Significance
The adenosine A1 receptor (A1AR) is an important target for cardioprotection, but current A1AR drugs are limited for this indication because of the occurrence of bradycardia as a major adverse effect mediated by the same receptor. To address this problem, we designed a ligand that simultaneously bridges two different sites on the A1AR; the hypothesis is that this bitopic mode would result in unique receptor conformations that will signal to desirable pathways while sparing those pathways mediating undesirable effects. This mechanism was validated in native rodent cells and isolated rat atria, providing proof of concept that the design of bitopic ligands may be a path forward to separating beneficial from harmful effects mediated by the same drug target.
Abstract
The concepts of allosteric modulation and biased agonism are revolutionizing modern approaches to drug discovery, particularly in the field of G protein-coupled receptors (GPCRs). Both phenomena exploit topographically distinct binding sites to promote unique GPCR conformations that can lead to different patterns of cellular responsiveness. The adenosine A1 GPCR (A1AR) is a major therapeutic target for cardioprotection, but current agents acting on the receptor are clinically limited for this indication because of on-target bradycardia as a serious adverse effect. In the current study, we have rationally designed a novel A1AR ligand (VCP746)—a hybrid molecule comprising adenosine linked to a positive allosteric modulator—specifically to engender biased signaling at the A1AR. We validate that the interaction of VCP746 with the A1AR is consistent with a bitopic mode of receptor engagement (i.e., concomitant association with orthosteric and allosteric sites) and that the compound displays biased agonism relative to prototypical A1AR ligands. Importantly, we also show that the unique pharmacology of VCP746 is (patho)physiologically relevant, because the compound protects against ischemic insult in native A1AR-expressing cardiomyoblasts and cardiomyocytes but does not affect rat atrial heart rate. Thus, this study provides proof of concept that bitopic ligands can be designed as biased agonists to promote on-target efficacy without on-target side effects.
G protein-coupled receptors (GPCRs) are the largest family of cell surface proteins and tractable drug targets (1, 2). Unfortunately, there remains a high attrition rate associated with traditional GPCR-based drug discovery that, in part, reflects an emphasis on the endogenous agonist binding (orthosteric) site as the predominant means of achieving selective GPCR drug targeting (3). Over the last decade, substantial breakthroughs have occurred in the exploitation of topographically distinct GPCR allosteric sites as a means for attaining greater selectivity, especially in those instances where there is high sequence similarity in the orthosteric site across GPCR subtypes (4–6). However, there are increasing examples where both the therapeutic effect and adverse effects are mediated by the same GPCR target (7). In these situations, the desired selectivity needs to be attained at the level of the intracellular signaling pathways linked to a given receptor subtype.
GPCRs are highly dynamic proteins, fluctuating between different conformations; these conformations can be linked to different cellular outcomes (8). Thus, chemically distinct ligands, interacting with either orthosteric or allosteric sites, have the potential to stabilize different interaction networks within a GPCR to promote a subset of signaling pathways linked to the receptor at the expense of others. This phenomenon has been termed biased agonism (7, 9, 10). The overall promise of biased agonism is the ability to design GPCR ligands that selectively engage therapeutically relevant signaling pathways while sparing pathways that contribute to undesirable side effects mediated by the same target.
The adenosine receptor (AR) family is an important class of physiologically and therapeutically relevant GPCRs that can benefit substantially from more selective drug targeting. Although all four AR subtypes are expressed in the mammalian heart (11, 12), the well-known protective effects of adenosine in this tissue are predominantly mediated by the adenosine A1 receptor (A1AR) subtype, especially under conditions of ischemia and reperfusion injury (13–17). Unfortunately, the transition of A1AR agonists into the clinic has been severely hindered because of high doses causing on-target bradycardia, atrioventricular block, and hypotension (13, 18). As a consequence, clinical trials of AR agonists have had limited success because of the suboptimal dose of agonist that can be used (19–22). It is possible that this problem may be overcome through the exploitation of biased agonism at the A1AR.
Although no study has identified biased orthosteric A1AR ligands, we recently showed that the 2-amino-3-benzoylthiophene allosteric modulator (VCP171) could promote biased signaling in the activity of the prototypical orthosteric agonist, R(-)N6-(2-phenylisopropyl) adenosine (R-PIA) (23). Thus, we hypothesized that the rational design of a bitopic ligand (i.e., a class of hybrid molecule containing both orthosteric and allosteric pharmacophores) (24–26) may be able to achieve high efficacy and biased agonism at the A1AR in a single molecule. Herein, we report proof of concept that it is possible to use this approach as a means to dissociate on-target efficacy from on-target side effects.
Results
Rationale for Design and Synthesis of Hybrid Orthosteric/Allosteric Ligands of the A1AR.
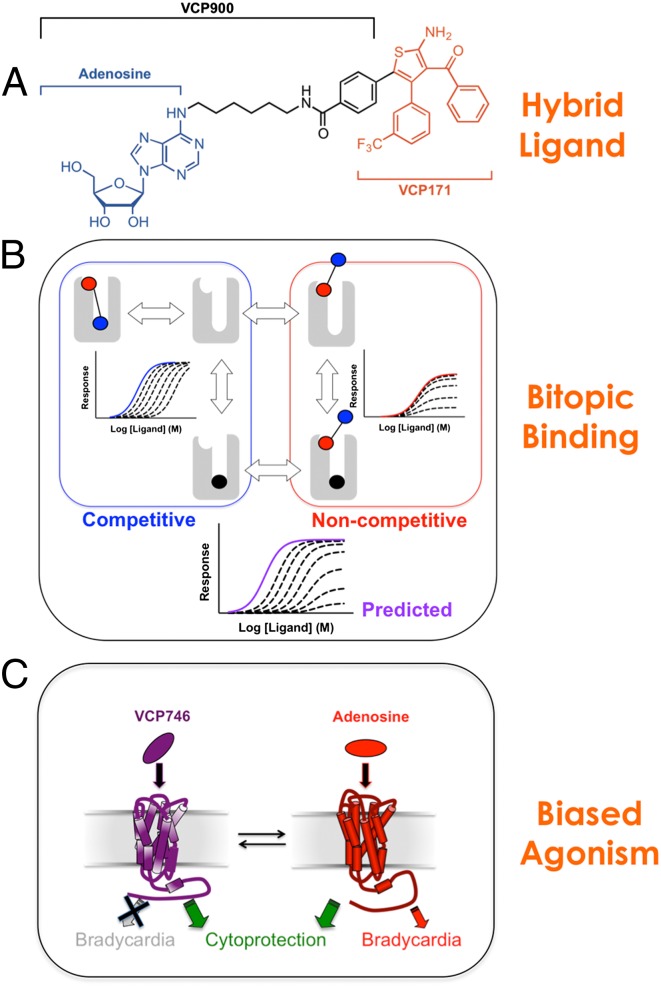
Fig. 1 summarizes the chemical biology framework underpinning the generation of hybrid orthosteric/allosteric ligands for the current study. We chose to link the endogenous agonist, adenosine, to the allosteric modulator, VCP171 (Fig. 1A and Fig. S1), with the hypothesis being that the adenosine moiety would confer high efficacy to the hybrids, whereas the VCP171 moiety would induce biased signaling (23). Although the location of the allosteric site on the A1AR has not been definitively determined, it is thought to comprise residues that are more extracellular to the orthosteric site (27–29). Assuming that the hybrid molecule binds concomitantly to both sites in this topography, Fig. 1B illustrates schematically the simplest expectations arising from such a bitopic mode of orientation with two assumptions. (i) A bitopic mode of engagement would represent the higher-affinity interaction and be largely indistinguishable from a reversible competitive interaction, because the orthosteric site is engaged (in addition to the allosteric site). (ii) If the orthosteric site is already occupied (e.g., in the presence of very high concentrations of orthosteric antagonist), then the bitopic ligand may adopt a different, lower-affinity orientation using only the allosteric site and thus, allow an allosteric interaction between the two molecules to occur (24–26, 30). If the allosteric moiety displays agonism, then the additional potential exists for a noncompetitive interaction when the ligands are interacting allosterically. As shown in Fig. S2, this situation is indeed the case when VCP171 (allosteric agonist) is tested against 8-cyclopentyl-1,3-dipropylxanthine (DPCPX; orthosteric antagonist) in an assay of A1AR-mediated ERK1/2 phosphorylation, whereas simple competition is observed when an orthosteric agonist (R-PIA) is used. Thus, it may be predicted that a bitopic ligand retaining these features would display a mixed mode of competitive (at lower concentrations of antagonist) and noncompetitive antagonism (at higher concentrations of antagonist). If biased agonism is attained, Fig. 1C illustrates an idealized pathophysiologically relevant outcome for cardiac A1ARs, where on-target cytoprotection is attained in the absence of on-target bradycardia.
Fig. 1.
Building biased bitopic agonists. (A) VCP746 is a rationally designed hybrid comprising the endogenous orthosteric agonist (adenosine) linked to a positive allosteric modulator (VCP171); VCP900 is adenosine plus the linker alone. (B) Theoretical behavior of a bitopic agonist. The black circle denotes the orthosteric antagonist (e.g., DPCPX), the blue circle denotes the orthosteric agonist, and the pink circle denotes the allosteric pharmacophore. The scheme on the left represents expectations for a simple competitive interaction, whereas the scheme on the right represents negative allosteric modulation by orthosteric antagonist of allosteric agonist signaling efficacy (as seen for the interaction between DPCPX and VCP171) (Fig. S2). (C) Scheme summarizing ideal bias properties of an A1AR therapeutic.
Hydrid Orthosteric/Allosteric Ligands Are Potent Agonists of the A1AR.
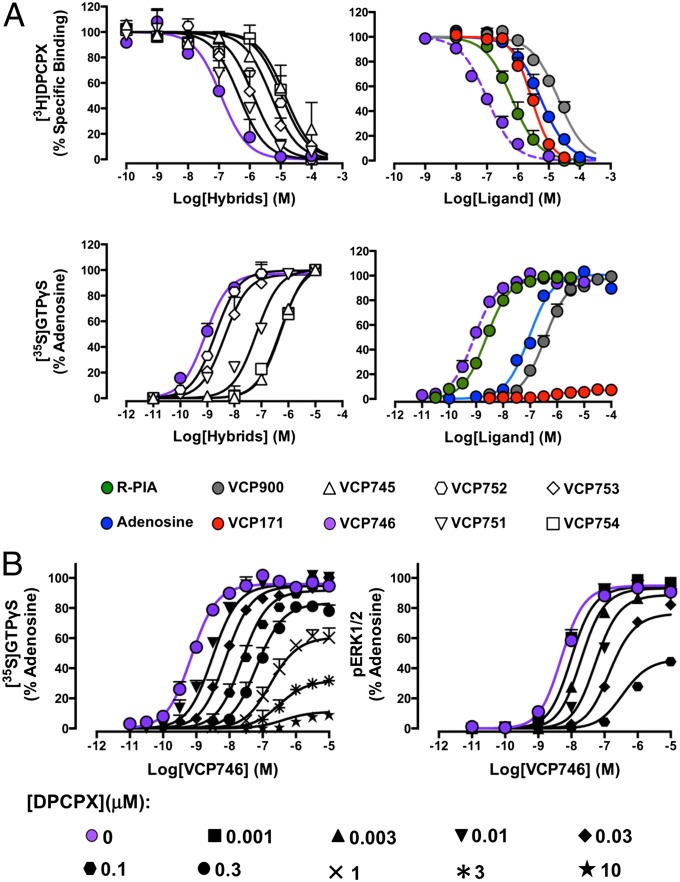
It is known that the introduction of bulky substituents at the N6 position of adenosine is often well-tolerated (31, 32). The introduction of bulky substituents at the four and five positions of the thiophene ring of 2-amino-3-benzoylthiophene modulators can also preserve or enhance their allosteric properties (23, 33–35). We, thus, generated six hybrid ligands by attaching the N6 of adenosine to the five position of the thiophene ring of VCP171 with a 4-benzamidoalkyl linker that ranged from 2 to 12 carbon atoms in length (Fig. S1). Using the antagonist radioligand, [3H]DPCPX (1 nM), as a probe of orthosteric site occupancy, we determined the affinity of the hybrids and comparator ligands in whole-cell binding assays performed on FlpIn-CHO cells stably expressing the human A1AR. As shown in Fig. 2A, all compounds inhibited the binding of [3H]DPCPX, indicating a competitive interaction or in the case of VCP171, high negative binding cooperativity (fixed to a value of logα = −2.5; values above this number yielded a worsening of the fit as judged by increases in the sum of squares); the estimated affinities for each of the ligands are shown in Table 1. We found a bell-shaped relationship between linker length and hybrid compound affinity (Fig. S3A). Hybrid VCP746, possessing a 6 carbon 4-benzamidohexyl linker between orthosteric and allosteric moieties, had the highest affinity (KI ∼ 60 nM), which was almost 100-fold higher than the affinity exhibited by either of its constituent molecules (adenosine or VCP171). To assess the effect of the linker itself on the observed pharmacology, we synthesized the orthosteric analog, VCP900, which comprised adenosine with an N6-(6-benzamidohexyl) substituent. VCP900 exhibited a slightly lower affinity for the receptor compared with adenosine, suggesting that the addition of the linker has a modest inhibitory effect on orthosteric ligand affinity. A comparison between VCP900 and VCP746, thus, highlights a striking overall improvement in affinity (200-fold) that is gained on incorporation of the allosteric moiety, VCP171.
Fig. 2.
(A) Pharmacological characterization of hybrid and comparator ligands in whole-cell radioligand binding assays using [3H]DPCPX and membrane-based functional assays of [35S]GTPγS binding in CHO cells stably expressing the human A1AR. Data represent the mean of three experiments ± SEMs performed in duplicate. (B) The functional interaction between VCP746 and DPCPX conforms to the expectations of a bitopic receptor model (Fig. 1) in assays of A1AR-mediated ERK1/2 phosphorylation or [35S]GTPγS binding. Data represent the mean of three experiments ± SEMs performed in duplicate.
Table 1.
Affinity (pKI or pKB), potency (pEC50), and maximal agonist effect (Emax) estimates of hybrid and comparator ligands in FlpIn-CHO cells stably expressing the A1AR
| Hybrid orthosteric/allosteric | Orthosteric | Allosteric (VCP171) | ||||||||
| VCP754 | VCP753 | VCP746 | VCP752 | VCP751 | VCP745 | R-PIA | Adenosine | VCP900 | ||
| Linker length | 2 | 4 | 6 | 8 | 10 | 12 | n/a | n/a | 6 | n/a |
| pKI* or pKB† | 5.16 ± 0.17 | 6.65 ± 0.13 | 7.23 ± 0.17 | 6.18 ± 0.14 | 5.61 ± 0.13 | 5.27 ± 0.12 | 6.47 ± 0.07 | 5.53 ± 0.05 | 4.97 ± 0.05 | 5.81 ± 0.04 |
| pEC50 | 6.37 ± 0.08 | 8.31 ± 0.08 | 9.05 ± 0.08 | 8.71 ± 0.08 | 7.20 ± 0.08 | 6.34 ± 0.08 | 8.60 ± 0.04 | 7.03 ± 0.06 | 6.45 ± 0.05 | 6.01 ± 0.30 |
| Emax | 103 ± 5 | 97 ± 5 | 98 ± 1 | 100 ± 2 | 100 ± 5 | 106 ± 3 | 98 ± 1 | 100 | 97 ± 2 | 7 ± 1 |
Values represent the means ± SEMs from three experiments performed in duplicate. n/a, not applicable.
Negative logarithm of the hybrid or orthosteric ligand equilibrium dissociation constant.
Negative logarithm of the allosteric modulator equilibrium dissociation constant.
Because the A1AR preferentially couples to Gi/o proteins, we subsequently used an assay of [35S]GTPγS binding as a proximal measure of receptor activity to determine the agonistic properties of the hybrid molecules. In this assay, adenosine and all of the hybrids were full agonists, albeit with varying potencies (Fig. 2A and Table 1). As with the binding assays, a bell-shaped relationship was noted between linker length and agonist potency (Fig. S3B), with VCP746 remaining the most potent. In contrast, the allosteric agonist, VCP171, was minimally active. Because the adenosine A3 receptor (A3AR) shares closest sequence homology to and signal transduction mechanisms with the A1AR and has also been implicated in cardiac physiology (36), we also assessed activity at the A3AR. In contrast to robust [35S]GTPγS binding promoted by adenosine, neither VCP746 or VCP171 exhibited any A3AR agonism; rather, a modest inhibitory effect was noted at the highest concentration used (Fig. S4). Collectively, these results indicate that it is possible to generate highly potent agonists for the A1AR by linking orthosteric and allosteric pharmacophores.
Validation of a Bitopic Binding Mode for VCP746 at the A1AR.
To confirm that the mechanism of action (VCP746) involved concomitant association with both orthosteric and allosteric sites on the A1AR, we performed cell-based interaction studies between the hybrid and DPCPX and compared the results with theoretical model predictions of bitopic ligand–receptor interaction (Fig. 1B). The interaction between DPCPX and VCP746 was assessed using both [35S]GTPγS and ERK1/2 phosphorylation assays, and the latter was chosen to ensure that the observed results were not caused by differences between membrane- and whole cell-based signaling assays. As shown in Fig. 2B, increasing concentrations of DPCPX induced a reduction in VCP746 potency with no change in the maximal response, which was followed by a depression of the Emax in both instances, supporting the hypothesis that the hybrid molecule is, indeed, interacting with the A1AR in a bitopic mode.
VCP746 Is a Biased Agonist.
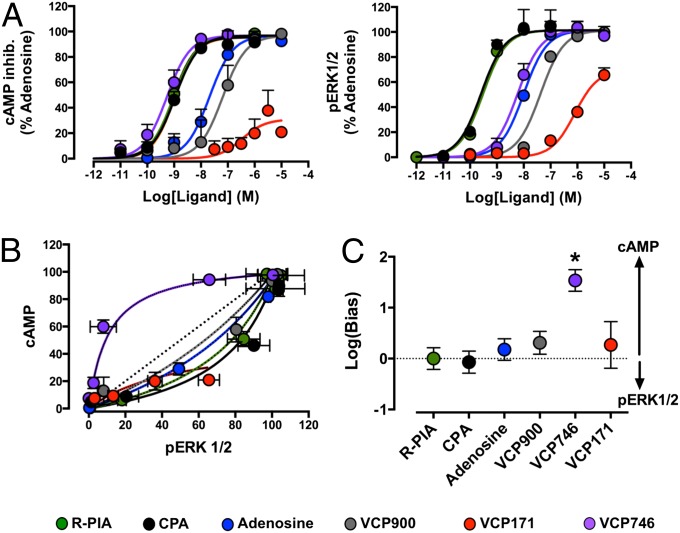
To assess the potential for our bitopic ligand to display biased agonism, we extended our assays to investigate effects on A1AR-mediated inhibition of forskolin-stimulated cAMP accumulation, which is a canonical pathway linked to A1AR–Gi/o–adenylate cyclase coupling (11), and included an additional reference orthosteric agonist, N6-cyclopentyladenosine (CPA), commonly used for studying A1AR pharmacology. The results from these experiments were then compared with results measuring A1AR-mediated ERK1/2 phosphorylation, and the latter was chosen as a noncanonical downstream convergent signaling pathway, because it can be both G protein-dependent and -independent. In both assays, the allosteric agonist (VCP171) had low efficacy, whereas the orthosteric agonists (R-PIA, CPA, adenosine, and VCP900) and the bitopic agonist (VCP746) displayed full agonism (Fig. 3A). However, VCP746 showed a difference in its relative potency between the two pathways—a classic hallmark of biased agonism. The divergence of VCP746 from prototypical orthosteric agonist pharmacology is best illustrated when the data are replotted in the form of a bias plot (Fig. 3B); relative to the orthosteric agonists, which show a preferential coupling to ERK1/2 phosphorylation, VCP746 is biased to the cAMP pathway, indicating that it is able to engender a different conformation of the A1AR. Application of an operational model of agonism (37, 38) to the data allowed for the quantification of the degree of the bias relative to the preferences of R-PIA (Fig. 3C), where it was found that VCP746 had an approximately 30-fold preference to cAMP relative to ERK1/2 (Fig. 3C and Table S1). Similar results were obtained comparing signaling in the [35S]GTPγS with signaling in the ERK1/2 pathway (Fig. S5). These findings represent the largest degree of biased agonism thus far identified at the A1AR.
Fig. 3.
VCP746 is a biased agonist. (A) Effects of selected ligands on A1AR-mediated inhibition of (Left) forskolin-stimulated cAMP accumulation or (Right) ERK1/2 phosphorylation (pERK1/2) in CHO FlpIn cells stably expressing the human A1AR. (B) Bias plots showing the effects of equimolar agonist concentrations at the two pathways. (C) Quantification of bias factors using R-PIA as the reference agonist. Data represent the mean of three experiments ± SEMs performed in duplicate. *P < 0.05; Student's t test.
(Patho)physiological Relevance of Biased Agonism by VCP746.
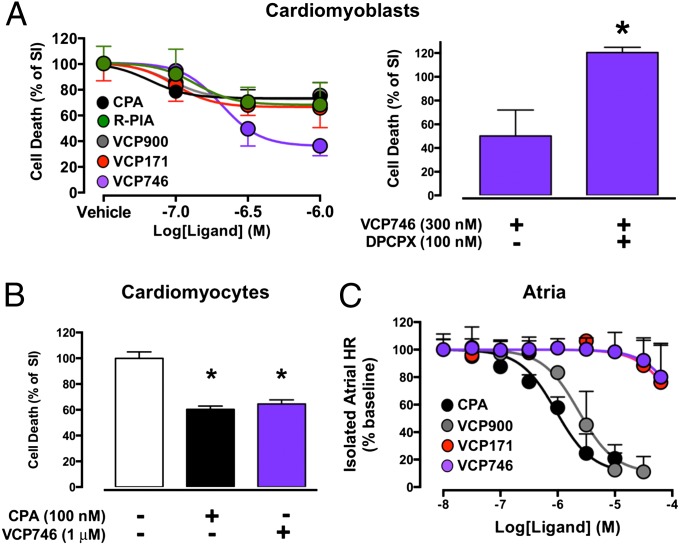
The findings from our recombinant cell assays indicated that VCP746 could promote a unique conformation of the A1AR relative to prototypical A1AR ligands. To ascertain the physiological relevance of biased agonism by VCP746, however, it is necessary to determine its pharmacology in native A1AR models. A highly desirable consequence of A1AR activation is protection against cardiac cell death associated with hypoxic and/or ischemic conditions, but this protective ability remains suboptimally exploited because of concomitant on-target adverse effects, such as bradycardia and hypotension. We, thus, subjected rat H9c2(2-1) embryonic cardiomyoblasts, which endogenously express the A1AR, to simulated ischemia (SI) at a pH 6.4 under 100% nitrogen gas atmosphere for 12 h at 37 °C and assessed cell viability using propidium iodide (PI) in the absence and presence of the orthosteric full agonists R-PIA, CPA, and VCP900, the allosteric ligand VCP171, or the bitopic ligand VCP746 (Fig. 4A) (39, 40). All ligands prevented H9c2(2-1) cell death compared with cells treated with vehicle alone, but the greatest effect was observed with VCP746. To confirm that this effect was mediated by the A1AR, the decrease in H9c2(2-1) cell death by VCP746 was prevented by the A1AR orthosteric antagonist DPCPX (Fig. 4A). Key findings from the cardiomyoblast studies were also confirmed in a second native A1AR-expressing cell system: neonatal cardiomyocytes (NCMs). In this model, both the canonical A1AR orthosteric agonist CPA (100 nM) and VCP746 (1 μM) prevented NCM cell death caused by 3 h of SI (Fig. 4B). Finally, we compared the effects of increasing concentrations of CPA with the effects mediated by VCP746, its orthosteric congener VCP900, or its allosteric moiety VCP171 on A1AR-mediated inhibition of isolated rat atrial rate (Fig. 4C). In contrast to the orthosteric CPA or VCP900, which caused an almost complete reduction in heart rate, neither VCP746 nor VCP171 had any substantial effect, even at a concentration of VCP746 that had maximal cytoprotective activity. Collectively, these results highlight that it is possible to achieve therapeutically relevant biased agonism through the rational design of bitopic ligands.
Fig. 4.
VCP746 is cytoprotective but does not cause bradycardia. (A) Effects of the indicated compounds on cell death under conditions of SI in native A1AR-expressing rat h9c2 caridomyoblasts. Data represent the means ± SEMs of three to nine experiments. (B) Effects of the indicated compounds on cell death under SI conditions in native A1AR-expressing rat neonatal ventricular cardiomyocytes. Data represent the means ± SEMs of three experiments. (C) Effects of the indicated compounds on rat atrial heart rate (HR). Data represent the means ± SEMs of three experiments. *P < 0.05 as determined by Student's t test (A) or one way ANOVA followed by Dunnett's posttest (B).
Discussion
This study provides proof of concept for the rational design of a biased ligand for the A1AR with improved pharmacological properties by concomitant targeting of orthosteric and allosteric binding sites on this receptor. Although VCP746 is not the first example of a bitopic ligand described for GPCRs, it is a previously unidentified example with a shown dissociation of potential on-target efficacy from on-target side effect liability. This finding is particularly pertinent to the targeting of A1ARs in the heart, where the engagement of powerful cytoprotective mechanisms by existing drug therapies is hampered by dose-limiting bradycardia and hypotensive episodes.
The paradigms of allosteric modulation and biased agonism are revolutionizing modern drug discovery. Both phenomena exploit the dynamic nature of GPCRs, where chemically distinct ligands stabilize different subsets of receptor conformations to yield new interactive properties of the receptor to other ligands (allosteric modulation) or intracellular effectors (biased agonism) (41). To date, biased ligands for GPCRs have been identified through two general approaches: compound screening at preselected pathways (often β-arrestin– vs. G protein-dependent) or retrospectively by taking established compounds with divergent pharmacologies and profiling them broadly to identify potential differentiating signatures (42–46). The first approach relies on mechanistic rationales to guide the choice of pathway(s) chosen for screening. Unfortunately, most diseases are polygenic in nature, and it is usually not known what pathway(s) is most relevant in the disease context. The second approach relies on preexisting data, indicating that different classes of ligand targeting a common GPCR yield functionally divergent outcomes in terms of on-target efficacy and/or adverse effects. The goal of this latter approach is to cluster compounds in terms of pathway-specific signatures that may prove predictive in subsequent screening studies or identify the actual pathways mediating the desired profiles. However, this approach can prove time-consuming, with no guarantee of a clear path. Our study presents a variation on these methods, in that we purposefully exploit the allosteric nature of GPCRs to maximize the likelihood that a biased ligand can be rationally engineered from the outset. This approach is especially noteworthy given that previous screening studies have been unable to identify biased ligands at the A1AR (47, 48). Moreover, assaying of the engineered biased ligand in native A1AR-expressing systems revealed unique properties, suggesting a favorable efficacy vs. side effect profile.
Recent studies have highlighted both the attractive potential of linking orthosteric and allosteric pharmacophores and the challenges associated with the development of such ligands (25, 26). Ideally, an optimal bitopic ligand should display improved affinity relative to its individual orthosteric and allosteric pharmacophores. This property was observed with our agonist, VCP746, at the A1AR and previously, the bitopic antagonist, THRX-160209, at the M2 muscarinic acetylcholine receptor (49) but not the bitopic agonists LUF6258 at the A1AR (28) or hybrid 2 at the M2 receptor (50). Possible reasons for the failure to improve affinity in these cases include a mismatch between the constituent orthosteric and allosteric pharmacophores (e.g., orthosteric agonist plus allosteric antagonist), a lack of concomitant engagement of both binding sites (e.g., suboptimal linker length), or a deleterious effect of the linker itself (26, 51, 52). We noted the latter with VCP900 (adenosine plus linker), which was less potent than adenosine itself. However, this finding actually meant that the observed gain in affinity and potency on addition of the allosteric moiety to yield the bitopic VCP746 was even more striking as a consequence. The bell-shaped relationship that we observed between hybrid compound affinity/potency and linker length may also have been suggestive of suboptimal engagement with both sites on the A1AR for the less potent compounds. It is for this reason that additional mechanistic studies were performed with VCP746 to provide further validation that the observed pharmacology was consistent with the expectations of a bitopic agonist model (Fig. 2).
As mentioned above, the other potential property of the bitopic ligand is that of biased agonism. Interestingly, prior studies at the M2 muscarinic receptor with hybrid 2 showed some evidence of bias in recombinant cell assays, although no physiological sequelae were identified (50). Irrespective, when taken together with our findings with VCP746, these results provide encouraging support for the hypothesis that bitopic agonists have a higher likelihood of yielding biased signaling. It is important to note, however, that the manifestation of biased agonism will always be cell/system-dependent, which is why it is necessary to use a reference ligand (e.g., R-PIA) when quantifying bias to cancel the impact of such system-dependent influences (53). Nonetheless, the system-dependent components of bias also mean that the pathways identified in a given cell background (e.g., CHO cell) may not necessarily be related to the mechanism(s) underlying the physiologically relevant bias. For instance, VCP746 is biased away from ERK1/2 phosphorylation relative to prototypical orthosteric agonists when assayed in our recombinant cell line. This result should not be taken as evidence that a bias to cAMP relative to ERK1/2 is the desired mechanism for cytoprotection with no on-target bradycardia, although it may represent a logical starting point for future mechanistic studies. In our view, the most appropriate role of recombinant cell studies in the detection and quantification of biased agonism is to serve as a robust and facile readout of the ability of test ligands to engender different conformational states of the receptor. After biased agonism has been identified, it should be validated in a more appropriate cellular/tissue model, which can also serve as the foundation of studies aimed at delineating the actual intracellular mechanisms underlying the bias. Ongoing work in our laboratory is addressing this important aspect of VCP746 pharmacology, but the major aim/outcome of the current study was the demonstration that it is actually possible to engineer bias in the absence of a defined cellular mechanism by exploiting the ability of allosteric sites to change the interactive properties of orthosteric sites.
In conclusion, we have designed, synthesized, and characterized a bitopic A1AR ligand that represents a previously unidentified high-affinity biased agonist for the A1AR that may provide a path forward to overcoming the current bottleneck in the preclinical progression of A1AR-targeting cytoprotective agents (namely, on-target side effects). Moreover, our study presents a general chemical biology framework that can potentially expedite preclinical workflows for discovering biased ligands at other GPCRs. For instance, one can envisage a scenario where different building blocks are used to design bitopic ligands, which are then profiled in recombinant lines to quickly assess for bias without any preconceived notions of what sort of pathway bias for which one should be looking. Depending on the outcome, ligands are then clustered, and representatives of each cluster are tested in more physiologically relevant systems. Identification of desired properties in the latter can be used to initiate mechanistic studies while still allowing the progression of already identified actives down the preclinical development path.
Materials and Methods
Chemical Synthesis.
A series of hybrid ligands, exhibiting both an orthosteric and an allosteric moiety, for the A1AR was designed and synthesized (Fig. 1 and Scheme S1). A detailed description of the synthesis is in SI Materials and Methods.
Cell Culture and Membrane Preparation.
FlpIn-CHO cells stably expressing the A1AR were generated and cultured as described previously (46). Membranes of A1ARs were generated as described previously (47). H9c2(2-1) rat cardiomyoblast cells were cultured as described previously (39, 40). NCMs were isolated and cultured as described previously (54). All experiments were performed under approval from the Monash University Animal Ethics Committee.
Radioligand Equilibrium Binding and Cell Signaling Assays in Intact Recombinant Cells.
Equilibrium radioligand binding, [35S]GTPγS binding, cAMP accumulation, and ERK1/2 phosphorylation assays were performed as described previously (23, 33) and in SI Materials and Methods.
SI Model in H9c2(2-1) Rat Cardiomyoblasts and Rat NCMs.
SI conditions were induced by removal of the growth DMEM and incubation of the cells at 37 °C under 100% nitrogen gas atmosphere for 12 [H9c2(2-1) cells] or 3 h (NCMs) in hypoxic SI buffer at pH 6.4 containing 137 mM NaCl, 3.5 mM KCl, 0.88 mM CaCl20.2H2O, 0.51 mM MgSO40.7H2O, 5.55 mM d-glucose, 4 mM Hepes, 10 mM 2-deoxy-d-glucose, and 20 mM dl-lactic acid plus 0.1% BSA. Fresh SI buffer was prepared for each experiment and sterile-filtered before experimentation. Agonists were dissolved in SI buffer and added to the wells at a range of concentrations.
Cell Viability Assay and Imaging.
Detection of nonviable cells resulting from ischemia was achieved by PI staining assay. For subsequent SI, cells were first washed with PBS and stained with 5 µM PI for 5 min followed by a PBS rinse two times before imaging. Images were taken using a confocal microscope (Nikon A1; Nikon Instruments) using 561-nm excitation laser. PI-positive cells were quantified using ImageJ (National Institute of Health Image; National Institute of Health). The normalized dead cell percentage was calculated by dividing the number of PI-positive cells per well by the average number of PI-positive cells in the SI-treated wells for that experiment.
Heart Rate Measurements in Isolated Rat Atria.
Rats were euthanized using CO2, and hearts were rapidly removed and placed in ice-cold Krebs solution of 120 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 0.5 mM EDTA, 22 mM NaHCO3, 11 mM glucose, and 2.5 mM CaCl2. Right atria were isolated and attached to a force transducer for recording of heart rate. After a 30-min equilibration period, concentration–response curves to VCP746, VCP171, and VCP900 were constructed. All experiments were performed under approval from the Monash University Animal Ethics Committee.
Data Analysis.
Computerized nonlinear regression was performed using Prism 6.0 (GraphPad Software) as described previously (37, 55, 56). All affinities, potencies, efficacies, and cooperativity parameters were estimated as logarithms. Results are expressed as means ± SEs unless otherwise stated. Statistical analyses were by Student t test or one-way ANOVA followed by Dunnett’s or Bonferroni’s posttest as appropriate. Values of P < 0.05 were considered statistically significant. More details on experimental methods and data analysis are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Michael Crouch and Ron Osmond for providing the ERK1/2 phosphorylation assay kit and Dr. Bing Wang for providing the NCMs. This work was funded by National Health and Medical Research Council of Australia (NHMRC) Program Grants 519461 and APP1055134 and Australian Research Council Discovery Grant DP110100687. L.T.M. is an Australian Research Council Discovery Early Career Research Award Fellow. P.M.S. and A.C are Principal Research Fellows of the NHMRC.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320962111/-/DCSupplemental.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7(4):339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Jacoby E, Bouhelal R, Gerspacher M, Seuwen K. The 7 TM G-protein-coupled receptor target family. ChemMedChem. 2006;1(8):761–782. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- 4.Christopoulos A. Allosteric binding sites on cell-surface receptors: Novel targets for drug discovery. Nat Rev Drug Discov. 2002;1(3):198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 5.May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2007;47(1):1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 6.Conn PJ, Jones CK, Lindsley CW. Subtype-selective allosteric modulators of muscarinic receptors for the treatment of CNS disorders. Trends Pharmacol Sci. 2009;30(3):148–155. doi: 10.1016/j.tips.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Violin JD, Lefkowitz RJ. β-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol Sci. 2007;28(8):416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: The impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62(2):265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leach K, Sexton PM, Christopoulos A. Allosteric GPCR modulators: Taking advantage of permissive receptor pharmacology. Trends Pharmacol Sci. 2007;28(8):382–389. doi: 10.1016/j.tips.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Mailman RB. GPCR functional selectivity has therapeutic impact. Trends Pharmacol Sci. 2007;28(8):390–396. doi: 10.1016/j.tips.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredholm BB, IJzerman AP, Jacobson KA, Klotz KN, Linden J. International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev. 2001;53(4):527–552. [PMC free article] [PubMed] [Google Scholar]
- 12.Headrick JP, Ashton KJ, Rose’meyer RB, Peart JN. Cardiovascular adenosine receptors: Expression, actions and interactions. Pharmacol Ther. 2013;140(1):92–111. doi: 10.1016/j.pharmthera.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, et al. Cardiac overexpression of A1-adenosine receptor protects intact mice against myocardial infarction. Am J Physiol Heart Circ Physiol. 2002;282(3):H949–H955. doi: 10.1152/ajpheart.00741.2001. [DOI] [PubMed] [Google Scholar]
- 14.Donato M, Gelpi RJ. Adenosine and cardioprotection during reperfusion—an overview. Mol Cell Biochem. 2003;251(1-2):153–159. [PubMed] [Google Scholar]
- 15.Minamino T. Cardioprotection from ischemia/reperfusion injury: Basic and translational research. Circ J. 2012;76(5):1074–1082. doi: 10.1253/circj.cj-12-0132. [DOI] [PubMed] [Google Scholar]
- 16.Reichelt ME, et al. Genetic deletion of the A1 adenosine receptor limits myocardial ischemic tolerance. Circ Res. 2005;96(3):363–367. doi: 10.1161/01.RES.0000156075.00127.C3. [DOI] [PubMed] [Google Scholar]
- 17.Marzilli M, Orsini E, Marraccini P, Testa R. Beneficial effects of intracoronary adenosine as an adjunct to primary angioplasty in acute myocardial infarction. Circulation. 2000;101(18):2154–2159. doi: 10.1161/01.cir.101.18.2154. [DOI] [PubMed] [Google Scholar]
- 18.Mustafa SJ, Morrison RR, Teng B, Pelleg A. Adenosine receptors and the heart: Role in regulation of coronary blood flow and cardiac electrophysiology. Handbook Exp Pharmacol. 2009;193(193):161–188. doi: 10.1007/978-3-540-89615-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopecky SL, et al. AmP579 Delivery for Myocardial Infarction REduction study A randomized, double-blinded, placebo-controlled, dose-ranging study measuring the effect of an adenosine agonist on infarct size reduction in patients undergoing primary percutaneous transluminal coronary angioplasty: The ADMIRE (AmP579 Delivery for Myocardial Infarction REduction) study. Am Heart J. 2003;146(1):146–152. doi: 10.1016/S0002-8703(03)00172-8. [DOI] [PubMed] [Google Scholar]
- 20.Kloner RA, et al. Impact of time to therapy and reperfusion modality on the efficacy of adenosine in acute myocardial infarction: The AMISTAD-2 trial. Eur Heart J. 2006;27(20):2400–2405. doi: 10.1093/eurheartj/ehl094. [DOI] [PubMed] [Google Scholar]
- 21.Kloner RA, Schwartz Longacre L. Introduction to proceedings of the NHLBI workshop: New Horizons in cardioprotection—a focused issue of Journal of Cardiovascular Pharmacology and Therapeutics. J Cardiovasc Pharmacol Ther. 2011;16(3-4):222. doi: 10.1177/1074248411413547. [DOI] [PubMed] [Google Scholar]
- 22.Braunwald E. Clinical efforts to reduce myocardial infarct size—the next step. J Cardiovasc Pharmacol Ther. 2011;16(3-4):349–353. doi: 10.1177/1074248411407637. [DOI] [PubMed] [Google Scholar]
- 23.Aurelio L, et al. Allosteric modulators of the adenosine A1 receptor: Synthesis and pharmacological evaluation of 4-substituted 2-amino-3-benzoylthiophenes. J Med Chem. 2009;52(14):4543–4547. doi: 10.1021/jm9002582. [DOI] [PubMed] [Google Scholar]
- 24.Valant C, Sexton PM, Christopoulos A. Orthosteric/allosteric bitopic ligands: Going hybrid at GPCRs. Mol Interv. 2009;9(3):125–135. doi: 10.1124/mi.9.3.6. [DOI] [PubMed] [Google Scholar]
- 25.Valant C, Robert Lane J, Sexton PM, Christopoulos A. The best of both worlds? Bitopic orthosteric/allosteric ligands of g protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2012;52(1):153–178. doi: 10.1146/annurev-pharmtox-010611-134514. [DOI] [PubMed] [Google Scholar]
- 26.Lane JR, Sexton PM, Christopoulos A. Bridging the gap: Bitopic ligands of G-protein-coupled receptors. Trends Pharmacol Sci. 2013;34(1):59–66. doi: 10.1016/j.tips.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Göblyös A, Ijzerman AP. Allosteric modulation of adenosine receptors. Purinergic Signal. 2009;5(1):51–61. doi: 10.1007/s11302-008-9105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narlawar R, et al. Hybrid ortho/allosteric ligands for the adenosine A(1) receptor. J Med Chem. 2010;53(8):3028–3037. doi: 10.1021/jm901252a. [DOI] [PubMed] [Google Scholar]
- 29.Peeters MC, et al. The role of the second and third extracellular loops of the adenosine A1 receptor in activation and allosteric modulation. Biochem Pharmacol. 2012;84(1):76–87. doi: 10.1016/j.bcp.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Valant C, et al. A novel mechanism of G protein-coupled receptor functional selectivity. Muscarinic partial agonist McN-A-343 as a bitopic orthosteric/allosteric ligand. J Biol Chem. 2008;283(43):29312–29321. doi: 10.1074/jbc.M803801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson RD, Secunda S, Daly JW, Olsson RA. Activity of N6-substituted 2-chloroadenosines at A1 and A2 adenosine receptors. J Med Chem. 1991;34(12):3388–3390. doi: 10.1021/jm00116a007. [DOI] [PubMed] [Google Scholar]
- 32.Middleton RJ, et al. New fluorescent adenosine A1-receptor agonists that allow quantification of ligand-receptor interactions in microdomains of single living cells. J Med Chem. 2007;50(4):782–793. doi: 10.1021/jm061279i. [DOI] [PubMed] [Google Scholar]
- 33.Valant C, et al. Delineating the mode of action of adenosine A1 receptor allosteric modulators. Mol Pharmacol. 2010;78(3):444–455. doi: 10.1124/mol.110.064568. [DOI] [PubMed] [Google Scholar]
- 34.Aurelio L, et al. The synthesis and biological evaluation of 2-amino-4,5,6,7,8,9-hexahydrocycloocta[b]thiophenes as allosteric modulators of the A1 adenosine receptor. Bioorg Med Chem Lett. 2011;21(12):3704–3707. doi: 10.1016/j.bmcl.2011.04.080. [DOI] [PubMed] [Google Scholar]
- 35.Valant C, et al. Synthesis and characterization of novel 2-amino-3-benzoylthiophene derivatives as biased allosteric agonists and modulators of the adenosine A(1) receptor. J Med Chem. 2012;55(5):2367–2375. doi: 10.1021/jm201600e. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5(3):247–264. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenakin T, Watson C, Muniz-Medina V, Christopoulos A, Novick S. A simple method for quantifying functional selectivity and agonist bias. ACS Chem Neurosci. 2012;3(3):193–203. doi: 10.1021/cn200111m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans BA, et al. Quantification of functional selectivity at the human α(1A)-adrenoceptor. Mol Pharmacol. 2011;79(2):298–307. doi: 10.1124/mol.110.067454. [DOI] [PubMed] [Google Scholar]
- 39.Urmaliya VB, et al. A novel highly selective adenosine A1 receptor agonist VCP28 reduces ischemia injury in a cardiac cell line and ischemia-reperfusion injury in isolated rat hearts at concentrations that do not affect heart rate. J Cardiovasc Pharmacol. 2010;56(3):282–292. doi: 10.1097/FJC.0b013e3181eb8563. [DOI] [PubMed] [Google Scholar]
- 40.Urmaliya VB, et al. Cardioprotection induced by adenosine A1 receptor agonists in a cardiac cell ischemia model involves cooperative activation of adenosine A2A and A2B receptors by endogenous adenosine. J Cardiovasc Pharmacol. 2009;53(5):424–433. doi: 10.1097/FJC.0b013e3181a443e2. [DOI] [PubMed] [Google Scholar]
- 41.Keov P, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors: A pharmacological perspective. Neuropharmacology. 2011;60(1):24–35. doi: 10.1016/j.neuropharm.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Aplin M, Christensen GL, Hansen JL. Pharmacologic perspectives of functional selectivity by the angiotensin II type 1 receptor. Trends Cardiovasc Med. 2008;18(8):305–312. doi: 10.1016/j.tcm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Schmid CL, et al. Functional selectivity of 6′-guanidinonaltrindole (6′-GNTI) at κ-opioid receptors in striatal neurons. J Biol Chem. 2013;288(31):22387–22398. doi: 10.1074/jbc.M113.476234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohinc BN, Gesty-Palmer D. Biased agonism at the parathyroid hormone receptor: A demonstration of functional selectivity in bone metabolism. Mini Rev Med Chem. 2012;12(9):856–865. doi: 10.2174/138955712800959125. [DOI] [PubMed] [Google Scholar]
- 45.Stallaert W, Dorn JF, van der Westhuizen E, Audet M, Bouvier M. Impedance responses reveal β₂-adrenergic receptor signaling pluridimensionality and allow classification of ligands with distinct signaling profiles. PLoS One. 2012;7(1):e29420. doi: 10.1371/journal.pone.0029420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen JA, et al. Discovery of β-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc Natl Acad Sci USA. 2011;108(45):18488–18493. doi: 10.1073/pnas.1104807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart GD, et al. Determination of adenosine A1 receptor agonist and antagonist pharmacology using Saccharomyces cerevisiae: Implications for ligand screening and functional selectivity. J Pharmacol Exp Ther. 2009;331(1):277–286. doi: 10.1124/jpet.109.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langemeijer EV, Verzijl D, Dekker SJ, Ijzerman AP. Functional selectivity of adenosine A1 receptor ligands? Purinergic Signal. 2013;9(1):91–100. doi: 10.1007/s11302-012-9334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinfeld T, Mammen M, Smith JAM, Wilson RD, Jasper JR. A novel multivalent ligand that bridges the allosteric and orthosteric binding sites of the M2 muscarinic receptor. Mol Pharmacol. 2007;72(2):291–302. doi: 10.1124/mol.106.033746. [DOI] [PubMed] [Google Scholar]
- 50.Antony J, et al. Dualsteric GPCR targeting: A novel route to binding and signaling pathway selectivity. FASEB J. 2009;23(2):442–450. doi: 10.1096/fj.08-114751. [DOI] [PubMed] [Google Scholar]
- 51.Mohr K, et al. Rational design of dualsteric GPCR ligands: Quests and promise. Br J Pharmacol. 2010;159(5):997–1008. doi: 10.1111/j.1476-5381.2009.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohr K, Schmitz J, Schrage R, Tränkle C, Holzgrabe U. Molecular alliance-from orthosteric and allosteric ligands to dualsteric/bitopic agonists at G protein coupled receptors. Angew Chem Int Ed Engl. 2013;52(2):508–516. doi: 10.1002/anie.201205315. [DOI] [PubMed] [Google Scholar]
- 53.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: Detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12(3):205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 54.Lekawanvijit S, et al. Does indoxyl sulfate, a uraemic toxin, have direct effects on cardiac fibroblasts and myocytes? Eur Heart J. 2010;31(14):1771–1779. doi: 10.1093/eurheartj/ehp574. [DOI] [PubMed] [Google Scholar]
- 55.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 56.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54(2):323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.