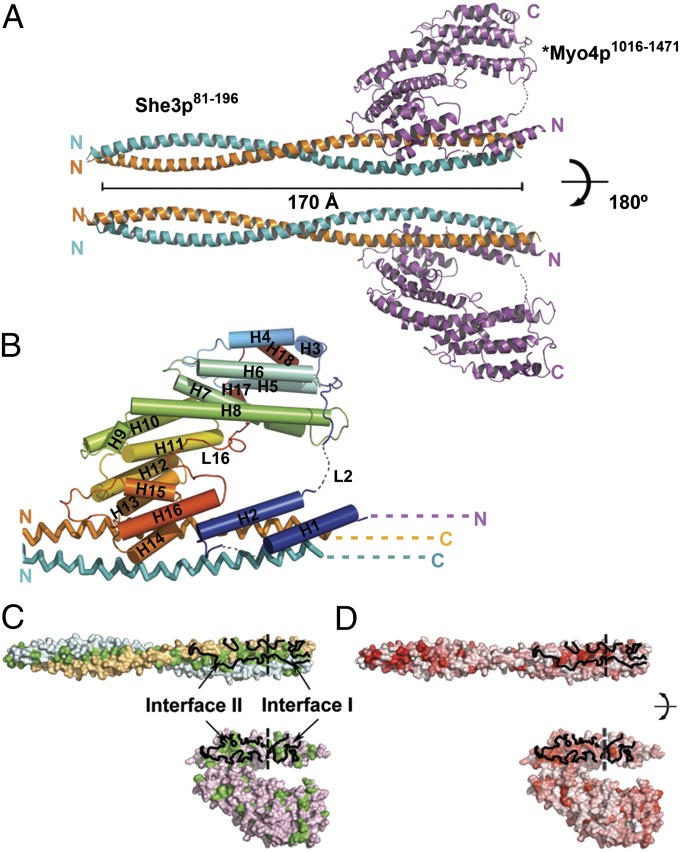
We obtained crystals of suitable diffraction quality from a complex that contained a slightly altered Myo4p918-1471 protomer, *Myo4p918-1471 (for a description, see Fig. S2A), and a protomer of She3p81-311. Importantly, *Myo4p918-1471 retained its in vivo trafficking activity (Fig. S2 C–E) indicating that, at least by this criterion, the introduced changes did not affect its function. The crystal structure of *Myo4p918-1471•She3p81-311 was refined to final Rwork and Rfree values of 22.4% and 26.8%, respectively, to a resolution of 3.6 Å (Table S1 and Materials and Methods) (PDB ID code 4LL8). The asymmetric unit of the crystal contains one *Myo4p protomer and two She3p protomers, consistent with the stoichiometry of our MALS measurements of protomers of similar size (Fig. 1B). Residues 197–311 of the She3p homodimers are not visible in the crystal and are presumed disordered (indicated by dashed lines in Figs. 1A and 2B and Fig. S2B). She3p (residues 81–196) forms a 170-Å-long pseudocoiled coil (Fig. 2A) with numerous hydrophobic residues at the buried interface of 5,930 Å2 (Fig. 2C and Fig. S3A). Notably, Cys-147 forms a disulfide bond between the two protomers of She3p (Fig. S4A). This disulfide bond may help in stabilizing posttranslational alignment of the two chains during its parallel assembly. The N-terminal halves of the two She3p protomers form a standard coiled coil (rmsd of 1.5 Å when superimposing residues 81–147). Interestingly, the C-terminal halves deviate slightly from an ideal coiled coil (rmsd of 1.6 Å when superimposing residues 148–196), thereby opening up a pseudocoiled-coil interface to expose an extensive hydrophobic surface for binding of *Myo4p (Fig. 2C, Upper). The *Myo4p918-1471 protomer interacts with the C-terminal third of the She3p81-196 pseudocoiled coil. *Myo4p residues 918–1,015 (Fig. S2A) and small regions of loops L1 and L2 of *Myo4p918-1471 were not visible and therefore are presumed disordered (indicated by dashed lines in Fig. 1, Fig. 2 A and B, and Fig. S2A). The remainder of *Myo4p (residues 1,016–1,471) consists of helices (H) and connecting loops (L) (Fig. 2B). The convoluted path of secondary structural elements starts with a helical hairpin (H1 and H2), changes direction via a long and partially disordered loop (L2), flows into a series of irregularly stacked helices (H3–H16), and reverses direction again with a long loop (L16) so that the last two helices (H17 and H18) wind up in the vicinity of H3 and H4 (Fig. 2B and Fig. S2A). In a surface representation, the crystallized *Myo4p protomer displays the shape of a “lobster-claw” with a minor prong (H1, H2, and H12–H16) and a major prong (H3–H11, H17, and H18) (Fig. 2 C and D).
Fig. 2.
Crystal structure of the 1:2 Myo4p•She3p heterotrimer. Crystals were obtained with a mutated Myo4p fragment, termed *Myo4p918-1471 (Fig. S2A). (A) Ribbon representation of the 1:2 *Myo4p•She3p complex, rotated by 180° in Upper and Lower. Two copies of She3p81-196 (yellow and blue) form a parallel pseudocoiled coil that measures 170 Å in length; *Myo4p1016-1471 (magenta) folds into a convoluted path of helices (H) and connecting loops (L) with the overall shape of lobster-claw, featuring a minor and a major prong; dashed lines represent disordered regions (Figs. S2A and S3A). (B) Enlarged view of the *Myo4p•She3p complex. Alternating helices and loops are rainbow-colored (from N-terminal blue to C-terminal red); helices are numbered H1–H18, and two prominent loops, L2 and L16, are indicated. The transition into disordered, N-terminal *Myo4p region (cyan N and dashes) and disordered, C-terminal She3p region (yellow and blue C and dashes) (Figs. S2A and S3A) are indicated. Note that *Myo4p features two binding interfaces (I and II); interface I comprises a helical hairpin (H1 and H2) and interface II stretches from H12 to L15 (Figs. S2A and S3A). Although both interfaces are separated by 264 residues in the primary structure of Myo4p (Fig. S2A), they are adjacent in the quaternary structure and reside together in the minor prong of the claw. (C and D) Surface representation of the *Myo4p•She3p interfaces in an open-book depiction. Interfaces are encircled in continuous black line, and interfaces I and II are separated by a dashed black line. Interfaces and surfaces in C indicate *Myo4p chain (pink), two chains of She3p (yellow and blue), and hydrophobic residues (green) and in D indicate evolutionarily highly conserved residues (red) [using a color gradient from white (0%) to red (100%) identity]. Note that interfaces are primarily hydrophobic and evolutionarily highly conserved (Figs. S2A and S3A).
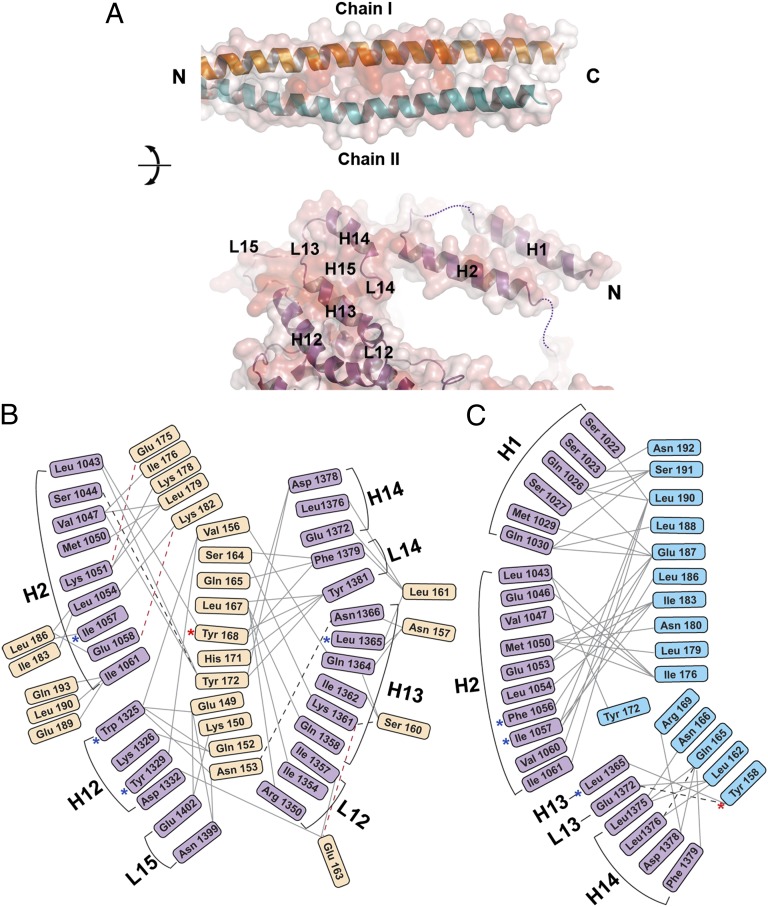
A closer look at the interface shows several noteworthy features (Figs. 2 and 3 and Fig. S2A). First, *Myo4p contributes two distinct binding interfaces. “Interface I” is located to residues 1,022–1,061 in helices H1 and H2. “Interface II” is located between residues 1,325 and 1,402 and is formed by regions of stacked helices and interconnecting loops, altogether extending from H12 until L15 (Fig. 2B and Fig. S2A). Although interfaces I and II are separated in the primary structure of Myo4p by 264 residues (Fig. S2A), in the 3D structure, both interfaces are located immediately adjacent to each other on the minor prong of the claw (Fig. 2 B–D). Second, residue-by-residue analyses of the interaction show that the two She3p helices contribute unequally to binding with *Myo4p such that chain I of She3p (Figs. 2 B–D and 3) interacts primarily with interface II of Myo4p, burying 2,346 Å2 of surface area, whereas chain II of She3p (Figs. 2B and 3) interacts principally with interface I of Myo4p, burying 1,466 Å2 of surface area. The intermolecular contacts are largely established by Van der Waals contacts with only a very few electrostatic interactions and hydrogen bonds (Fig. 3 B and C and Fig. S4). Moreover, the residues in the interacting regions of chain I and chain II of the She3p pseudocoiled coil and of interface I and interface II of Myo4p are well conserved (Fig. 2D and Figs. S2A and S3A). Third, a comparison of the structure of the nonliganded, shorter wtMyo4p (Myo4p1092-1471) (PDB ID code 4LL6) with the same region of the larger form of *Myo4p after She3p binding indicated small conformational changes at the Myo4p–She3p interface (Fig. S4 B and C). Although we did not refine a wtMyo4p918-1471•She3p81-311 structure (Materials and Methods), the overall structural features of wt- and *Myo4p were similar, including a largely disordered and evolutionarily poorly conserved loop, L2, of which 15 residues were removed in the mutant form (Fig. S2A). Detailed experimental and refinement parameters are listed in Table S1.
Fig. 3.
Residue-by-residue contacts in the *Myo4p-She3p heterotrimer. (A) Interfaces between She3p coiled-coil region and *Myo4p are shown in an open-book depiction, and their transparent surface representations are colored according to sequence conservation, from white (0%) to red (100%) identity. (Upper) The two chains of She3p are labeled as chain I (orange) and II (blue). (Lower) Helices and loops of *Myo4p are marked and numbered; disordered parts of loops are indicated by dashed lines (Figs. S2A, S3A, and S4B). (B and C) Interactome of residues of *Myo4p and the two She3p chains: chain I (Glu-149–Gln-193) (Left), and She3p chain II (Tyr-158–Asn-192) (Right). Solid gray lines represent hydrophobic Van der Waal interactions, dashed black lines indicate hydrogen bonds, and red dashed lines depict electrostatic interactions. Note that two She3p chains interact asymmetrically with *Myo4p (Figs. S2A, S3A, and S4). Mutations reported previously are indicated by blue asterisks, and mutations studied in this work are indicated by red asterisks (25).
Mutational Analyses of the Myo4p•She3p Interactome.
Heuck et al. (17) have previously identified several double mutations in Myo4p [(F1056R, I1057R), (W1325D, Y1329D), and (L1365G, I1367G)], which affect interactions with She3p or ASH1 mRNA localization. With the exception of Ile-1367, our crystal data confirm that the reported Myo4p residues interact with She3p (Fig. S5A). Therefore, these mutant residues could potentially weaken interaction with She3p (Fig. 3 B and C and Fig. S5A). We introduced two additional mutations in residues of She3p that interact with Myo4p, namely She3p Y158E and Y168E (Fig. 3 B and C) and assayed either mutant’s ability to rescue in vivo trafficking of Myo4p to the daughter-cell cytoplasm in a SHE3 deletion strain. Either of the two mutants diminished trafficking activity by about one half (Fig. S5 B–F).
Structure of N-Terminally Extended She3p Coiled Coil.
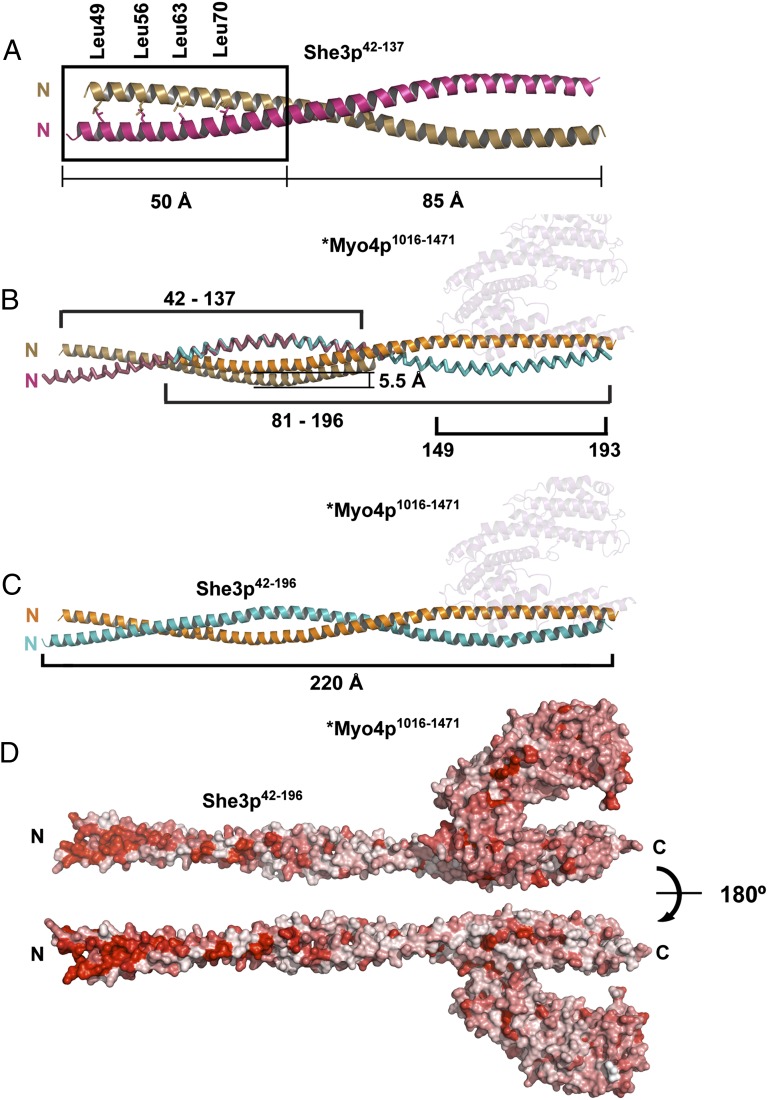
We also solved the crystal structure of another fragment of She3p, She3p42-137. This fragment (Fig. 4) overlaps with, and N-terminally extends, the She3p81-196 fragment that was crystallized as part of Myo4p•She3p (Fig. 2). The crystal structure of She3p42-137 (Materials and Methods) was refined to final Rwork and Rfree values of 22.8% and 26.5% (Table S1) (PDB ID code 4LL7), respectively, to a resolution of 2.3 Å. Two She3p42-137 protomers form a coiled coil of 135 Å in length (Fig. 4A) that opens up at the C terminus. Dimerization was also confirmed by MALS (Fig. S3B). The coiled-coil interface is primarily hydrophobic and contains a classical leucine zipper motif, comprising Leu-49, -56, -63, and -70 at the N-terminal half (Fig. 4A). When joining the two overlapping She3p structures at residue 81 (Fig. 4B), the combined pseudocoiled coil measures 220 Å in length (Fig. 4C). The Myo4p binding site is located at the C-terminal third of the combined pseudocoiled coil (Fig. 4C). Notably, there is a high degree of evolutionary conservation of surface residues in the N-terminal half of the pseudocoiled coil (Fig. 4D). We propose this highly conserved She3p region as a potential binding site for a yet unidentified ligand of tubular ER (tER) for Myo4p•She3p-mediated transport (Discussion).
Fig. 4.
Crystal structure of an upstream section of the She3p coiled coil reveals a highly conserved surface region. (A) She3p42-137 was separately crystallized (Fig. 1 and Fig. S3 A and B) and formed a 135-Å-long, parallel coiled coil with a classical Leucine Zipper at its N-terminal end (the four Leucines of the zipper are indicated as sticks). The two She3p42-137 helices are colored brown and pink. The upstream Leucine Zipper region of She3p is boxed. (B) Overlap of the coiled-coil region of She3p42-137 (A) and the She3p moiety of *Myo4p1016-1471•She3p81-196 (Fig. 2A). In their superimposed form, one of the She3p42-137 ribbons is displaced by 5.5 Å. The position of *Myo4p1016-1471 is indicated in pale cyan. (C) A 220-Å-long composite structure of She3p42-196 in complex with *Myo4p1016-1471 (pale cyan) is indicated in the color code of Fig. 2A. (D) Surface representation of a composite *Myo4p1016-1471•She3p42-196 complex. Sequence conservation is colored from white (0%) to red (100%) identity. Note high degree of surface conservation at the N-terminal Leucine zipper region of She3p. We propose this region of She3p to be a candidate for binding to a not yet identified ligand of tubular endoplasmic reticulum (tER) (Fig. 6B).
Coupling of Two Myo4p•She3p Heterotrimers by a Single She2p Tetramer and Zipcode Oligonucleotide.
Both zipcoded mRNA and cognate She2p tetramers have long been recognized to be essential for in vitro assembly of a transport-competent motor complex (15, 25, 34, 35). Our crystal and biophysical data here clearly show that Myo4p and She3p form a 1:2 heteotrimer instead of the purported 1:1 heterodimer.
She2p and/or zipcode are likely to couple two Myo4p•She3p heterotrimers to generate the transport-competent motor complex with two myosin molecules obligatory for processive movements along an actin cable. To test this supposition, we carried out stepwise in vitro assembly at physiological salt concentration. For these in vitro reconstitution experiments, we used a complex of wtMyo4p918-1471•full-length (FL) She3pFL as starting material, followed by incubation with She2pFL and/or a synthetic oligonucleotide representing ASH1 mRNA zipcodes E2B (E2B-44), E3 (E3-51), or a shortened E3 (E3-33). Assembly into higher-order complexes was assayed by size-exclusion chromatography and MALS (Fig. 5 and Fig. S6).
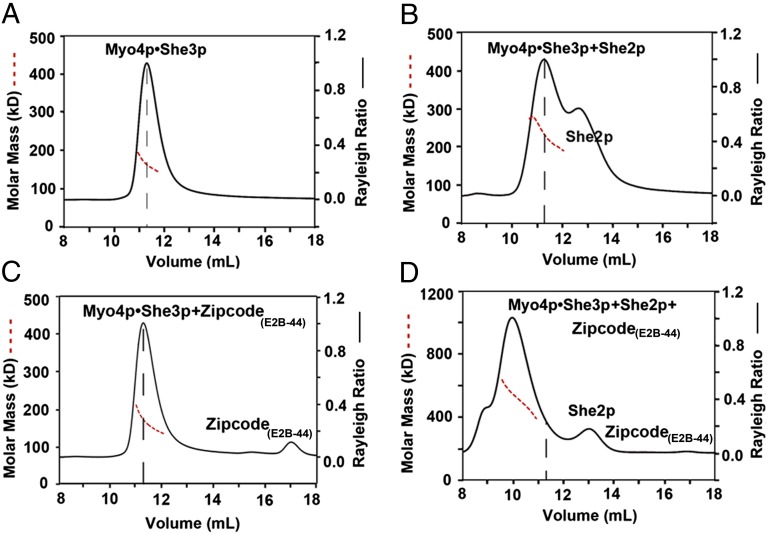
Fig. 5.
Coupling of two Myo4p•She3p heterotrimers by zipcode oligonucleotide and She2p. Indicated components were incubated and subsequently analyzed by size-exclusion chromatography coupled to MALS, plotted as Fig. 1B. (A) Peak of wtMyo4p918-1471•She3pFL (340 µg of protein at 1.7 mg/mL) elutes with a molar mass of 165.6 ± 0.3 kDa (dashed red line), close to the expected molar mass of 162.9 kDa for a 1:2 heterotrimer; the peak position in the elution profile is marked with a vertical dashed line (black) in this panel and the following panels. (B) As A, but after incubation with full-length She2pFL (680 µg of protein at 1.7 mg/mL in a molar ratio of Myo4p:She3p:She2p of 1:2:4); peak elutes with a molar mass of 237.4 ± 0.5 kDa, indicating a dynamic equilibrium between Myo4p•She3p and a She2p tetramer. (C) As A, but after incubation with oligonucleotide E2B-44 (340 µg of protein at 1.7 mg/mL and 29.2 µg of E2B-44 at 0.146 mg/mL; molar ratio of Myo4p:She3p:E2B-44 1:2:1). The peak elutes at a molar mass of 163.8 ± 0.65 kDa (dashed red line) but shows a slightly higher molar mass at the leading edge of the peak, consistent with some zipcode oligonucleotide binding to Myo4p•She3p heterotrimer. (D) As in B, but after incubation with 44-nt-long, zipcode RNA (E2B-44) (total 680 µg of protein mixture at 1.7 mg/mL with 101 µg of E2B-44 at 0.252 mg/mL; molar ratio of Myo4p:She3p:She2p:E2B-44 is 1:2:4:3); peak elutes at a molar mass of 467.2 ± 1.2 kDa, consistent with coupling of two Myo4p•She3p heterotrimers by one She2p tetramer and a single E2B-44 oligonucleotide in a molar ratio of 2:4:4:1 (calculated molar mass: 457.1 kDa). However, because of the small mass contribution of the oligonucleotide, we cannot rule out that more than one oligo bound.
By itself, the wtMyo4p918-1471•She3pFL complex eluted as a single peak with a molar mass of 165.6 ± 0.3 kDa, close to the calculated molar mass of 162.9 kDa for a 1:2 Myo4p918-1471•She3pFL heterotrimer (Fig. 5A). Therefore, just like the two protomers of She3p81-311, two full-length equivalents of She3p also engage in heterotrimer formation with one Myo4p918-1471 protomer. Notably, there was no detectable formation of stable higher-order complexes, indicating that the Myo4p918-1471•She3pFL complex by itself does not form higher-order conjugates, at least not under the physiological salt condition and the protein concentrations that were tested here (Fig. 5A). These data argue strongly against a model in which a complex of two molecules of Myo4p and an unidentified number of She3p molecules can form in an unassisted fashion in the cytoplasm (25, 36).
When Myo4p918-1471•She3pFL was incubated with She2pFL (a tetramer with molar mass of 117.2 kDa), we observed a considerable broadening of the Myo4p918-1471•She3pFL peak, increasing its molar mass from 165.6 ± 0.3 kDa to 237.4 ± 0.5 kDa (Fig. 5 A and B). These data suggested that She2pFL tetramer could associate with the Myo4p918-1471•She3pFL heterotrimer, but only in an unstable form.
When Myo4p918-1471•She3pFL was incubated with a 44-nt-long oligonucleotide representing E2B zipcode of ASH1 mRNA (termed E2B-44), there was a slight increase in the molar mass at the leading edge of the peak (Fig. 5C), compatible with a previously reported nonspecific binding of She3p to RNA (25). We conclude that, in our in vitro reconstitution system, neither zipcode nor She2p, when added alone, can assemble higher-order Myo4p918-1471•She3pFL complexes that would contain the two Myo4p molecules obligatory for processive movement on an actin cable.
However, when Myo4p918-1471•She3pFL heterotrimers were incubated together with tetramer of She2pFL and zipcode oligonucleotide E2B, we observed the appearance of a new “heavy” peak with a molar mass of 467.2 kDa ± 1.2 kDa (Fig. 5D). The mass of this new peak is consistent with an assembly product where two Myo4p918-1471•She3pFL heterotrimers were joined together by one She2p tetramer bound to one 44-nt-long zipcode, as is illustrated in the model shown in Fig. 6A. Such a complex of Myo4p•She3p•She2p•zipcode in a molar ratio of 2:4:4:1 has a calculated molar mass of 457.1 kDa, close to the experimentally observed molar mass of 467.2 kDa for the heavy peak. We conclude that binding of a single zipcode oligonucleotide to a single She2p tetramer is sufficient to couple two Myo4p918-1471•She3pFL heterotrimers (Fig. 6A). These data established that an in vitro reconstituted motor complex contains four instead of the previously predicted two molecules of She3p.
Fig. 6.
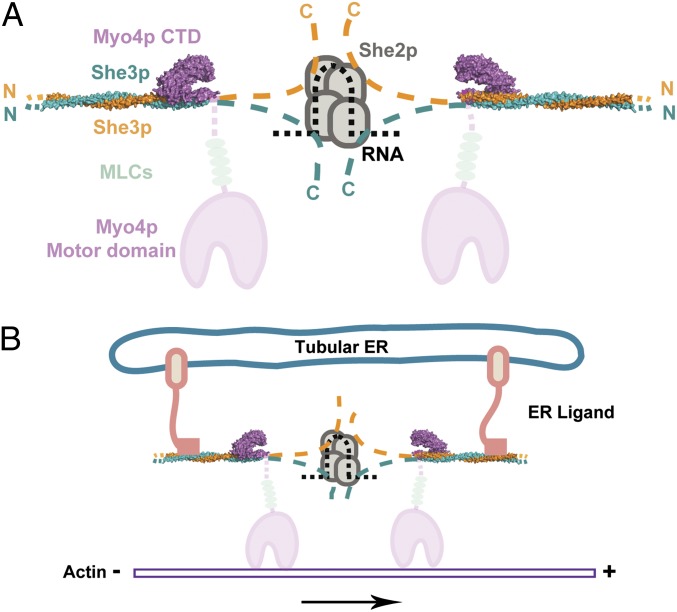
Model for cargo-mediated coupling of two Myo4p•She3p heterotrimers. (A) A centrally located complex of a single She2p tetramer (four overlapping ovals in gray) cooperatively couples a single zipcode element of mRNA (black dashes) and a dedicated region each of She3p moieties (orange and blue dashes) emanating from two surrounding *Myo4p1016-1471•She3p42-196 heterotrimers. The structures reported in this paper for the 1:2 Myo4p•She3p heterotrimer (Fig. 4D) are colored, in blue and orange for the She3p pseudocoiled coil, and in cyan for the claw representing the C-terminal domain (CTD) of Myo4p. Disordered and noncrystallized regions of She3p ( Figs. 1 and 2) are schematically indicated by dashes, and their N and C termini are marked with N and C, respectively. As for the Myo4p molecule, its C-terminal claw region (Figs. 2 and 4) transits into an about 100-residue-long upstream segment (faint cyan dashes) that is disordered in our crystal structure (Figs. 1A and 2 and Fig. S2), continues to the lever arm region with five bound light chains (LC) (indicated by gray ovals), and ends with the N-terminal motor domain (in faint cyan). Note that, in an assembled motor complex, the claws of the two Myo4p molecules face each other (for implications, see Discussion). (B) Two ligands (termed ER ligands), representing yet to be identified integral membrane protein(s) of tubular endoplasmic reticulum, are proposed to couple two Myo4p•She3p heterotrimers by attaching to a highly conserved surface feature of the She3p pseudocoiled-coil region located upstream of the Myo4p claw binding site (Fig. 4D).
In addition, we tested two other synthetic oligonucleotides: one representing the E3 zipcode of ASH1 mRNA, 51 nt long (E3-51) (10, 25), and the other representing a shortened E3 zipcode, only 33 nt long (E3-33), lacking a She2p-binding motif (10, 25). As previously observed for E2B (Fig. 5D and Fig. S6A), the E3-51 oligonucleotide, representing zipcode E3-51, stably linked two Myo4p•She3p heterotrimers in concert with the She2p tetramer (Fig. S6B). In contrast, the shortened oligonuceotide E3-33 failed to do so (Fig. S6C). These data further pinpoint the critical role of zipcode oligonucleotide—one oligonucleotide bound to one She2p teteramer being sufficient—to assemble a transport-competent motor complex by pairing two Myo4p•She3p heterotrimers and thereby providing the two myosin molecules obligatory for transport competence.
Discussion
Our crystal data here show that the C-terminal domain (CTD) of one Myo4p molecule adheres to a pseudocoiled-coil region of two She3p molecules to form a 1:2 Myo4p•She3p heterotrimer (Figs. 2 and 3). The 1:2 Myo4p•She3p heterotrimer also exists in solution as shown by size-exclusion chromatography coupled to multiangle light scattering (MALS) (Figs. 1B and 5A). Collectively, these data overturn the view that Myo4p and She3p associate with each other as a 1:1 heterodimer and are doing so via a dedicated helical region in each molecule (13, 23, 26). They also contradict the idea that Myo4p and She3p molecules can associate with each other to form a complex of two Myo4p molecules and an unspecified number of She3p molecules (25).
Our crystal structure (Figs. 2 and 3) revealed extensive molecular interactions in the 1:2 Myo4•She3p heterotrimer. Two adjacent regions of chain I and II of She3p form a slightly opened up coiled coil (Fig. 2C) and thereby provide an asymmetric platform for binding Myo4p CTD (Fig. 3A). The Myo4p CTD displays an overall structure reminiscent of a lobster claw, with its minor prong adhering to the She3p platform. The convoluted path of the Myo4p CTD creates two distinct binding interfaces, designated interface I and interface II. These binding interfaces, although separated by 264 residues in the primary structure, are adjacent to each other on the minor prong of the lobster claw (Fig. 2). A detailed residue-by-residue analysis of interaction between Myo4p CTD and She3p (Fig. 3A) reveals that binding is primarily through hydrophobic Van der Waals forces, with only a few hydrogen and electrostatic bonds. These interactions suggest that, in interphase cells, most of the Myo4p and She3p molecules are kinetically partitioned into a stable Myo4p•She3p heterotrimer located in the soluble fraction of the cytoplasm and that an envisioned higher-order complex of these two molecules with two Myo4p molecules (25) is unlikely to exist until heterotrimers encounter zipcoded- and She2p-marked mRNPs.
To directly assess formation of higher-order complexes, we incubated wtMyo4p918-1471•Full Length She3pFL heterotrimers with either She2pFL tetramer and/or synthetic oligonucleotides representing various zipcodes of ASH1 mRNA. We used three types of synthetic oligonucleotides representing two zipcodes of ASH1 mRNA: a 44-nt-long E2B, a 51-nt-long E3, and a shortened E3, E3-33, that is unable to bind to She2p tetramer (25). We assayed for assembly using size-exclusion chromatography coupled to MALS. Our data indicate (Fig. 5) that wtMyo4p911-1471•She3pFL heterotrimers interact with She2pFL tetramer, but only form unstable complexes (Fig. 5B). Only in the additional presence of oligonucleotides representing zipcode E2B or E3-51 of ASH1 mRNA were higher-order complexes assembled. The molar mass of these complexes is consistent with coupling of two heterotrimers by one She2pFL tetramer and one zipcode (Fig. 5D and Fig. S6 A and B). This assembled complex contains the two Myo4p molecules required for processive movement on an actin cable. As expected, the shortened E3-33 zipcode oligonucleotide was unable to stably couple wtMyo4p911-1471•She3pFL heterotrimers (Fig. S6C).
We incorporated our data here into a model (Fig. 6A): Two claw-like Myo4p CTDs, each attached to the pseudocoiled-coil region of She3p, are indirectly paired by a dedicated downstream region of She3p, each of which latches on to a monomer of the single She2p tetramer bound to a single zipcode element of mRNA. Our model in Fig. 6A is consistent with the stoichiometry that we observed in our in vitro reconstitution experiments (Fig. 5 and Fig. S6). It is also consistent with a stepwise assembly pathway: An idle Myo4p•She3p heterotrimer in the cytoplasm might be conjugated (Fig. 6A) as it encounters a She2p tetramer bound to a single zipcode of mRNA. The presence of multiple zipcodes in a single mRNA molecule (such as of ASH1 mRNA, which contains four zipcode elements) may trigger the assembly of four motor complexes to yield a millipede-like motion that is likely to be more effective because less prone to derail. Because the two “claws” of the paired Myo4p CTD•She3p heterotrimer face each other (Fig. 6A), we predict that any multimotor complexes are likely to contain multiples of two Myo4p molecules. It should be noted that, for other intrinsically monomeric myosins, cargo loading has been shown to yield the dimerization necessary for movement (37, 38).
An interesting tangent of our studies here concerns the strikingly conserved surface of an upstream coiled-coil region of She3p (Fig. 4D). We propose this highly conserved She3p coiled coil as a candidate binding site that represents the genetically mapped binding site on She3p for a hitherto unidentified ligand on the surface of tubular ER that can undergo transport from the mother to the daughter cell (28, 32, 39). As indicated in the model in Fig. 6B, two Myo4p•She3p heterotrimers are proposed to be cross-linked by putative tER ligands into a transport-competent motor complex. Coupling two Myo4p•She3p heterotrimers by either She2p•zipcode or tER ligand(s) may occur separately or in concert, in agreement with reported observations of discrete transport or cotransport of these two different cargos (29, 32). Again, as for ASH1 mRNA, multiple ligands arranged consecutively along the surface of tubular ER could recruit multiple motor complexes, each consisting of two cross-linked Myo4p•She3p heterotrimers, yielding a more processive mode of transport in a millipede-like fashion.
Materials and Methods
Cloning.
For bacterial expression, DNA fragments encoding She3p42-137, Flag-She2pFL, and various C-terminal fragments of Myo4p (Fig. S1) and of a mutant Myo4p, *Myo4p918-1471 (Fig. S2A), were cloned into modified pGEX-KG vector between NdeI and XhoI sites to create GST fusion proteins. Other cDNAs encoding She3p fragments and full-length She3pFL-His6 were inserted into modified pMR101 (ATCC) vector between NdeI and XhoI sites. For yeast expression, DNA fragments encoding wtShe3p, She3pY158E, and She3pY168E were cloned in vector pF415met-mCherry [modified from vector pF415met with mCherry (Clontech) fluorescent protein fusion at the N terminus] between SmaI and SalI sites.
Protein Interaction Analysis.
Fragments of Myo4p and She3p were coexpressed in Escherichia coli (see Protein Expression, Purification, and Crystallization). Harvested E. coli cells were resuspended as follows: 1 g of cells in 5 mL of buffer A [10 mM Hepes (pH 8.0) and 1 M NaCl]. The cells were lysed by sonication, the sonicate was centrifuged for 20 min at 30,000 × g, and the supernatant was loaded onto 50 μL of Glutathione Sepharose 4B resin (GE Healthcare) preequilibrated with buffer A. After washing the beads with 2 mL of buffer A, bound proteins were eluted with SDS sample buffer, separated by SDS/PAGE, and stained with Coomassie Blue.
Protein Expression, Purification, and Crystallization.
Protein fragments either were expressed individually (GST-She3p42-137, GST-Flag-She2pFL, or GST-Myo4p1098-1471) or were coexpressed [(GST-*Myo4p918-1471 and She3p81-311), (GST-wtMyo4p918-1471 and She3pFL), and (GST-wtMyo4p918-1471 and She3p81-311)] in LB medium under the appropriate antibiotics selection, using E. coli BL21 (DE3) Gold cells (Agilent Technologies). When the OD600 reached between 0.5 and 0.8, expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and cells were grown for another 5 h at 37 °C (GST-Myo4p1098-1471; GST-wtMyo4p918-1471 and She3pFL) or overnight at 20 °C (GST-She3p42-137; GST-Flag-She2pFL; GST-*Myo4p918-1471 and She3p81-311).
After harvesting, cells were processed by sonication using buffer B [10 mM Hepes (pH 8.0), 200 mM NaCl, and 10 mM DTT] for Myo4p1098-1471 and *Myo4p918-1471•She3p81-311, buffer C 10 mM Hepes (pH 8.0), 100 mM NaCl, and 10 mM DTT for GST-Flag-She2pFL, and buffer D 10 mM Hepes (pH 8.0), 5% (vol/vol) glycerol, 200 mM NaCl, and 10 mM DTT for GST-She3p42-137. Glutathione Sepharose 4B-bound proteins were eluted by thrombin digestion except in the case of Myo4p1098-1471, which was eluted by trypsin digestion. Eluted Myo4p1098-1471; GST-Flag-She2pFL and *Myo4p918-1471•She3p81-311 were further purified by anion-exchange chromatography using 5 mL of HiTrap Q HP column (GE Healthcare), followed by size-exclusion chromatography using Superdex 75 16/60 (GE Healthcare) (for Myo4p1098-1471) and Superdex 200 16/60 (GE Healthcare) (for GST-Flag-She2pFL and *Myo4p918-1471•She3p81-311). She3p42-137 was purified by cation-exchange chromatography using HiTrap SP HP column (GE Healthcare), followed by size-exclusion chromatography using HiLoad 16/60 Superdex 75 column. wtMyo4p918-1471•She3p81-311 and wtMyo4p918-1471•She3pFL were purified with the same procedure as *Myo4p918-1471•She3p81-311. Purified proteins were concentrated by centricon to 10–12 mg/mL and stored at 4 °C in buffer E [10 mM Hepes (pH 8.0), 500 mM NaCl, 10 mM DTT, and 5% (vol/vol) glycerol] before crystallization or size-exclusion chromatography and multiangle light scattering (MALS). All selenomethionine (Se-Met)-labeled proteins were produced using a previously published protocol (40), and individual proteins were purified as described for native proteins.
We first crystallized native and Se-Met Myo4p1098-1471 in 10% (vol/vol) Tacsimate (Hampton Research) (pH 7.0), 25% (wt/vol) PEG 1K, and 10 mM DTT at 20 °C, and solved the structure of Se-Met derivative by multiple-wavelength anomalous diffraction (MAD). Using these data, we refined the structure of the native protein to 2.3 Å with Rwork 22.5% and Rfree 27.8% (Table S1).
She3p42-137 was crystallized in 100 mM Mes 2-(N-morpholino)ethanesulfonic acid (pH 6.5), 450 mM (NH4)2SO4, and 10% (vol/vol) 2-propanol. Crystals were soaked in the same buffer supplemented with 2 mM K2PtCl4 (to improve resolution) and 3 mM DyCl3 (to produce derivatives as well as to improve resolution). Because the native crystal [without Platinum (Pt)] diffracted only up to 2.9 Å and the electron density quality of the N-terminal region was insufficient to build a model, we solved the structure of the derivative crystal [with both Dysprosium (Dy) and Pt at wavelength of 1.1050 Å] to 2.3 Å with Rwork 22.8% and Rfree 26.5% (Table S1).
The complexes of Myo4p918-1471•She3p81-311 and *Myo4p918-1471•She3p81-311 were crystallized in 100 mM Hepes (pH 7.0), 8% (vol/vol) Tacsimate, 12% (wt/vol) PEG 3,350, and 10 mM DTT. Both structures were solved by molecular replacement using the coordinates of Myo4p1098-1471 and She3p42-137. A dataset from Se-Met *Myo4p918-1471•She3p81-311 was collected to calculate the difference Fourier map, which confirmed the register of the final structure. The resolutions of Myo4p918-1471•She3p81-311 and *Myo4p918-1471•She3p81-311 were 4.5 Å and 3.6 Å, respectively. The overall electron density did not show significant differences between the two; therefore, only the 3.6-Å structure was refined (Table S1).
Data Collection and Structure Determination.
Diffraction data were acquired at the beamlines X-12C and X-29 at the National Synchrotron Light Source (NSLS) and processed using HKL2000 (41). The electron density map of Myo4p1098-1471 was calculated by SOLVE/RESOLVE (42), of She3p42-137 by SHELX (43) combined with SOLVE/RESOLVE, and of Myo4p918-1471•She3p81-311 and *Myo4p918-1471•She3p81-311 by Phaser (44, 45) from CCP4 (46). Buccaneer (46) was used for initial model building. CNS (47) and Refmac (46) were used for refinement. Interactive structure building was carried out in Coot (48). Structure figures were generated by PyMOL (49). Ramachandran plot was calculated using procheck from the CCP4 package, and the Molprobity Score was obtained from the Molprobity server (50).
Multiangle Light Scattering.
The following components were used: Myo4p918-1471•She3p1-311, His6-She3p1-227, Myo4p918-1471•She3pFL-His6, Flag-She2pFL, and synthetic oligoribonucleotides E2B-44, E3-51, and E3-33. The 44-nt-long E2B-44 represents a shortened form of published E2B; this shortened E2b retains the binding capacity to the She2p tetramer. E3-51 and E3-33 have been previously described (10, 25). After incubation for 30 min on ice with gel filtration buffer containing 10 mM Hepes (pH 7.5), 150 mM NaCl, 2 mM MgCl2, and 1 mM DTT, various mixtures were analyzed at room temperature by size-exclusion chromatography coupled to multiangle light scattering (MALS) (51), at amounts, concentrations, and molar ratios indicated in figure legends. Samples were injected into a Superdex 200 10/30 GL size-exclusion chromatography column (GE Healthcare) equilibrated with gel filteration buffer. The synthetic RNAs E2B-44 (5′-AUCUCCAUCUCCCUCCACACCGACGAAAAGUGGCAAGAUGAGAU-3′), E3-33, and E3-51 (25) were purchased from Integrated DNA Technology. The chromatography system was connected in series with an 18-angle light scattering detector (DAWN HELEOS) and refractive index detector (Optilab rEX; Wyatt Technology). Data were collected every 1 s at a flow rate of 0.25 mL/min at 25 °C. Data analysis was carried out using the program ASTRA (Wyatt Technology).
Yeast Strains Construction and Imaging.
pF415met-mCherry vector and the plamids carrying genes encoding wtShe3p, She3pY158E, and She3pY168E were transformed into MYO4-EGFP::HIS5, she3Δ:KAN, mat A, BY4741 strain and grown in complete synthetic medium (CSM) devoid of Leucine (CSM-Leu) to OD600 of 0.6 at 30 °C. Twenty-five milliliters of cells were harvested by centrifugation and washed once with CSM devoid of both Leucine and Methione (CSM-Leu-Met), and then grown in the same medium for 35 min to induce the expression of proteins. The cells were harvested by centrifugation and washed once in 1 mL of CSM-Leu medium followed by another centrifugation for 3 min at 700 × g. The pellet was resuspended in ∼800 µL of CSM-Leu medium.
The cDNA of *Myo4-EGFP with HIS5 selection marker were amplified and inserted into myo4Δ:KAN, mat A, BY4741 strain and grown in CSM devoid of Histidine to OD600 of 0.6 at 30 °C. Twenty-five milliliters of cells were washed once with the same media and pelleted by centrifugation for 5 min at 1,800 × g. Cells were resuspended in 1 mL of CSM medium devoid of Histidine.
Two microliters of the yeast-cell suspension were used for live-cell imaging. Imaging was performed using an Axio Imager.Z1 (Zeiss) fluorescence microscope. Experiments were repeated three times with more than 200 dividing cells each time. The concentration of Myo4p-EGFP was determined by fluorescence intensity measurement (ImageJ).
Supplementary Material
Acknowledgments
We thank Annie Héroux, Howard Robinson, and Alexei Soares (National Synchrotron Light Source) for beamline support; Sozanne Solmaz for assistance with multiangle light scattering; and Erik Debler, Alok Sharma, and Songhai Shi for critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4LL6, 4LL7, and 4LL8).
See Commentary on page 4351.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401428111/-/DCSupplemental.
References
- 1.Blower MD. Molecular insights into intracellular RNA localization. Int Rev Cell Mol Biol. 2013;302:1–39. doi: 10.1016/B978-0-12-407699-0.00001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326(5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepard KA, et al. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci USA. 2003;100(20):11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389(6646):90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 6.Long RM, et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277(5324):383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol. 1999;9(6):337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- 8.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10(6):1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 9.Jambhekar A, et al. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102(50):18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivier C, et al. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25(11):4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84(5):687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 12.Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119(4):491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Krementsova EB, et al. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J Cell Biol. 2011;195(4):631–641. doi: 10.1083/jcb.201106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller M, et al. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 2009;15(11):2002–2012. doi: 10.1261/rna.1753309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonsalvez GB, et al. RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA. 2003;9(11):1383–1399. doi: 10.1261/rna.5120803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodges AR, Krementsova EB, Trybus KM. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J Biol Chem. 2008;283(11):6906–6914. doi: 10.1074/jbc.M708865200. [DOI] [PubMed] [Google Scholar]
- 17.Heuck A, et al. The structure of the Myo4p globular tail and its function in ASH1 mRNA localization. J Cell Biol. 2010;189(3):497–510. doi: 10.1083/jcb.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Blobel G. UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc Natl Acad Sci USA. 2010;107(50):21382–21387. doi: 10.1073/pnas.1013038107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24(17):1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napetschnig J, et al. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci USA. 2009;106(9):3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19(23):6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci USA. 2000;97(10):5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bookwalter CS, Lord M, Trybus KM. Essential features of the class V myosin from budding yeast for ASH1 mRNA transport. Mol Biol Cell. 2009;20(14):3414–3421. doi: 10.1091/mbc.E08-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodges AR, Bookwalter CS, Krementsova EB, Trybus KM. A nonprocessive class V myosin drives cargo processively when a kinesin- related protein is a passenger. Curr Biol. 2009;19(24):2121–2125. doi: 10.1016/j.cub.2009.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller M, et al. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9(4):e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sladewski TE, Bookwalter CS, Hong MS, Trybus KM. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol. 2013;20(8):952–957. doi: 10.1038/nsmb.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genz C, Fundakowski J, Hermesh O, Schmid M, Jansen RP. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J Biol Chem. 2013;288(45):32384–32393. doi: 10.1074/jbc.M113.486431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aronov S, et al. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(9):3441–3455. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estrada P, et al. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J Cell Biol. 2003;163(6):1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landers SM, Gallas MR, Little J, Long RM. She3p possesses a novel activity required for ASH1 mRNA localization in Saccharomyces cerevisiae. Eukaryot Cell. 2009;8(7):1072–1083. doi: 10.1128/EC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19(20):5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid M, Jaedicke A, Du TG, Jansen RP. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol. 2006;16(15):1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Heuck A, et al. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc Natl Acad Sci USA. 2007;104(50):19778–19783. doi: 10.1073/pnas.0706780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. J Cell Biol. 2010;189(4):755–767. doi: 10.1083/jcb.200912011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruse C, et al. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol. 2002;159(6):971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niedner A, Müller M, Moorthy BT, Jansen RP, Niessing D. Role of Loc1p in assembly and reorganization of nuclear ASH1 messenger ribonucleoprotein particles in yeast. Proc Natl Acad Sci USA. 2013;110(52):E5049–E5058. doi: 10.1073/pnas.1315289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138(3):537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 38.Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331(6018):757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- 39.Fundakowski J, Hermesh O, Jansen RP. Localization of a subset of yeast mRNAs depends on inheritance of endoplasmic reticulum. Traffic. 2012;13(12):1642–1652. doi: 10.1111/tra.12011. [DOI] [PubMed] [Google Scholar]
- 40.Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat Struct Mol Biol. 2006;13(6):540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11(Pt 1):49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 43.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49. Schrodinger, LLC (2010) The PyMOL Molecular Graphics System (Schrödinger, LLC, Cambridge, MA), Version 1.3r1.
- 50.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knobloch JE, Shaklee PN. Absolute molecular weight distribution of low-molecular-weight heparins by size-exclusion chromatography with multiangle laser light scattering detection. Anal Biochem. 1997;245(2):231–241. doi: 10.1006/abio.1996.9984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.