Significance
For complex biological processes, the formation of protein complexes is a strategy for coordinating the activities of many enzymes in space and time. It has been hypothesized that growth of the bacterial cell wall involves stable synthetic complexes, but neither the existence of such complexes nor the consequences of such a mechanism for growth efficiency have been demonstrated. Here, we use single-molecule tracking to demonstrate that the association between an essential cell wall synthesis enzyme and the cytoskeleton is highly dynamic, which allows the cell to buffer growth rate against large fluctuations in enzyme abundance. This indicates that dynamic association can be an efficient strategy for coordination of multiple enzymes, especially those for which excess abundance can be harmful to cells.
Keywords: Pencillin binding proteins, bacterial cell wall, multienzyme complexes, superresolution microscopy
Abstract
Assembly of protein complexes is a key mechanism for achieving spatial and temporal coordination in processes involving many enzymes. Growth of rod-shaped bacteria is a well-studied example requiring such coordination; expansion of the cell wall is thought to involve coordination of the activity of synthetic enzymes with the cytoskeleton via a stable complex. Here, we use single-molecule tracking to demonstrate that the bacterial actin homolog MreB and the essential cell wall enzyme PBP2 move on timescales orders of magnitude apart, with drastically different characteristic motions. Our observations suggest that PBP2 interacts with the rest of the synthesis machinery through a dynamic cycle of transient association. Consistent with this model, growth is robust to large fluctuations in PBP2 abundance. In contrast to stable complex formation, dynamic association of PBP2 is less dependent on the function of other components of the synthesis machinery, and buffers spatially distributed growth against fluctuations in pathway component concentrations and the presence of defective components. Dynamic association could generally represent an efficient strategy for spatiotemporal coordination of protein activities, especially when excess concentrations of system components are inhibitory to the overall process or deleterious to the cell.
For the wide variety of organisms that have large internal osmotic pressures, the cell wall is essential for maintaining cellular integrity and morphology (1). As a consequence, the bacterial cell wall is an important antibiotic target. Much research has been performed to elucidate the chemical composition of the cell wall and identify the enzymes required for its synthesis, yet we still know little about the dynamics of cell wall assembly in vivo. The cell wall is composed of peptidoglycan, a network of long glycan strands that are cross-linked by short peptides. During growth, existing bonds in the peptidoglycan network are broken, and new glycans are polymerized, inserted, and cross-linked. This process depends on a host of enzymes, most notably the penicillin-binding proteins (PBPs) (2). It is presumed that the spatial and temporal coordination between these proteins is responsible for the maintenance of cell shape during growth and division, although the strategies used to robustly achieve such a task that spans both molecular and cellular scales have not yet been elucidated.
Recent live-cell imaging studies in the rod-shaped bacteria Escherichia coli (3) and Bacillus subtilis (4, 5) showed that clusters of MreB, a bacterial actin homolog that is essential for rod-like shape, move in linear tracks oriented in the circumferential direction. These dynamics are coupled to cell wall synthesis; exposure to cell wall-targeting antibiotics or depletion of cell wall precursors halts MreB motion (3–5). In E. coli, MreB motion was stopped by exposure to high concentrations of the β-lactam antibiotic mecillinam, which inhibits the transpeptidase PBP2 (3), suggesting a model in which PBP2 activity is necessary for MreB motion, possibly by directing new cell wall insertion. In B. subtilis, tracking of MreB-associated proteins (MreC/D) and PBPs provided support for the hypothesis that coordination between MreB and the wall synthesis machinery is achieved through colocalization into large, moving, multienzyme complexes (4, 5). However, biochemical isolation of such a complex has remained elusive (6), raising the possibility that this picture is incomplete, particularly in E. coli, in which dynamic imaging of the PBPs has not yet been achieved.
PBP2 is thought to be the major enzyme responsible for covalently cross-linking new glycan strands into the cell wall during growth (7). PBP2 is essential; cells enlarge and eventually lyse upon depletion of PBP2 (8). In E. coli, PBP2 was previously observed to localize as dispersed puncta and at septa during division (9). PBP2 concentration affects cell size (10), and overexpression is lethal (11). Although MreB binds to the cytoplasmic face of the inner membrane, PBP2 is a transmembrane protein with a cytoplasmic N-terminus and a periplasmic domain required for transpeptidase activity. Given the requirement of a functional PBP2 for MreB motion and peptidoglycan synthesis, we sought to determine whether it could be a component of a cell wall-synthesizing, multienzyme complex moving with MreB.
In this study, we applied single-particle tracking photoactivated localization microscopy (sptPALM) (12), a technique based on the limited photoactivation of fluorescent proteins, to quantify the dynamics of MreB and PBP2 in E. coli. In contrast to MreB, we find that PBP2 exhibits rapid, diffusive motions that do not depend on PBP2 catalytic activity. These data suggest a model in which PBP2 transiently associates with sites of cell wall synthesis, and hence can act in a distributed manner and need not be rate limiting for growth as was previously hypothesized (3). In support of this model, growth was unaffected for more than two doublings during depletion of PBP2. Finally, we show that both growth rate and MreB speed decrease during mecillinam treatment in a dose-dependent manner, indicating that a catalytically active PBP2 molecule is required during the incorporation of a glycan strand.
Results
Single-Molecule Tracking of MreB Circumferential Motion in E. coli.
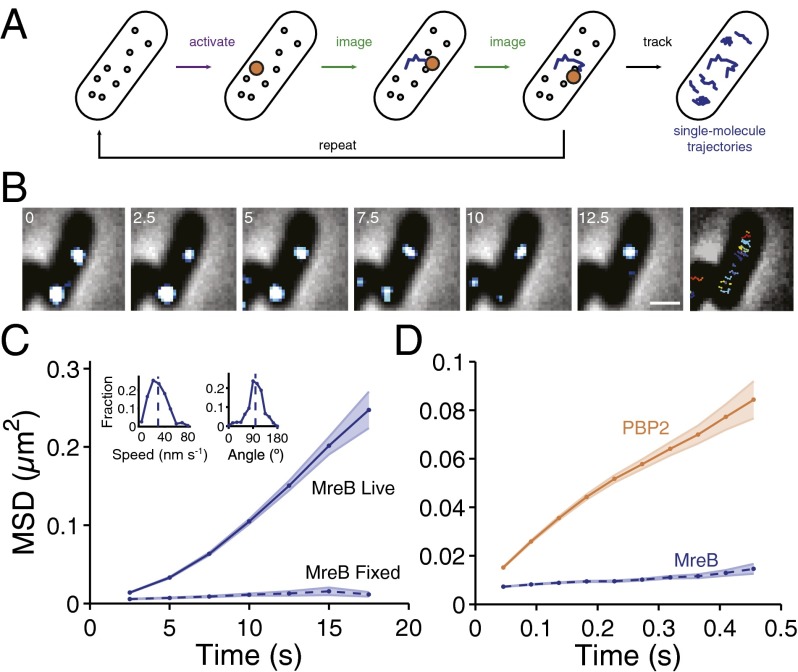
We tagged E. coli MreB with PAmCherry (13) as an internal sandwich fusion (14) and expressed this fusion (MreBsw-PAmCherry) as the sole copy at the native chromosomal locus. E. coli cells expressing MreBsw-PAmCherry fusions were viable and remained rod-shaped, although cells were slightly wider (Fig. S1 A–C), as previously observed for mCherry sandwich fusions (14). We imaged single cells using total internal reflectance fluorescence (TIRF) microscopy through several hundred iterations of a cycle in which a single molecule was activated, followed by time-lapse imaging of the molecule’s dynamics every 2.5 s until it photobleached (Fig. 1A, Fig. S2, and Materials and Methods). Our sptPALM measurements revealed directed, circumferential motion of single MreB molecules (Fig. 1 B and C), similar to previous measurements of diffraction-limited clusters of MreB in E. coli (3) and B. subtilis (4, 5). For other cell wall synthesis proteins (15), particularly those in low abundance, these results indicate that the circumferential motion of single molecules is a requisite signature for colocalization with MreB.
Fig. 1.
Single-molecule dynamics reveal that the cell wall synthesis enzyme PBP2 undergoes fast, diffusive motion, unlike the directed motion of the MreB cytoskeleton. (A) Schematic of sptPALM. Photoactivatable fluorescent protein fusions are used to monitor the movement of individual proteins in a crowded environment. A small population of molecules is activated by UV laser light, and then imaged over time to track molecular positions. (B) TIRF images of MreBsw-PAmCherry single molecules in live E. coli TKL039 (MG1655 mreBsw-PAmCherry) cells taken every 2.5 s, overlaid on phase-contrast images. The solid lines in the last panel are representative MreB molecular tracks. (Scale bar: 1 μm.) (C) Mean squared displacement (MSD) of MreB molecules in live (n = 382 molecules) and fixed (n = 105) E. coli TKL039 cells. The shaded area represents SEM. (Inset) The distribution of MreB track speeds (28.6 ± 15.4 nm⋅s−1, SD) at 37 °C and angles relative to the cell midline (99.1 ± 29.4°, SD). Most trajectories were approximately perpendicular to the long axis of the cell, with a slight right-handed bias quantitatively consistent with previous studies of left-handed twisting during growth (42). (D) MSD of MreB molecules (n = 1,018) in TKL039 cells and PBP2 molecules (n = 854) in TKL112 (∆mrdA Pmrda-PAmCherry-mrdA) imaged at high frame rates. The shaded area represents SEM. The linear MSD of PBP2 indicates that PBP2 molecules move diffusively, with a diffusion constant of D = 0.06 ± 0.006 µm2/s.
Rapid, Diffusive Motion of PBP2 Molecules.
To determine the dynamics of PBP2 in E. coli, we constructed an N-terminal fusion of PAmCherry to PBP2 expressed from the endogenous promoter and verified that this fusion rescued cells from depletion of the native, unlabeled PBP2 (Fig. S1 D–G). As the sole source of PBP2, either expressed from a low-copy plasmid or integrated at the native chromosomal locus, PAmCherry-PBP2 complemented wild-type growth rate with slightly higher average cell width (Fig. S1 F and G). Bocillin gels and Western blots demonstrated that the fusion remained stable and intact inside the cell (Materials and Methods; Figs. S3 and S4A).
We observed striking qualitative and quantitative differences between the motions of MreB and PBP2. PBP2 molecules moved much faster and more erratically than MreB. Reconstructing the trajectory of a PBP2 molecule required imaging on a timescale 100 times faster (∼10 ms) than that required to observe linear MreB trajectories (Materials and Methods; Fig. S5). Moreover, the mean squared displacement (MSD) of PBP2 motion was linear (Fig. 1D), indicating diffusive behavior with an apparent diffusion constant of 0.06 ± 0.006 µm2/s. The difference in the mobilities of MreB and PBP2 excluded the possibility that MreB and PBP2 function together in a long-lived synthesis complex.
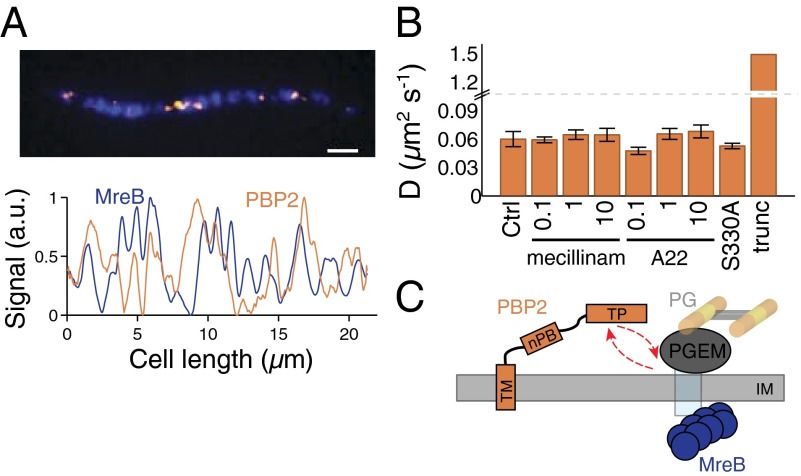
To directly quantify the colocalization of MreB and PBP2, we constructed a strain expressing both MreBsw-sfGFP and PAmCherry-PBP2. Taking into account the likelihood of random association of diffraction-limited foci, individual cells exhibited a weak, although significant, correlation between the fluorescence signals from these fusion proteins (Fig. S6; Materials and Methods), suggesting that PBP2 spends at most a small fraction of its time interacting with MreB (Fig. 2A).
Fig. 2.
Rapid PBP2 diffusion is independent of catalytic activity, and buffers cell growth rate from fluctuations in PBP2 concentration. (A) Representative image of MreB and PBP2 colocalization in a TKL233 (mreBsw-sfGFP ∆mrdA PmrdA-PAmCherry-mrdA) cell filamented with cephalexin. (Upper) MreBsw-sfGFP and PAmCherry-PBP2 were coimaged via TIRF. (Lower) The fluorescence profiles over the length of the cell, averaged over the transverse direction. MreB and PBP2 profiles have a small, but significant correlation [two-sample t test: experimental data vs. scrambled data, t(76) = 2.79, P = 0.0068; Materials and Methods; Fig. S6]. (B) The apparent PBP2 diffusion constant is unaffected by antibiotic treatment. SEM is shown (n = 5 experiments). Mecillinam binds the active site of PBP2, A22 disrupts MreB polymerization, and an S330A mutation in PBP2 ablates the catalytic active site. Concentrations are given in micrograms per milliliter. The diffusion constant increases roughly 25-fold when the transpeptidase domain is truncated. (C) Model for PBP2 interaction with MreB. PBP2 moves rapidly within the inner membrane (IM), forming transient interactions with sites of peptidoglycan (PG) synthesis through its transpeptidase (TP) domain, either with the peptidoglycan elongation machinery (PGEM), or with the wall itself. PBP2 also contains transmembrane (TM) and nonpenicillin binding (nPB) domains, which may mediate interactions with other morphogenetic proteins.
The Transpeptidase Domain but Not Catalytic Activity Influences PBP2 Motion.
PBP2 transpeptidase activity is essential for cell wall growth; mutations or drugs that interfere with transpeptidation are lethal (8, 16). One such drug is the antibiotic mecillinam, which specifically inhibits PBP2 by acting as a substrate mimic that irreversibly locks the active site of PBP2 (Fig. S3) (16). To determine whether PBP2 dynamics are dependent on its catalytic activity, we tracked PBP2 motion over a range of mecillinam concentrations (Fig. 2B). In each case, we observed diffusive motion with no observable difference in the diffusion constant. Similarly, a mutation that disrupts the PBP2 active site (S330A) (11) did not impact PBP2 diffusivity (Fig. 2B; Materials and Methods). These data indicated that the process of transpeptidation does not measurably inhibit PBP2 motion.
Our observed diffusion constant of PBP2 is substantially lower than measurements for transmembrane proteins of similar size (2–5 µm2/s) (17). Given the small correlation between MreB and PBP2 localization, it was possible that PBP2 diffusion is affected by its transient interaction with MreB or indirectly through an MreB-associated component such as MreC (Fig. S6). However, disruption of MreB polymerization with the antibiotic A22 (18) had little effect on PBP2 diffusivity (Fig. 2B). In addition, disrupting the activity of PBP1a, which binds PBP2 in vitro (19), either through inhibition with cefsulodin or deletion, had no effect on diffusivity (Fig. S7). To test whether PBP2 diffusion could be increased by removing potential sites of interactions with other periplasmic components (Fig. 2C), we constructed a PAmCherry-PBP2 fusion with the entire periplasmic, transpeptidase domain truncated. We observed a large increase in diffusivity (Fig. 2B and Fig. S8), approximately to the levels expected based on other similarly sized transmembrane proteins (17). Importantly, this increase was substantially more than would be predicted from the Stokes–Einstein relation (D ∼ 1/R) based solely on the decrease in mass of PBP2 due to truncation, and is likely an underestimate, as molecules moving at that speed cannot be well localized with single-molecule fluorescence. These data indicate that PBP2 is slowed by interactions through its transpeptidase domain, possibly with other proteins such as MreC (20) and/or the wall itself through its transpeptidase domain. Nonetheless, these interactions must have sufficiently rapid kinetics that PBP2 motion appears diffusive on the fast timescale of our measurements. Based on these observations, we hypothesized a model in which PBP2 rapidly binds and dissociates from active sites of wall synthesis through the transpeptidase domain (Fig. 2C). Importantly, the independence of PBP2 motion from MreB in addition to PBP2’s lack of directed motion provide further evidence that E. coli cell wall assembly is not mediated by a stable, MreB-associated, multienzyme complex containing PBP2.
Growth Rate Is Maintained During Depletion of PBP2.
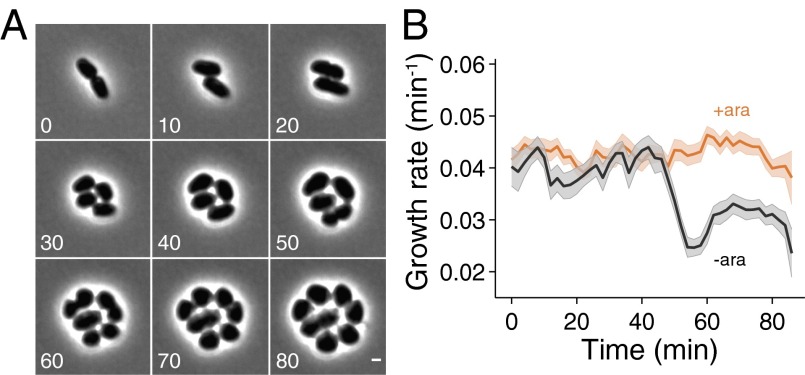
Transient interactions between MreB and PBP2 may explain how E. coli maintains robust cell growth despite only expressing ∼100 PBP2 molecules per cell (21). If PBP2 were part of a stable multienzyme complex, then previous calculations have indicated that ∼100 PBP2 molecules would be required to maintain a 20-min doubling time (3), and reductions in PBP2 levels would lead to a decrease in growth rate. In contrast, based on our measured diffusive motion of PBP2 and previous experimentally determined estimates for the density of peptide cross-links in the cell wall (22), we estimated that as few as ∼30–40 PBP2 molecules would be sufficient to move among all active sites of synthesis spread across the cell surface (Materials and Methods), indicating that PBP2 abundance should not be limiting for growth in wild-type cells. Therefore, we hypothesized that the combination of high diffusivity, rapid catalytic activity, and transient association could passively buffer growth rate against fluctuations in PBP2 concentration. To measure growth rate during changes in PBP2 concentration, we imaged cells during PBP2 depletion through several divisions. The depletion of PBP2 was confirmed by Western blot, with levels equivalent to the chromosomal fusion 15 min after the removal of inducer (Fig. S4 A and B). The rate of depletion qualitatively agreed with the loss of rod-shape morphology (Fig. 3A). During the course of depletion (Fig. S4 A and B), cells gradually became wider and rounder over the course of several divisions (Fig. 3A), consistent with previous observations (11). Despite these morphological changes indicative of substantial PBP2 depletion, we detected no slowing of the growth rate, defined as the areal strain rate (1/A dA/dt), through two rounds of division (Fig. 3B). It was only after this interval that growth rate became drastically compromised, consistent with our theoretical prediction of the robustness of growth rate to large reductions in the number of PBP2 molecules (Materials and Methods); regardless of the initial abundance before depletion, the rapid drop in growth rate distinguishes our model from one in which growth is directly proportional to PBP2 abundance. Quantitatively similar behavior was observed in cells during PAmCherry-PBP2 depletion (Fig. S4C), and during PBP2 depletion in minimal medium (Fig. S4D) and in filamentous cells (Fig. S4E; Materials and Methods), providing further indication that growth rate is robust to substantial changes in PBP2 levels.
Fig. 3.
Growth rate is unaffected by substantial PBP2 depletion. (A) Time-lapse microscopy of E. coli TKL141 (∆mrdA Para-mrdA) cells growing during PBP2 depletion. Time is specified in minutes. Even though cells lose their rod-shaped morphology, they continue to grow at a rate comparable to wild-type cells until two to three divisions have taken place. (B) Growth rate during PBP2 depletion is quantitatively unaffected over more than two doubling times. Ara+ represents the nondepleted control, and Ara− represents cells undergoing PBP2 depletion. The shaded area represents SE. (Scale bar: 1 µm.)
Mecillinam Treatment Reduces Growth Rate and MreB Speed in a Dose-Dependent Manner.
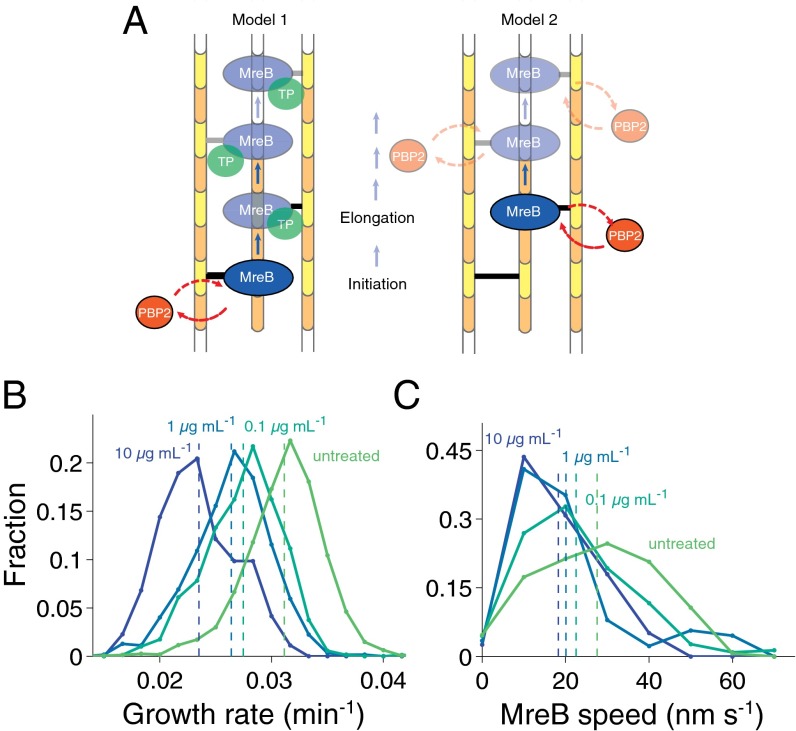
Because overexpression of catalytically inactive PBP2 is lethal (11), we expected that mecillinam treatment would be functionally distinct from depletion due to the interference of inactive PBP2 with the cell wall synthesis machinery. Although both treatments result in a lower concentration of active PBP2 molecules, we expect that mecillinam-bound PBP2 could block the action of active enzymes. Inhibition of PBP2 activity with high levels of mecillinam resulted in a concentration-dependent reduction in the average growth rate (Fig. 4B and Fig. S9A), consistent with a requirement for active PBP2 to either (i) initiate peptidoglycan insertion events (with subsequent cross-linking along the glycan strands carried out by another enzyme), or (ii) cross-link glycan strands throughout the process of new cell wall incorporation (Fig. 4A). To differentiate between these possibilities, we measured MreB speeds during mecillinam treatment. If PBP2 were required only for initiation, the speeds of processive MreB molecules should be maintained and the reduction in growth rate would be due to decreased initiation, i.e., there would be fewer processive MreB molecules but their speed would be unaffected. If PBP2 activity is required throughout strand insertion, MreB motion would be contingent on the arrival of an active PBP2, and thus mecillinam would decrease MreB speeds. Supporting the second hypothesis, single-molecule measurements showed mecillinam concentration-dependent decreases in MreB speed (Fig. 4C). The fraction of MreB molecules moving in a directed fashion also decreased (Fig. S9B), perhaps indicating a concomitant decrease in initiation or reflecting a decrease in average glycan strand length. These quantitative measurements linking perturbations of PBP2 activity to reductions in growth rate and MreB speeds confirm that the activity of PBP2 molecules is required throughout the process of insertion of glycan strands, and provide further evidence that MreB motion is directly coupled to cell wall synthesis.
Fig. 4.
PBP2 activity is required throughout MreB-directed peptidoglycan synthesis. (A) Potential models for the role of PBP2 activity during cell wall synthesis. (Left) PBP2 is required for initiating strand synthesis by the rest of the cell wall synthesis machinery, but is not required for further synthesis-dependent MreB motion. (Right) Diffusing PBP2 molecules rapidly associate and disassociate with MreB-associated proteins to drive the progress of strand incorporation and MreB motion. (B) Cell growth rate decreases under mecillinam treatment in a concentration-dependent manner. Exponential phase cells (MG1655) were placed on agarose pads containing the indicated concentration of mecillinam and imaged for 15 min. To calculate growth rate, cell contours were extracted and cell length over time was fit to an exponential function. (C) MreB speed in TKL039 cells also decreases under mecillinam treatment in a concentration-dependent manner, indicating a requirement for active PBP2 molecules during the insertion of glycan strands.
Discussion
Our single-molecule measurements demonstrate that PBP2 undergoes fast, diffusive dynamics, and indicate that PBP2 activity is coupled to MreB motion and cell wall synthesis. These data are consistent with a model in which individual glycan strands are synthesized through the interchange of diffusing PBP2 molecules, predicting that growth rate can be maintained in the face of fluctuations in PBP2 concentration without requiring large excess. During PBP2 depletion, growth rate was maintained until a sharp decrease after several doubling times. Our diffusivity measurements suggest that mecillinam-bound PBP2 molecules are still recruited to the site of synthesis, but their inability to catalyze cross-linking decreases the rate of insertion due to the requirement for a functional PBP2. Thus, our model of diffuse PBP2 activity predicts that the fraction of cross-linked peptidoglycan should be independent of mecillinam concentration, with the rate of cell wall growth slowing to match the rate of glycan cross-linking. We confirmed this prediction using ultra performance liquid chromatography (UPLC) (Fig. S10; Materials and Methods). Therefore, because cross-linking affects cell wall elasticity (23), E. coli cells also appear to buffer the mechanical strength of the cell wall against mecillinam treatment, despite the resultant changes in cell shape that may be due to the spatial pattern of material or subtle changes in glycan strand length (22).
Our study also indicates that the coordination of cell wall synthesis does not necessitate the colocalization of the proteins involved. In fact, transient associations are beneficial for buffering growth against fluctuations in enzyme abundance. At a PBP2 abundance of ∼100 enzymes per cell (21), one would expect 1/√N ∼ 10% fluctuations, yet growth rate remains consistent through cell division (24). In addition, transient association loosens the requirement for an MreB complex to spatially and temporally order the steps of cell wall synthesis. This line of reasoning is supported by the observation that growth rate is unaffected by A22 treatment (25), despite disruption of MreB spatial organization. In vitro interactions between E. coli PBP2 and PBP1a (19) and between Helicobacter pylori PBP2 and MreC (20) have been identified, and in the latter case these proteins appear to form a complex in vivo. Although perturbation of PBP1a does not affect PBP2 mobility (Fig. S8), it remains possible that some components of the cell wall synthesis machinery interact and move together, but not stably with MreB. Interestingly, studies in B. subtilis suggest that the converse is also possible: coordination can occur through an MreB-based complex (4, 5). However, in B. subtilis, PBP2A and PBP2H engage in diffusive motion when MreB is depleted (4). Whether these differences can be attributed to a specialization for Gram positive-specific cell wall growth remains to be resolved.
Although PBP2 activity is required throughout MreB-mediated cell wall synthesis, the origin of directed MreB movement is yet unknown. It is presumed that MreB dynamics are related to the known biochemical steps in peptidoglycan synthesis, but a detailed mechanism by which they couple to or drive MreB motion is unclear. Observing the dynamics of the cell wall synthesis components will clarify their role in the mechanochemical cycle underlying MreB dynamics, and the use of single-molecule techniques will be essential. In our study, sptPALM was critical for gaining insight into PBP2 dynamics rather than relying on epifluorescence colocalization to determine whether MreB and PBP2 interact in a stable complex. The importance of single-molecule techniques has been highlighted in other studies of multienzyme processes, such as the observed exchange of polymerases during DNA replication (26). Our results indicate that similar dynamic association occurs in peptidoglycan synthesis and has an important role in maintaining the rate of this spatially distributed process in the presence of concentration fluctuations, especially important given the low abundances of the enzymes involved. More generally, random links between the processing elements of distributed, multistep processes have been proposed to suppress fluctuations in processing times (27); diffusive PBP2 motion between sites of peptidoglycan synthesis may be a biological realization of this principle. Given the numerous cellular processes and timescales involved in cell growth, dynamic association between key players may be central to strategies for optimizing spatiotemporal coordination during growth.
Materials and Methods
Strain Construction.
The strains and plasmids used in this study are described in Tables S1 and S2, respectively. Plasmids were constructed using enzymatic assembly methods (28). Expression plasmids were constructed using a low-copy plasmid with a pSC101 origin (pRM102) (29), and coding sequences for the relevant genes were amplified from E. coli MG1655 with the appropriate homology regions for assembly (Table S3). Gene deletions and fluorescent protein fusions were introduced into the chromosome using allelic exchange methods with suicide plasmids (30). The desired sequences for integration were amplified by PCR and cloned into pDS132 (30). MFDpir (31) cells were transformed with the resulting plasmids and used for conjugative transfer into the recipient strain. Resulting merodiploids were selected on lysogeny broth (LB) (32) plates supplemented with chloramphenicol. Strains that had lost the integrated plasmid (and sacB gene) by homologous recombination were selected on LB plates containing 5% (wt/vol) sucrose. All chromosomal modifications were confirmed by amplification and sequencing of the targeted region.
Bocillin Labeling.
To verify that PAmCherry-PBP2 fusions were stable and remained intact, bocillin gels were carried out (Fig. S3). Overnight cultures were diluted 1:200 in LB and grown to an optical density at 600 nm (OD600) of 0.5, pelleted, and resuspended in PBS. One millimolar phenylmethanesulfonylfluoride (Sigma), 10 µM Bocillin-FL (Invitrogen), and 500 µg/mL polymixin B (Fisher Scientific) were added to the cells and incubated at 37 °C for 1.5 h. Cells were pelleted and washed three times with cold PBS, resuspended in Laemmli Sample Buffer (Bio-Rad), and boiled at 95 °C for 10 min, and total protein content was quantified using a DC protein assay (Bio-Rad) according to manufacturer's instructions. Each sample was normalized to 10 µg of total protein and separated on a 4–15% precast polyacrylamide gel. The gel was rinsed with water 10 times and then visualized on a Typhoon 9410 Variable Mode Imager (GE Healthcare) with 488-nm excitation.
Immunoblotting.
Overnight cultures from strains containing the PAmCherry-PBP2 fusion were diluted 1:100 in EZ-RDM plus 0.2% glucose and supplemented with 0.1% arabinose when appropriate. Cultures were grown to an approximate OD600 of 0.7, and then 1 mL of culture was pelleted and resuspended in 100 µL of 4% (wt/vol) SDS. For Para-PAmCherry-mrdA depletion experiments, exponential phase cultures were rinsed three times with media lacking arabinose and samples were drawn at the indicated time points. Whole-cell lysates were prepared by incubating at 95 °C for 10 min, and total protein content was quantified with a DC protein assay (Bio-Rad) according to manufacturer’s instructions. Lithium dodecyl sulfate (LDS) sample buffer (4×) was added to each sample and incubated at 95 °C for 10 min, and then 12.5 µg of total protein from each sample were loaded onto the gel. Proteins were transferred to a PVDF membrane (Immobilon, Millipore), first stained with rabbit anti-mCherry antibodies (ab167453; Abcam), and then stained with IRDye 800CW goat anti-rabbit antibodies (Li-Cor) for detection on a Li-Cor Odyssey imaging system according to manufacturer’s instructions.
Microscopy.
Single-particle tracking was performed on a TIRF microscope built with a Ti-E Eclipse stand (Nikon Instruments). The objectives used were either an Apo TIRF 100× (N.A. 1.49) or a Plan Apo Lambda 100× DM (N.A. 1.45) (Nikon), depending on whether phase-contrast images were acquired concurrently. CUBE diode 405-nm and Sapphire OPSL 561-nm lasers (Coherent) were combined into an optical fiber and into a TIRF illuminator (Nikon) attached to the microscope stand. Shuttering of the laser illumination was controlled by an acoustooptic tunable filter (AA Optoelectronics) before the fiber coupler. Images were acquired with an iXon3+ 887 EMCCD (Andor Technology) camera, and synchronization between components was achieved using μManager (33) with a microcontroller (Arduino).
Single-Particle Imaging.
Cells expressing fluorescently labeled proteins were grown to saturation overnight in the rich medium EZ-RDM (Teknova) (34) with 0.2% glucose and then diluted 1:100 in fresh medium and incubated with shaking at 37 °C for 2 h. Cells were spotted onto 1% agarose pads with EZ-RDM plus 0.2% glucose and covered with argon plasma-cleaned coverslips. For drift correction, Tetra-speck 100-nm fluorescent beads (Invitrogen) were added at a dilution of 1:1,000. To measure MreB speeds, we used an imaging sequence consisting of a 50-ms exposure with an activation laser (405 nm at ∼0.05 kW/cm2) followed by capture of 10 images with an imaging laser (561 nm at ∼0.5 kW/cm2) every 2.5 s, which was repeated for a total of 500–1,000 cycles. To capture the more rapid PBP2 dynamics, we simultaneously exposed the cells to both activation and imaging lasers (405 nm at ∼0.05 kW/cm2 and 561 nm at ∼1 kW/cm2) for 10-ms exposures every 45.3 ms (except for the truncation mutant, which required a shorter imaging interval; Fig. S8). In single-particle tracking experiments under antibiotic treatments, agarose pads were cast with the appropriate antibiotic, and cells were directly spotted from liquid culture.
Single-Particle Analysis.
Images were analyzed computationally to generate single-particle tracks using the u-track package (35) in MATLAB (MathWorks). Gaussian mixture-model fitting was used for particle detection and the routines were modified to use multicore processors. The fitted particle locations were corrected for drift using the cross-correlation between bright fluorescent beads after smoothing using the “spaps” MATLAB function. Tracks were calculated using the “costMatLinearMotion” cost function in u-track. Only MreB tracks of a certain length (5–15 time points) were retained for further analysis. To identify the subset of MreB molecules that showed directed motion, the MSD of each individual track was fit to the sum of linear and quadratic terms [MSD(t) = c + 4Dt + at2], and particles that fit well (R2 > 0.9) with a > 0 were considered to undergo directed motion with velocity v = a1/2. The MreB molecules that could not be fit well were either moving diffusively or too ambiguous to be classified as directed motion according to the limits set by the temporal and spatial resolution of our measurements. The angle of motion was calculated by fitting the coordinates of the track to a line and measuring the angle as this linear fit crossed the cell midline. For PBP2 molecules, tracks with a length of 3–15 time points were considered for further analysis. An estimate for the diffusion constant was determined from a linear fit to the first four points of the mean squared displacement.
Colocalization of PBP2 and MreB.
E. coli MG1655 ∆mrdA pKC137(PmrdA-PAmCherry-mrdA) cells were transformed with a plasmid expressing MreBsw-mVenus (pRMmreBmVenus) or transduced with P1 lysate made from cells containing MreBsw-sfGFP::kan. These cells were filamented with 35 µg/mL cephalexin and prepared on 1% agarose pads with EZ-RDM plus 0.2% glucose. The population of PAmCherry-PBP2 molecules was photoactivated with a 3-s burst from the 405-nm laser, and 50-ms exposure images were acquired with 488-nm and 561-nm lasers. Cell outlines were calculated using a custom MATLAB package, and the average fluorescence along the length of the cell was calculated. A moving average of 5 pixels (530 nm) was used to smooth the signal, and an average over 100 pixels was used to correct for background, after which the signal was normalized. The Pearson correlation coefficient was calculated between these two normalized signals. To estimate the error bars, the order of the signal was randomized once per cell and the correlation coefficient was calculated on the randomized signal.
Estimate of the Number of PBP2 Molecules Required for Growth.
The doubling of an E. coli cell involves a length increase from ∼2 to 4 µm, with a commensurate surface area increase of ∼6 µm2. Each pair of disaccharides and associated cross-link corresponds to ∼10 nm2 in surface area (23). Therefore, given the high degree of in-plane cross-linking, ∼6 × 105 cross-links are added per doubling. Taken together, for a doubling time of 20 min, ∼500 cross-links per second need to be added during elongation.
Previous studies have estimated that there are ∼100 PBP2 molecules per E. coli cell (21). To estimate whether each PBP2 could participate in five cross-linking events per second, we assume that cell wall material is synthesized in a spatially uniform fashion across the cylindrical portion of the cell, which has an average length of 3 µm and an average surface area of ∼9 µm2. Thus, each of the 500 new cross-links corresponds to a region with area of ∼0.02 µm2. Assuming that the transpeptidation reaction is fast relative to the speed of PBP2 motion across this area, a PBP2 molecule has to achieve an MSD of at least 0.1 µm2 within 1 s to visit all five cross-linking sites. Our measured diffusion constant of ∼0.06 µm2/s corresponds to an MSD of 0.24 µm2 in 1 s based on the relationship MSD(t) = 4Dt for 2D diffusion, and is therefore sufficient for coverage of cross-linking across the entire cell surface during cell elongation.
Single-Cell and Colony-Growth Imaging.
Exponentially growing cells were spotted onto 1% agarose pads and then imaged every 30 s in a temperature-controlled chamber (HaisonTech) at 37 °C. For experiments involving mecillinam treatment, cells were transferred from liquid culture with no antibiotic directly onto agarose pads with the appropriate concentration of mecillinam and imaged for 15 min. Cell contours were automatically extracted from phase-contrast images using a custom MATLAB package. The cell length over time was fit to an exponential [L(t) = L0 exp(kt)] to calculate the growth rate k.
To analyze the growth rates of single cells in a microcolony during PBP2 depletion, cells grown in media supplemented with 0.1% arabinose were rinsed three times with media lacking arabinose and then spotted onto agarose pads lacking arabinose. Cells were imaged every 30 s for 2 h. These cells were segmented and analyzed using the MATLAB package MicrobeTracker (36). Growth rate was defined as the fractional change in cell outline area over time and averaged with a sliding window of eight frames. Growth rates were averaged over all cells for a given treatment at each time point, and SEs were calculated based on the number of cells measured at each time point.
To analyze the growth rates of single filamentous cells during PBP2 depletion, cells were grown with 0.1% arabinose and 7 µg/mL cephalexin for 30 min, rinsed without arabinose, and then imaged on agarose pads with 7 µg/mL cephalexin. These cells were segmented and analyzed using a custom MATLAB package, and growth rate was measured as the fractional change in area as described above with an average sliding window of 15 frames.
Purification of Sacculi and UPLC Analysis of Peptidoglycan Composition.
Overnight cultures of E. coli MG1655 were diluted 1:100 in LB and supplemented with 0, 0.01, 0.1, and 1 µg/mL mecillinam. The cultures were grown at 37 °C to an OD600 of 0.7. The cultures were then harvested by centrifugation at 5,000 × g for 10 min at room temperature and resuspended in 3 mL of LB. Cell suspensions were lysed by boiling in SDS, and SDS-insoluble material was collected by several rounds of ultracentrifugation at 400,000 × g. Samples were prepared for UPLC analysis as previously described (37) and injected onto a Waters H Class UPLC system equipped with a BEH C18 1.7-µm column (Waters), using elution conditions previously described (38, 39). Peaks were quantified and identified as particular muropeptide species, from which the cross-linking density and strand length were calculated (40, 41).
Supplementary Material
Acknowledgments
We thank Ned Wingreen, Josh Shaevitz, Alex Dunn, David Ehrhardt, and Sven van Teeffelen for their feedback. We also thank Piet de Boer and Felipe Bendezu for providing strains, and Tom Bernhardt and Monica Markovski for assistance with bocillin labeling. This work was supported by a Siebel Scholars Graduate Fellowship (to T.K.L.), support from a National Institutes of Health (NIH) Biotechnology Training Grant (to T.K.L.), a Stanford Interdisciplinary Graduate Fellowship (to C.T.), a Bio-X Senior Postdoctoral Fellowship (to R.D.M.), NIH Ruth L. Kirschstein National Research Service Award 1F32GM100677-01A1 (to J.H.), a Stanford School of Medicine Dean's Postdoctoral Fellowship (to J.H.), support from the Stanford School of Engineering Chinese Undergraduate Visiting Research Program (to E.G.), NIH Director’s New Innovator Awards DP2OD004389 (to Z.G.) and DP2OD006466 (to K.C.H.), and National Science Foundation CAREER Award 1149328 (to K.C.H.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4355.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313826111/-/DCSupplemental.
References
- 1.Young KD. Bacterial shape: Two-dimensional questions and possibilities. Annu Rev Microbiol. 2010;64:223–240. doi: 10.1146/annurev.micro.112408.134102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32(2):234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108(38):15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333(6039):225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 5.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333(6039):222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: New insights from localization studies. Microbiol Mol Biol Rev. 2005;69(4):585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura TBP, et al. Penicillin-binding protein 2 is essential in wild-type Escherichia coli but not in lov or cya mutants. J Bacteriol. 1989;171(6):3025–3030. doi: 10.1128/jb.171.6.3025-3030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Den Blaauwen T, Aarsman MEG, Vischer NOE, Nanninga N. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol Microbiol. 2003;47(2):539–547. doi: 10.1046/j.1365-2958.2003.03316.x. [DOI] [PubMed] [Google Scholar]
- 10.Philippe N, Pelosi L, Lenski RE, Schneider D. Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J Bacteriol. 2009;191(3):909–921. doi: 10.1128/JB.01419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legaree BA, Adams CB, Clarke AJ. Overproduction of penicillin-binding protein 2 and its inactive variants causes morphological changes and lysis in Escherichia coli. J Bacteriol. 2007;189(14):4975–4983. doi: 10.1128/JB.00207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5(2):155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 13.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6(2):153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28(3):193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci USA. 2006;103(29):10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratt BG. The mechanism of action of mecillinam. J Antimicrob Chemother. 1977;3(Suppl B):13–19. doi: 10.1093/jac/3.suppl_b.13. [DOI] [PubMed] [Google Scholar]
- 17.Ramadurai S, et al. Lateral diffusion of membrane proteins. J Am Chem Soc. 2009;131(35):12650–12656. doi: 10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- 18.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120(3):329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Banzhaf M, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85(1):179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- 20.El Ghachi M, et al. Characterization of the elongasome core PBP2 : MreC complex of Helicobacter pylori. Mol Microbiol. 2011;82(1):68–86. doi: 10.1111/j.1365-2958.2011.07791.x. [DOI] [PubMed] [Google Scholar]
- 21.Dougherty TJ, Kennedy K, Kessler RE, Pucci MJ. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J Bacteriol. 1996;178(21):6110–6115. doi: 10.1128/jb.178.21.6110-6115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol. 2011;81(2):340–353. doi: 10.1111/j.1365-2958.2011.07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105(49):19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, et al. Robust growth of Escherichia coli. Curr Biol. 2010;20(12):1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuson HH, et al. Measuring the stiffness of bacterial cells from growth rates in hydrogels of tunable elasticity. Mol Microbiol. 2012;84(5):874–891. doi: 10.1111/j.1365-2958.2012.08063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Natl Acad Sci USA. 2011;108(9):3584–3589. doi: 10.1073/pnas.1018824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korniss G, Novotny MA, Guclu H, Toroczkai Z, Rikvold PA. Suppressing roughness of virtual times in parallel discrete-event simulations. Science. 2003;299(5607):677–679. doi: 10.1126/science.1079382. [DOI] [PubMed] [Google Scholar]
- 28.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 29.Quan S, et al. Adaptive evolution of the lactose utilization network in experimentally evolved populations of Escherichia coli. PLoS Genet. 2012;8(1):e1002444. doi: 10.1371/journal.pgen.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51(3):246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Ferrières L, et al. Silent mischief: Bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol. 2010;192(24):6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186(3):595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol. 2010;Chapter 14:Unit 14.20. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80(3):612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown PJ, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2012;109(5):1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuru E, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51(50):12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caparrós M, Pisabarro AG, de Pedro MA. Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol. 1992;174(17):5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ottolenghi AC, Caparrós M, de Pedro MA. Peptidoglycan tripeptide content and cross-linking are altered in Enterobacter cloacae induced to produce AmpC beta-lactamase by glycine and D-amino acids. J Bacteriol. 1993;175(5):1537–1542. doi: 10.1128/jb.175.5.1537-1542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pisabarro AG, de Pedro MA, Vázquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161(1):238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc Natl Acad Sci USA. 2012;109(10):E595–E604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.