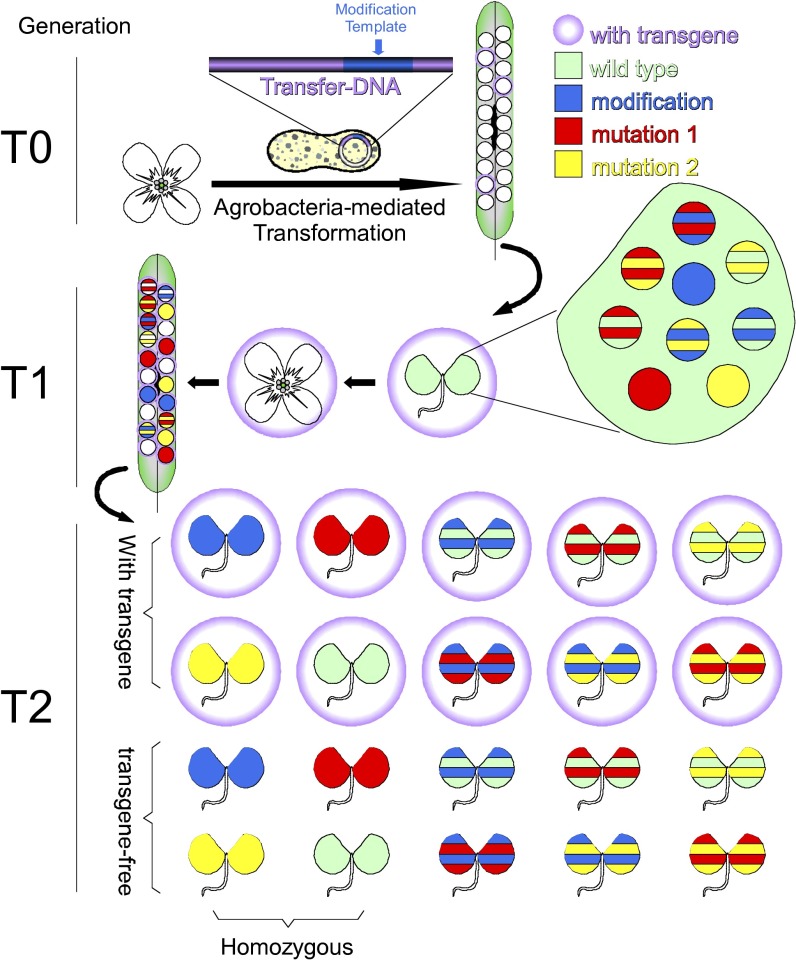
Targeted gene editing could offer tremendous advantages over traditional plant breeding to create new cultivars with advantageous combinations of alleles, especially when stacking of important traits is needed for crop improvement. Traditional methods of combining desirable alleles of different genes involve time-consuming crosses and selections, and in some cases such as combining closely linked loci, traditional methods are impossible to implement. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) relies on a small guide RNA (gRNA) molecule to direct specific cleavages of DNA sequences, and it has been widely tested for targeted gene editing in many organisms including plant species such as Arabidopsis, sorghum, rice, wheat, and tobacco. CRISPR/Cas can potentially be used to directly and precisely modify genes in the top commercial crop lines within a short period without affecting the existing valuable traits. However, before CRISPR/Cas can be widely used for crop improvement, several fundamental questions need to be addressed. What types of mutations can the CRISPR/Cas system generate in plants? Are the edited genomes stable and heritable? How often does unintended off-target mutagenesis occur and how do we decrease the off-target effects? In PNAS, Feng et al. (1) report a detailed analysis of CRISPR/Cas-mediated gene editing in Arabidopsis to address the aforementioned questions by following several generations of transgenic plants that harbor the CRISPR/Cas system, which was introduced into Arabidopsis by agrobacteria-mediated transformation. They studied seven genes at 12 distinct target sites to elucidate the pattern, specificity, and heritability of mutations induced by CRISPR/Cas. They clearly demonstrated that CRISPR/Cas could be used to generate transgene-free Arabidopsis plants with specific and heritable mutations within two generations (Fig. 1).
Fig. 1.
Efficient production of transgene-free Arabidopsis plants with desired modifications at the targeted loci. Arabidopsis flowers in the T0 generation are infiltrated with Agrobacteria that carry the transgenes for the CRISPR/Cas system and/or a template for modification. T1 plants carrying the transgenes are selected using an appropriate selection marker. Transgene-free plants with the desired modifications (solid blue colored seedling without a purple circle) can be obtained in the T2 generation. A purple circle indicates seeds, seedlings, or flowers that carry transgenes in the genome. Green, blue, red, and yellow refer to the cells that carry WT copy, desired modification, mutation 1, and mutation 2 at the targeted gene locus, respectively. There can be additional mutations in the population that are not shown in the figure. Solid colors indicate homozygous for the particular alleles. Stripped colors refer to heterozygous for the particular gene.
The predominant mutations generated by CRISPR/Cas in Arabidopsis are 1-bp deletions/insertions (1). This may reflect the most common way for a cell to imperfectly repair the double strand breaks (DSBs) through nonhomologous end-joining repair. Cleavage of the target DNA by Cas/gRNA predominantly generates 1-bp overhangs (2). The overhangs can be blunted by removing or filling in one nucleotide on each DNA strand before the breaks are joined by DNA ligase, resulting in the repaired DNA with 1-bp deletion or insertion. Other types of mutations occur at much lower frequency. Small deletions less than 20 bp long are the second most common mutation type. Larger deletions up to 100 bp are sometimes detected, albeit even more infrequently.
It was an open question whether all target sites in Arabidopsis were prone to modifications by CRISPR/Cas or if some of the sites were resistant. Feng et al. reported a 100% success rate for all 12 of the target sites tested, suggesting that CRISPR/Cas is able to generate mutations in Arabidopsis regardless of the gene structure and chromatin status (1). The results are also in line with other studies using CRISPR/Cas-based genome editing in plants and in animals (3–6). However, the efficiency of gene editing does vary among different target sites. The percentage of T1 plants with no detected mutations varies from 8% to 70% depending on the target sites (1). The presence of functional Cas9/gRNA does not always produce mutations in T1 plants. However, modification events are detected in most of the T2 plants derived from those WT-like T1 plants, suggesting that modification by CRISPR/Cas is a progressive process (1). The conditions under which CRISPR/Cas-mediated modifications take place in plants still remain unclear.
To use CRISPR/Cas for crop improvement, mutations generated by CRISPR/Cas have to be stable and heritable. Previous CRISPR studies in plant systems focused on testing the feasibility of the CRISPR/Cas system and were performed only on cultured cells or on the first generation of the transgenic lines (4, 5, 7). It has been shown that CRISPR/Cas can efficiently edit genes in somatic cells in Arabidopsis because mutations are easily detected in plant cells or in the T1 transgenic plants with Cas9/gRNA. Feng et al. investigate whether the editing events induced by CRISPR/Cas could take place in the ancestral cells of the germline or within the germline itself (1). The CRISPR/Cas system appears nonfunctional in the germline of the infiltrated T0 plants because plants with homozygous or biallelic mutations at the target sites were not detected in T1 plants (1). However, the inflorescence meristem, floral meristem, and/or germlines of the T1 plants are efficiently modified by the CRISPR/Cas system, because 22% of the T2 plants analyzed carried uniform mutations as being homozygous. More importantly, the mutations observed in T1 and T2 plants can be efficiently transmitted to the next generations. Furthermore, the mutations generated by CRISPR/Cas are stable and are not subject to further modifications by CRISPR/Cas (1). It is quite convincing that CRISPR/Cas can generate stable and heritable mutations, which can become homozygous in T2 plants (Fig. 1).
It is highly desired that the CRISPR/Cas system be removed after the target gene editing is accomplished during crop improvement. Plants that have the desired traits and that are transgene free will meet less resistance in the process of gaining regulatory approval for commercial applications. Transgene-free plants can also avoid further nonspecific modifications by the CRISPR/Cas system. Fortunately, it is not difficult to get rid of the CRISPR/Cas transgenes. In fact, transgene-free plants can be readily obtained in the T2 generation because the Cas9/gRNA construct can be easily segregated out (1) (Fig. 1). There is no need to conduct crosses to remove the transgenes, and thus segregation of valuable traits can
The work by Feng et al. addresses several key questions regarding CRISPR/Cas-mediated genome editing in plants.
be avoided. CRISPR/Cas promises rapid generation of nontransgenic crops with improved traits. With the recent development of ribozyme-flanked guide RNAs (8), it has become feasible to produce multiple gRNAs using a single construct to edit multiple genes simultaneously, providing an effective way to stack several traits and to remove the transgenes within a very short period.
Off-target effects are a major concern for CRISPR/Cas-mediated gene editing. In vitro biochemical studies and in vivo studies in animal cells have revealed significant off-target cleavages on DNA sites not perfectly matching the 20-bp complimentary region of the gRNA (2, 9–11). Therefore, it was surprising that the Cas9/gRNA construct targeting one site in the GA1 gene did not cause any off-target mutations (1). However, more extensive investigation is still needed regarding the specificity of the CRISPR/Cas system. This can be accomplished by testing more gRNAs-mediated genome editing, as well as in other plants. If off-target effects are found in other gRNA-mediated genome editing and/or in other plants, there are new methods to minimize the off-target effects. Using truncated gRNAs and using the Cas9-nickase/dual-gRNA system can greatly reduce off-target mutagenesis (12, 13).
Sometimes a valuable trait is conferred by a specific allele of a gene, and introducing this trait into a plant requires particular modifications such as a specific base pair change or an addition of a stretch of specific DNA to a desired location. This can in theory be achieved by DNA repair through homology-dependent recombination (HDR) with the presence of a repair template, after DSB of the target DNA is generated by Cas9/gRNA. The feasibility of this approach in plants was demonstrated by using CRISPR/Cas to modify a mutated and nonfunctional β-glucuronidase (GUS) reporter gene with the presence of a correct GUS gene template. About one-third of the transgenic T1 plants (16 of 44) showed obvious GUS staining (1). The staining is of a mosaic pattern, suggesting that the HDR events do not occur uniformly. Two T2 populations of those 16 T1 plants with GUS staining segregated some plants with uniform GUS staining, suggesting that the repaired GUS gene was homozygous and heritable (1). The results demonstrate that CRISPR/Cas system can be used to generate specialty alleles of a gene for crop improvement. It is also conceivable that CRISPR/Cas can be used to place markers such as a GFP gene in a desired location, providing valuable tools for basic research.
In summary, CRISPR/Cas is a powerful tool for efficient and specific gene editing in plants. The work by Feng et al. (1) addresses several key questions regarding CRISPR/Cas-mediated genome editing in plants, laying a solid foundation for using CRISPR/Cas in crop improvement.
Footnotes
The authors declare no conflict of interest.
See companion article on page 4632.
References
- 1.Feng Z, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao J, et al. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23(10):1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 6.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing [published online ahead of print December 30, 2013] J Integr Plant Biol. 2013 doi: 10.1111/jipb.12152. 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 9.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs [published online ahead of print January 26, 2014] Nat Biotechnol. 2014 doi: 10.1038/nbt.2808. 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]