ABSTRACT
Signaling through vertebrate Hedgehog (Hh) proteins depends on the primary cilium. In response to Hh signals, the transcriptional activator of the pathway, Gli2, accumulates at the ciliary tip, raising the possibility that ciliary localization is important for Gli2 activation. To test this hypothesis, we used the Floxin system to create knock-in Gli2 alleles in embryonic stem cells (ESCs) to allow methodical testing of which domains and residues are essential for the ciliary localization of Gli2. The Gli2 zinc fingers, transcriptional activation domain, repressor domain, phosphorylation cluster and a Sufu binding motif were each dispensable for ciliary localization. Mutating residues that are required for Gli2 sumoylation and nuclear trafficking also did not abrogate ciliary localization. By contrast, several other domains restricted Gli2 nuclear localization, and a central region, distinct from previously characterized domains, was required for ciliary localization. In addition to an inability to localize to cilia, Gli2 lacking this central domain was unable to activate target genes. Thus, our systematic analysis in ESCs reveals that distinct regions of Gli2 regulate its nuclear and ciliary localization. The identification of a domain essential for both ciliary localization and transcriptional activity suggests that ciliary localization of Gli2 is required for its activation.
KEY WORDS: Ciliary localization, Embryonic stem cell, Floxin genetic engineering, Gli transcription factors, Hedgehog signaling, Primary cilia
INTRODUCTION
Hedgehog (Hh) signaling plays important roles in embryonic patterning and adult tissue homeostasis (Ingham et al., 2011). Misregulation of Hh signaling can cause congenital defects, such as holoprosencephaly, and cancers, including basal cell carcinoma and medulloblastoma (McMahon et al., 2003; Pasca di Magliano and Hebrok, 2003). In the absence of Hh ligand, the Hh receptor Patched1 (Ptch1) inhibits the activity of another transmembrane protein, Smoothened (Smo). In this off state, the transcriptional effectors of Hh signaling, Gli2 and Gli3, are proteolytically processed into repressor forms (GliR), as is the Drosophila Gli ortholog, Ci (Aza-Blanc et al., 1997; Pan et al., 2006; Tempé et al., 2006). In the presence of Hh, Ptch1 inhibition of Smo is relieved, and full length Gli2 and Gli3 are converted into transcriptional activators (GliA). In many tissues, Gli3R is the predominant repressor of Hh target genes and Gli2A is the predominant activator (Ding et al., 1998; Hui and Angers, 2011; Mo et al., 1997; Motoyama et al., 1998). Ultimately, the transcriptional output of Hh signaling is determined by the relative activity of GliR and GliA (Eggenschwiler and Anderson, 2007).
Intriguingly, vertebrate Hh signaling requires the primary cilium, a microtubule-based organelle that projects from the surface of most cells (Goetz and Anderson, 2010). Mice lacking functional cilia display tissue patterning defects attributable to the loss of both GliR and GliA (Huangfu and Anderson, 2005; Huangfu et al., 2003; Liu et al., 2005). Underscoring the link between primary cilia and Hh signaling, several components of the pathway localize to the cilium. Both Ptch1 and Smo localize in a ligand-dependent manner, with Ptch1 exiting and Smo entering the cilium in the presence of Hh (Corbit et al., 2005; Rohatgi et al., 2007). Moreover, mutant forms of Smo that fail to localize to the cilium cannot activate the pathway, strongly suggesting that Smo functions at the cilium (Aanstad et al., 2009; Corbit et al., 2005). Thus, the cilium is proposed to be a specialized sub-cellular site that coordinates Hh signal transduction.
The Gli proteins also localize to the cilium in a partially Hh-dependent manner: in the absence of Hh ligand, Gli proteins are present at the ciliary tip, but accumulate there in response to Hh stimulation (Haycraft et al., 2005; Kim et al., 2009; Wen et al., 2010; Zeng et al., 2010). Although it is clear that the cilium is required for GliA activity and Gli3 processing, it is not clear whether Gli localization to the cilium is essential for this regulation.
In vertebrates, the three Gli proteins, Gli1, Gli2 and Gli3, share conserved C2H2-type zinc fingers that bind target DNA (Kinzler et al., 1988). Gli2 and Gli3 possess an N-terminal repressor domain and a C-terminal activator domain that contribute to their bi-functionality (Sasaki et al., 1999). The activator domain of Gli3 is cleaved to create Gli3R through a process enhanced by protein kinase A (PKA) phosphorylation and a processing determinant domain (PDD), a region of ∼200 residues that controls the proteolytic processing of Gli2 and Gli3 (Pan et al., 2006; Pan and Wang, 2007; Tempé et al., 2006; Wang and Li, 2006). Compared to Gli3, a smaller fraction of Gli2 is processed, perhaps because Gli2 is degraded in response to PKA-promoted ubiquitylation by SCFβTRCP/Slimb (a Skp1−Cullin−F-box complex containing the F-box protein βTRCP) (Bhatia et al., 2006; Pan et al., 2006; Pan and Wang, 2007; Pan et al., 2009). In contrast to Gli2 and Gli3, Gli1 lacks a repressor domain, is not proteolytically processed and is a Hh transcriptional target that functions as a positive feedback on the pathway (Dai et al., 1999).
Proteins that regulate Gli transcriptional activity also localize to the cilium or its foundation, the basal body. PKA localizes to the basal body (Barzi et al., 2010; Tuson et al., 2011) and promotes the processing of Gli3 and the degradation of Gli2. Mice lacking PKA activity exhibit excess activation of Hh signal transduction in the neural tube, indicative of increased GliA activity. The loss of PKA increases the number of cilia exhibiting Gli2 localization, whereas the activation of PKA by forskolin reduces Gli2 ciliary localization (Tukachinsky et al., 2010; Tuson et al., 2011; Zeng et al., 2010). Thus, PKA inhibits Gli2 translocation to the cilium and restrains its transcriptional activity (Tuson et al., 2011). However, Gli2 alleles bearing either phospho-mimetic or phospho-dead residues at conserved PKA phosphorylation sites retain their ability to enter the cilium and are sensitive to PKA activation by forskolin, suggesting that PKA regulates Gli2 ciliary localization independently of these sites (Zeng et al., 2010).
Sufu, a negative regulator of Gli transcriptional activation, localizes to the primary cilium in a Gli-dependent manner (Haycraft et al., 2005; Zeng et al., 2010). Sufu can bind to Gli1 at two sites, one of which, the SYGH motif, is conserved among all three Gli proteins (Dunaeva et al., 2003). Sufu negatively regulates the Gli proteins by tethering them to the cytoplasm and suppressing Gli transcriptional activity in the nucleus (Barnfield et al., 2005; Chen et al., 2009; Cheng and Bishop, 2002; Ding et al., 1999; Dunaeva et al., 2003; Kogerman et al., 1999; Merchant et al., 2004). Hh promotes the dissociation of Sufu from full-length Gli2 and Gli3 (Humke et al., 2010; Tukachinsky et al., 2010) and, upon release, GliA can enter the nucleus to control the transcriptional outcome of Hh target genes. Hh-mediated dissociation does not occur in cells either lacking cilia or with activated PKA, suggesting that the dissociation of Sufu-Gli complexes occurs at the cilium in response to PKA inactivation (Humke et al., 2010; Tukachinsky et al., 2010).
Many questions about the activation of Gli proteins remain, including how they are trafficked into the cilium, how they accumulate upon Hh stimulation and how they move from the cilium to the nucleus. Moreover, it remains unclear whether PKA inactivation and dissociation from Sufu occur at the primary cilium or whether other ciliary events are required for Gli activation and nuclear entry. With these questions in mind, we sought to further define the mechanisms that control the subcellular localization of Gli2, and how the control of subcellular localization, in turn, regulates Gli2 activity.
In this study, we used an efficient knock-in approach called the Floxin system to genetically tailor the Gli2 locus in mouse embryonic stem cells (ESCs) (Singla et al., 2010). We created a panel of GFP-tagged Gli2 alleles that systematically delete or modify domains or residues implicated in Gli2 function. We found that several domains are crucial for the exclusion of Gli2 from the nucleus, and identified a central region of Gli2 that plays an important role in ciliary translocation, protein stability and transcriptional activity. Thus, distinct regions of Gli2 regulate localization to distinct subcellular sites, and a domain required for ciliary localization is essential for Gli2 transcriptional activity, suggesting that ciliary localization is required for Gli2 function.
RESULTS
A knock-in Gli2–GFP allele recapitulates wild-type Gli2 localization and activity
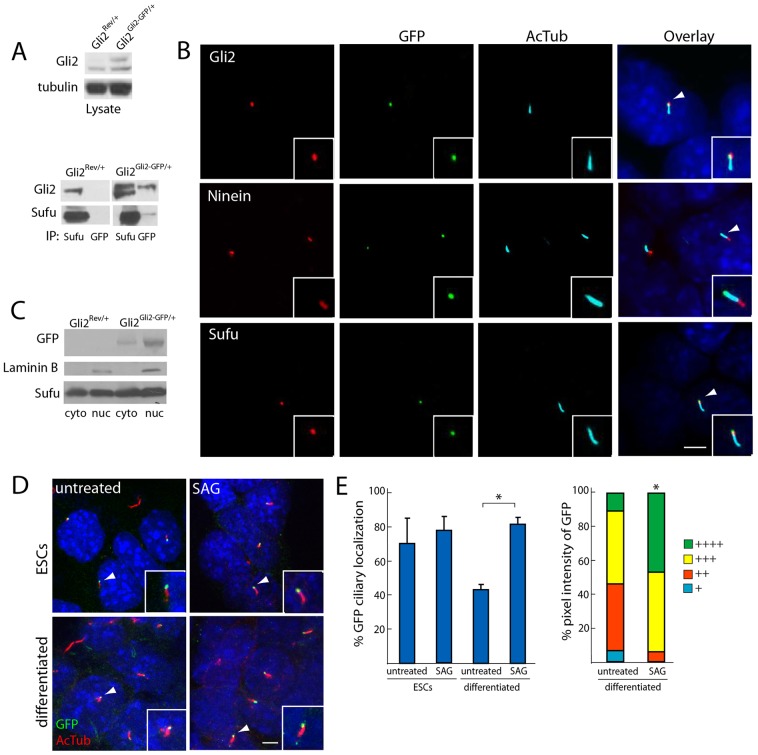
We obtained a Floxin-compatible gene trap ESC line, XG045 (Gli2gt/+). The gene trap is located within the intron after the exon containing the Gli2 start codon, producing a fusion of the first 46 codons of Gli2 to β-geo (supplementary material Fig. S1). We reverted the gene-trap mutation to create Gli2Rev/+ ESCs. We used the Floxin system to insert a cDNA encoding the Gli2 exons downstream of the gene trap with a C-terminal GFP tag to create Gli2Gli2−GFP/+ ESCs. Insertion was confirmed by neomycin resistance, β-galactosidase activity and PCR genotyping (data not shown).
We compared the expression of Gli2–GFP to that of wild-type Gli2 in Gli2Rev/+ and Gli2Gli2−GFP/+ ESCs by immunoblot. Gli2–GFP was expressed at levels comparable to wild-type Gli2 (Fig. 1A). To test whether Gli2–GFP recapitulates known Gli2 interactions, we assessed the binding of Gli2–GFP to Sufu. Gli2–GFP co-immunoprecipitated Sufu, and Sufu reciprocally co-immunoprecipitated both wild-type Gli2 and the higher-molecular-mass Gli2–GFP from Gli2Gli2−GFP/+ ESCs (Fig. 1A). Thus, by expressing Gli2–GFP under endogenous regulatory control, we recapitulated wild-type expression and interaction with Sufu.
Fig. 1.
Gli2–GFP recapitulates wild-type Gli2 localization and ciliary dynamics. (A) Whole-cell lysates from Gli2Rev/+ and Gli2Gli2–GFP/+ ESCs were probed with an anti-Gli2 antibody. Gli2–GFP was expressed at equivalent levels to wild-type Gli2. The lysate was also immunoprecipitated with either anti-GFP or anti-Sufu, and was immunoblotted for Gli2 and Sufu. Sufu co-immunoprecipitates Gli2 in Gli2Rev/+ ESC lysate and co-immunoprecipitates both Gli2–GFP and Gli2 in Gli2Gli2–GFP/+ ESC lysate. Anti-GFP antibody immunoprecipitates both Gli2–GFP and Sufu only from Gli2Gli2–GFP/+ ESC lysate. (B) Gli2–GFP localizes to the ciliary tip of Gli2Gli2–GFP/+ ESCs. GFP localizes to the opposite end of cilia (as identified by anti-acetylated tubulin, AcTub) compared to the basal body marker Ninein, and exhibits co-localization with the ciliary tip protein Sufu. Insets, the individual cilium indicated by the white arrowhead. Scale bar: 5 µm. (C) Gli2Rev/+ and Gli2Gli2–GFP/+ ESC lysates were separated into nuclear and cytoplasmic fractions and probed for GFP, Sufu and Laminin B, a component of the nuclear envelope. GFP and Sufu were detected in both the nuclear and cytoplasmic fractions, whereas Laminin B was only detected in the nuclear fraction. (D) Following treatment with DMSO or SAG (1 µM for 1 hour), Gli2Rev/+ and Gli2Gli2–GFP/+ ESCs were stained for GFP and AcTub. Ciliary localization of Gli2–GFP was detected in both undifferentiated and differentiated cells, and in untreated and SAG-treated cells (insets, the individual cilium indicated by the white arrowhead). Scale bar: 5 µm. (E) Undifferentiated and differentiated Gli2Gli2–GFP/+ ESCs were quantified for the number of cilia exhibiting GFP localization (three independent experiments, n = 100 per condition) and ciliary GFP pixel intensity (see Materials and Methods, and Wen et al., 2010; three independent experiments, n = 40 per condition). + denotes lowest pixel intensity, whereas ++++ denotes highest pixel intensity. SAG treatment of differentiated cells increased both the proportion of cilia with localization of Gli2–GFP (left, *P<0.05, Student's t-test) and the intensity of ciliary Gli2–GFP (right, *P<0.05, Chi-squared test). Error bars represent s.d. throughout.
Gli2 localizes to the tip of the primary cilium in many cell types (Haycraft et al., 2005; Kiprilov et al., 2008; Wen et al., 2010). We observed that Gli2 and other Hh signal transduction components, including Sufu and Smo, also localize to primary cilia in mouse ESCs (supplementary material Fig. S2A). Similarly, the majority of Gli2Gli2−GFP/+ ESCs exhibited GFP at the ciliary tip, as defined by co-staining with the basal-body marker Ninein and colocalization with Sufu (Fig. 1B). Although we did not detect nuclear localization of Gli2–GFP by immunofluorescence, nuclear-cytoplasmic fractionation of Gli2Gli2−GFP/+ ESCs revealed that Gli2–GFP does localize to the nucleus (Fig. 1C). Other researchers have similarly not detected nuclear immunofluorescence of endogenous Gli2, suggesting that it might be below the detection limit of standard approaches (Wen et al., 2010).
Gli2 accumulates at the ciliary tip upon Hh stimulation in mouse embryonic fibroblasts (MEFs) (Kim et al., 2009; Wen et al., 2010). Therefore, we ascertained whether Gli2–GFP displayed a similar accumulation in Gli2Gli2−GFP/+ ESCs by measuring the percentage of cells exhibiting GFP at cilia, as well as the pixel intensity of ciliary GFP with and without stimulation with SAG, a Smo agonist (Chen et al., 2002; Wen et al., 2010). In response to SAG, Gli2Gli2−GFP/+ cells that were differentiated by the withdrawal of LIF exhibited an increased prevalence of GFP ciliary localization and a higher percentage of high-pixel-intensity ciliary GFP (Fig. 1D,E). SAG-mediated ciliary localization was more prevalent in differentiated cells compared to undifferentiated cells and, similarly, differentiated cells are more responsive to Hh pathway activation (supplementary material Fig. S2B). Hh responsiveness to SAG, as ascertained by transcript levels of Gli1 and Ptch1, is equivalent for Gli2gt/+, Gli2Rev/+ and Gli2Gli2−GFP/+ cells, consistent with the haplosufficiency of Gli2 (supplementary material Fig. S2C). Together, these data demonstrate that the Floxin system can tailor the Gli2 locus to create genetically tractable ESC models of Gli2 subcellular localization. Therefore, we used the Floxin system to systematically determine how specific domains and post-translational modifications regulate the subcellular dynamics of Gli2 (supplementary material Fig. S2D).
Processed forms of Gli2 do not localize to the cilium
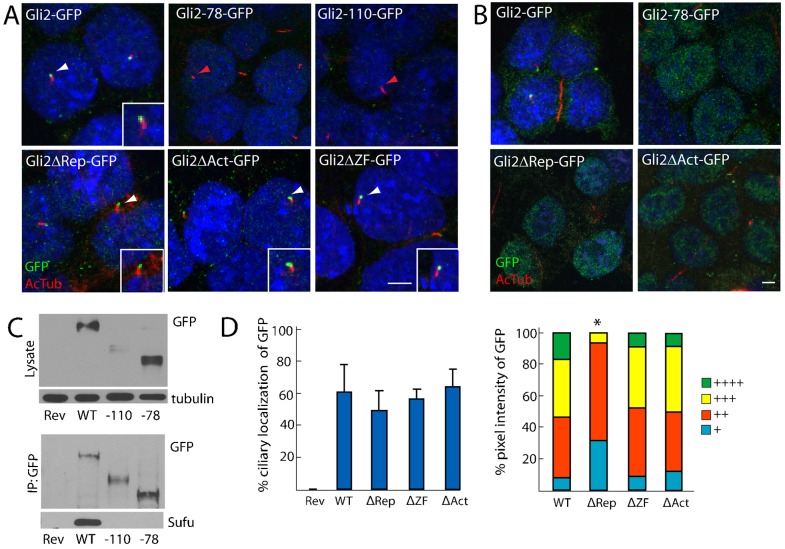
Like Gli3, Gli2 can be proteolytically processed to generate an N-terminal fragment of ∼78 kDa, termed Gli2-78 (Pan et al., 2006). Gli2-78 can repress the expression of Hh target genes, presumably because it lacks the C-terminal transcriptional activation domain but retains the N-terminal repressor domain and DNA-binding zinc fingers (Sasaki et al., 1999). Because processed Gli3 fails to localize to cilia (Wen et al., 2010), we investigated whether Gli2-78 localizes to cilia. Using the Floxin system, we inserted the exons downstream of the gene trap to the approximate processing site in frame with a C-terminal GFP tag (Pan et al., 2006). Unlike full-length Gli2–GFP, but similar to processed Gli3, Gli2-78–GFP did not localize to the ciliary tip (Fig. 2A). Instead, Gli2-78–GFP showed increased localization to the nucleus (Fig. 2B). This observation suggests that the repressor form of Gli2 is targeted to the nucleus, and that the Gli2 C-terminus both inhibits nuclear localization and is required for ciliary localization. Immunoblotting analysis indicated that the level of Gli2-78–GFP is comparable to that of full length Gli2–GFP (Fig. 2C).
Fig. 2.
Proteolytically processed forms of Gli2 do not localize to cilia but the activator, repressor and zinc finger domains are dispensable for the ciliary localization of Gli2. (A) Gli2–GFP ESCs were stained for AcTub (red) and GFP (green). The processed forms of GFP-tagged Gli2 (Gli2-78 and Gli2-110) do not localize to cilia (red arrowheads), whereas GFP-tagged Gli2 lacking the N-terminal repressor domain, C-terminal activator domain or central zinc finger domain (Gli2ΔRep, Gli2ΔAct or Gli2ΔZF, respectively) robustly localize to the cilium (white arrowheads). Insets, the individual cilium indicated by the white arrowhead. Scale bar: 5 µm. (B) Gli2-78, Gli2ΔRep and Gli2ΔAct localize more robustly to the nucleus than wild-type Gli2–GFP. Scale bar: 5 µm. (C) Lysates from Gli2Rev/+ (Rev), Gli2Gli2–GFP/+ (WT), Gli2Gli2-110–GFP/+ (-110) and Gli2Gli2-78–GFP/+ (-78) ESCs were blotted for GFP and tubulin, or were immunoprecipitated with anti-GFP and blotted for GFP and Sufu. There is less Gli2-110 protein than wild-type or Gli2-78 protein. Neither Gli2-78 nor Gli2-110 co-immuoprecipitate Sufu. (D) Gli2ΔRep–GFP, Gli2ΔAct–GFP and Gli2ΔZF–GFP ciliary localization prevalence and pixel intensity were compared to Gli2–GFP. + denotes lowest pixel intensity, whereas ++++ denotes highest pixel intensity. *P<0.05 (Chi-squared test, as compared to WT).
In addition to full-length Gli2–GFP, GFP immunoprecipitation from Gli2Gli2−GFP/+ ESC lysate revealed a faster-migrating peptide that is consistent with the C-terminal by-product of Gli2 processing, referred to as Gli2-110 (supplementary material Fig. S3A). However, no similarly sized peptide immunoprecipitated with Sufu, suggesting that Gli2-110 does not interact with Sufu (supplementary material Fig. S3A). To create a Gli2 allele that produces a GFP-tagged equivalent of Gli2-110 (Gli2-110–GFP), the exons from downstream of the approximate processing site were inserted at the Gli2 locus, in frame with GFP, using the Floxin system. Like Gli2-78–GFP, Gli2-110–GFP did not localize to cilia (Fig. 2A). Gli2-110–GFP protein levels were lower than those of wild-type Gli2–GFP, despite comparable mRNA levels, suggesting that Gli2-110 is less stable than full-length Gli2 or Gli2-78 (Fig. 2C; supplementary material Fig. S3B). The instability of Gli2-110–GFP might reflect the ability of the Gli2 carboxy-terminus to bind to multiple E3 ligases, including SCFβTRCP/Slimb, SPOP-Cul3 and Numb–Itch (Bhatia et al., 2006; Chen et al., 2009; Di Marcotullio et al., 2006; Zhang et al., 2009).
Neither Gli2-78–GFP nor Gli2-110–GFP robustly interacted with Sufu, suggesting that Sufu preferentially binds to full-length Gli2 (Fig. 2C). These data are similar to results obtained for Gli3 and are consistent with the model that Sufu does not prevent processed forms of Gli from entering the nucleus (Humke et al., 2010; Tukachinsky et al., 2010).
The activator, repressor and DNA-binding domains of Gli2 are dispensable for ciliary localization
Because the processed forms of Gli2 failed to localize to the cilium, we investigated which domains of Gli2 are required for ciliary localization. We initially examined the previously defined functional domains of Gli2: the transcriptional repressor domain, the activator domain and the DNA-binding domain (supplementary material Fig. S2D). To examine whether these domains are required for ciliary localization, we used Floxin to knock in GFP fusions with Gli2 that lack the N-terminal transcriptional repressor domain (Gli2ΔRep–GFP; lacking amino acids 47–271) or the C-terminal activation domain (Gli2ΔAct–GFP; lacking amino acids 1184–1544) (Sasaki et al., 1999). We similarly knocked in an allele that encodes Gli2 lacking the five zinc fingers that are necessary for DNA binding (Gli2ΔZF–GFP; lacking amino acids 417–569) (Pavletich and Pabo, 1993). Gli2ΔRep–GFP, Gli2ΔAct–GFP and Gli2ΔZF–GFP were all present in the same proportion of cilia as full-length Gli2–GFP (Fig. 2A,D). However, Gli2ΔRep–GFP displayed significantly lower ciliary GFP intensity than wild-type Gli2–GFP, suggesting that the repressor domain promotes Gli2 ciliary localization (Fig. 2D). Both Gli2ΔRep–GFP and Gli2ΔAct–GFP also displayed increased nuclear localization relative to full-length Gli2, similar to Gli2-78–GFP, indicating that the repressor and activator domains independently inhibit nuclear import or retention (Fig. 2B). Thus, although the processed forms of Gli2, Gli2-78–GFP and Gli2-110–GFP, failed to translocate to the cilium, neither the N-terminal repressor region, the C-terminal activator region nor the central zinc fingers themselves were essential for ciliary localization.
Sufu interaction, PKA phosphorylation and sumoylation are not required for Gli2 ciliary localization
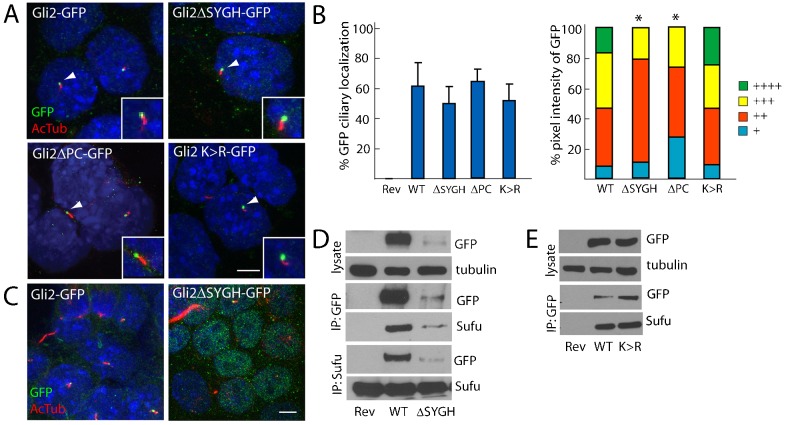
Between the repressor and activator domains, Gli2 contains a SYGH motif through which Gli2 interacts with Sufu and a phosphorylation cluster that includes residues phosphorylated by PKA, GSK3β and CKI (Bhatia et al., 2006; Dunaeva et al., 2003; Pan et al., 2006). Phosphorylation of the phosphorylation cluster promotes the degradation of Gli2 by SCFβTRCP/Slimb and enhances sumoylation, a post-translational modification that inhibits Gli2 transcriptional activity (Bhatia et al., 2006; Cox et al., 2010; Han et al., 2012; Pan et al., 2006). Two putative sumoylation sites flank each end of the zinc finger domain (Cox et al., 2010). Because Gli activation is postulated to involve dissociation from Sufu, inhibition of PKA phosphorylation and sumoylation, we used Floxin to knock in alleles that encode Gli2 lacking the SYGH motif, lacking the phosphorylation cluster or substituting arginines for the four putatively sumoylatable lysines (supplementary material Fig. S2D). We tested whether these domains and residues regulate Gli2 ciliary localization.
Gli2ΔSYGH–GFP (lacking amino acids 268–271) localized to a similar proportion of cilia as full-length Gli2–GFP, albeit with reduced intensity, which is potentially attributable to lower protein levels (Fig. 3A,B,D). These data are consistent with previous studies that show that Sufu is not required for the ciliary localization of Gli2 and that Sufu stabilizes Gli2 (Chen et al., 2009; Tukachinsky et al., 2010). Gli2ΔSYGH–GFP also showed enriched localization to the nucleus (Fig. 3C; supplementary material Fig. S3C), consistent with a role for Sufu in preventing the nuclear entry of Gli2 (Ding et al., 1999; Kogerman et al., 1999). Despite being enriched in the nucleus, the expression of Gli2ΔSYGH–GFP did not misactivate Hh transcriptional targets, in marked contrast to expression of Gli2ΔRep–GFP (supplementary material Fig. S3D).
Fig. 3.
The interaction with Sufu, phosphorylation and sumoylation are not required for the ciliary localization of Gli2. (A) GFP-tagged Gli2, Gli2ΔSYGH, Gli2ΔPC and Gli2 K>R ESCs were stained for cilia (AcTub, red) and GFP (inset of individual cilium, white arrowhead). Scale bar: 5 µm. (B) The ciliary localization prevalence and pixel intensity of Gli2ΔSYGH–GFP (ΔSYGH), Gli2ΔPC–GFP (ΔPC) and Gli2 K>R–GFP (K >R) were compared to those of Gli2–GFP (WT). + denotes lowest pixel intensity, whereas ++++ denotes highest pixel intensity. *P<0.05 (Chi-squared test, as compared to WT). (C) Gli2ΔSYGH–GFP exhibited increased nuclear localization compared to Gli2–GFP, similar to Gli2-78–GFP, Gli2ΔRep–GFP and Gli2ΔAct–GFP. (D) Gli2Rev/+ (Rev), Gli2Gli2–GFP/+ (WT) and Gli2Gli2ΔSYGH–GFP/+ (ΔSYGH) ESC lysates were blotted for GFP and tubulin. The lysate was immunoprecipitated with anti-GFP or anti-Sufu, and blotted for GFP and Sufu. Gli2ΔSYGH–GFP protein levels were less than those of Gli2–GFP. Gli2ΔSYGH–GFP interacts with Sufu. (E) Gli2Rev/+ (Rev), Gli2Gli2–GFP/+ (WT) and Gli2Gli2K>R–GFP/+ (K>R) ESC lysates were probed for GFP and tubulin, or were immunoprecipitated with anti-GFP and immunoblotted for GFP and Sufu. Gli2 K>R–GFP protein levels are equivalent to Gli2–GFP and both can interact with Sufu.
Deletion of the SYGH motif did not prevent Gli2 from interacting with Sufu: Gli2ΔSYGH–GFP immunoprecipitated Sufu and Sufu reciprocally immunoprecipitated Gli2ΔSYGH–GFP (Fig. 3D). Because Gli2–GFP does not immunoprecipitate Gli2, homotypic Gli2 interactions do not account for the SYGH-independent interaction with Sufu (Fig. 1A), indicating that the SYGH motif does not mediate all of the interaction of Gli2 with Sufu. Gli2ΔSYGH–GFP interacted with Sufu in both cytoplasmic and nuclear fractions (supplementary material Fig. S3C). Apart from its SYGH motif, Gli1 interacts with Sufu through a C-terminal domain (Merchant et al., 2004). Our findings reveal that Gli2, like Gli1, interacts with Sufu through multiple domains, and that the SYGH binding site is important for nuclear exclusion and for the stabilization of Gli2.
Gli2ΔPC–GFP (lacking amino acids 767–852, the region which contains the phosphorylation cluster) was present in the same proportion of cilia as full length Gli2–GFP and activates Hh transcription targets similarly to wild-type Gli2 (Fig. 3A,B; supplementary material Fig. S3D). However, we observed diminished amounts of ciliary Gli2ΔPC–GFP, suggesting that this region promotes the efficient localization of Gli2 to the cilium. Given that phospho-dead or phospho-mimetic substitution of the PKA sites does not affect the ability of Gli2 to reach the cilium (Tuson et al., 2011; Zeng et al., 2010) but that loss of the phosphorylation cluster does, phosphorylation by other kinases, such as CKI and GSK3, might promote Gli2 ciliary localization.
As Gli2 can be sumoylated in vitro (Cox et al., 2010; Lee et al., 2011) (supplementary material Fig. S3E), we substituted each of the four putatively sumoylatable lysines with unsumoylatable arginines to create Gli2K>R–GFP (Gli2K375R, K398R, K630R, K716R–GFP). Gli2K>R–GFP possessed similar ciliary localization to full-length Gli2–GFP, retained its ability to interact with Sufu and was not highly enriched in the nucleus (Fig. 3A,B,E). Consistent with previous findings, Gli2-containing individual K>R substitutions displayed greater transcriptional activity than wild-type Gli2 and multiple mutations increased this hyperactivity (Han et al., 2012) (supplementary material Fig. S3E). Thus, our data suggest that the increased activity seen in Gli2K>R–GFP is independent of altered Sufu interaction, ciliary localization or nuclear accumulation. We did not detect several Sumo pathway components at or near the primary cilium, including Sumo1–3, the desumoylation proteases Senp2, 3, 5 or the E3 Sumo ligase Pias (data not included), although the E2 Sumo-conjugating enzyme Ubc-9 localizes to cilia in Caenorhabditis elegans when fused to GFP (Li et al., 2012). Thus, sumoylation might be a repressive post-translational modification that limits Gli2 transcriptional activity.
Nuclear import signals do not mediate Gli2 ciliary entry, but nuclear export signals might play a role in Gli2 ciliary exit
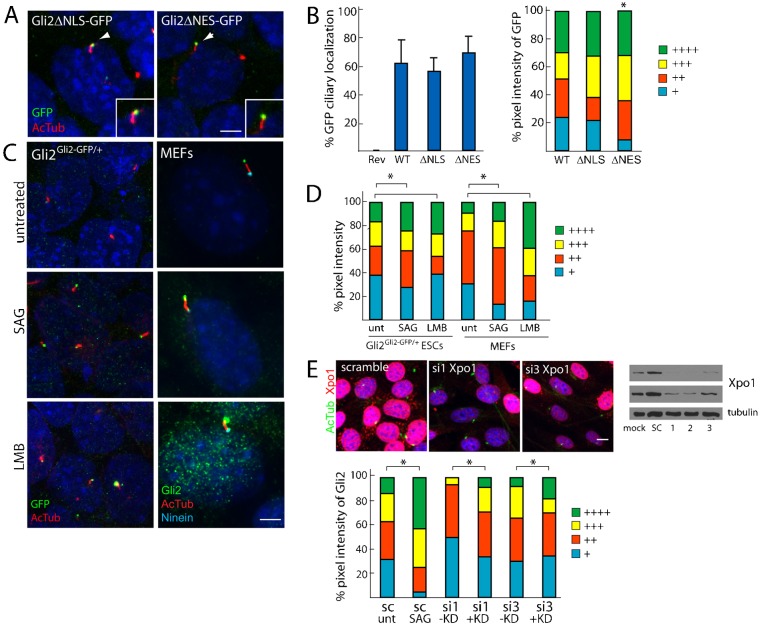
Recent studies have revealed that the mechanism of ciliary import of Kif17 and RP2 shares similarities with that of nuclear import (Dishinger et al., 2010; Hurd et al., 2011). Because nuclear localization signal (NLS)-like sequences within Kif17 and RP2 promote ciliary localization through interaction with importins, we examined whether putative nuclear localization signals were also involved in Gli2 entry into the cilium. Similarly, we examined whether putative leucine-rich nuclear export signals (NESs) affected Gli2 exit from the cilium (supplementary material Fig. S2D).
We used Floxin to knock in alleles that encode Gli2 lacking a previously described NLS or several predicted NESs fused to GFP (Gli2ΔNLS–GFP, lacking amino acids 715–725; Gli2ΔNES–GFP, lacking amino acids 229–241, 887–895 and 1037–1051). Although the percentages of cells displaying ciliary localization of GFP were not altered, Gli2ΔNES–GFP cells exhibited an increase in the intensity of ciliary GFP compared to full-length Gli2–GFP cells (Fig. 4A,B). Notably, Gli2ΔNES–GFP did not exhibit enriched nuclear localization, suggesting that these predicted NESs are not required for Gli2 nuclear export.
Fig. 4.
Nuclear import signals do not mediate ciliary localization of Gli2 but nuclear export signals might play a role in Gli2 ciliary exit. (A) Gli2Gli2ΔNLS–GFP/+ and Gli2Gli2ΔNES–GFP/+ ESCs were stained for cilia (AcTub, red) and GFP (insets show the individual cilium indicated by white arrowheads). Scale bar: 5 µm. (B) The ciliary localization prevalence and pixel intensity of Gli2ΔNLS–GFP (ΔNLS) and Gli2ΔNES–GFP (ΔNES) were compared to those of Gli2–GFP (WT). + denotes lowest pixel intensity, whereas ++++ denotes highest pixel intensity. *P<0.05 (Chi-squared test, as compared to WT). (C) Gli2Gli2–GFP/+ ESCs and MEFs were treated with DMSO, SAG (1 µM, 1 hour) and Leptomycin B (LMB, 20 nM, 90 minutes), an inhibitor of Exportin1 (Xpo1). Cells were stained for cilia (AcTub), GFP or Gli2 (to mark Gli2–GFP in ESCs and Gli2 in MEFs) and Ninein (to mark the basal body). (D) Quantification of panel C where the pixel intensities for GFP and Gli2 are assessed for differentiated Gli2Gli2–GFP/+ ESCs and MEFs, respectively. *P<0.05 (Chi-squared test). (E) NIH3T3 fibroblasts were transfected for 72 hours with siRNA against Exportin1 (si1,2,3) or a scramble control (sc), and were stained for cilia (AcTub, green) and Xpo1 (red). Scale bar: 10 µm. The knockdown efficiency was also assessed by an Xpo1 and tubulin immunoblot of the total cell lysate (top panel, 5-second exposure; middle panel, 30-second exposure). Ciliary Gli2 pixel intensity was assessed in cells showing diminished Xpo1 immunofluorescence (+KD) and normal Xpo1 levels (−KD) (three independent experiments, n = 50 per condition). *P<0.05 (Chi-squared test).
To further test whether the ciliary exit of Gli2 could function similarly to nuclear export, we treated differentiated Gli2–GFP ESCs and fibroblasts with leptomycin B, an inhibitor of Exportin1. We found that, similar to Hh stimulation, leptomycin B treatment led to increased pixel intensity of Gli2–GFP and endogenous Gli2 at the ciliary tip, as well as a subtle increase in nuclear Gli2–GFP (Fig. 4C,D). siRNA-mediated inhibition of Exportin1 increased ciliary Gli2 to a varying degree depending on the siRNA construct, suggesting that Exportin1 might play a role in Gli2 ciliary export (Fig. 4E). However, as we were unable to detect an interaction between Gli2–GFP and Exportin1, it remains to be elucidated whether nuclear export directly mediates Gli2 exit from the cilium. Alternatively, Exportin1 and leptomycin B might affect the distribution of Gli2 interactors that affect the ciliary exit of Gli2.
A central region of Gli2 is necessary for efficient ciliary localization
Because our analyses demonstrated that the majority of Gli2 is dispensable for ciliary localization, particularly the regions that regulate Gli2 transcriptional activity, mediate interaction with Sufu or are sites of known post-translational modifications, we searched for a region of Gli2 that is required for ciliary localization. Given that Gli2ΔRep–GFP, Gli2ΔAct–GFP and Gli2ΔZF–GFP reach the cilium, but that Gli2-78–GFP does not, we sought a region of Gli2 required for ciliary localization outside of the activation or zinc finger domains. Specifically, to screen the remaining portions of the protein, we created a Gli2 allele that lacks the region between the zinc finger domain and the phosphorylation cluster (amino acids 562–707), which spans the PDD (Pan and Wang, 2007). We also created an allele that removed the region between the phosphorylation cluster and the activation domain (amino acids 852–1183), a region previously described as a secondary activation domain (Sasaki et al., 1999).
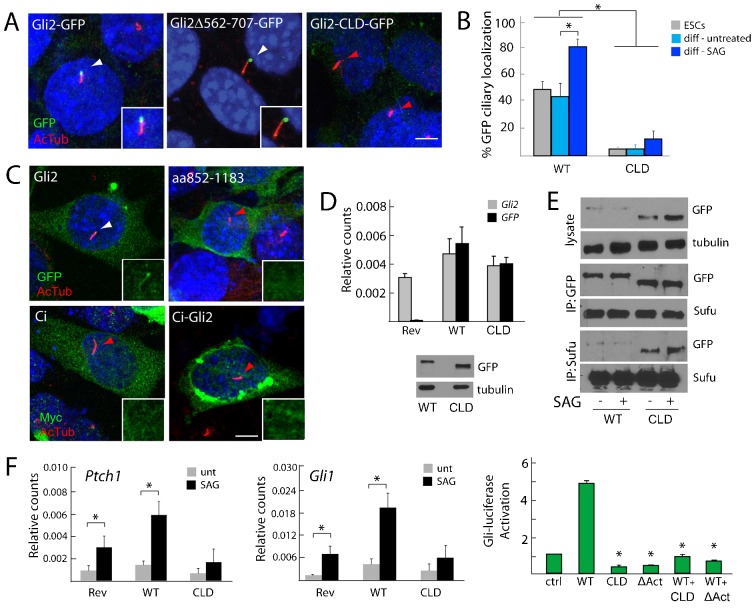
Gli2Δ562-707–GFP localized to the cilium robustly (Fig. 5A). By contrast, a very small percentage of cilia displayed Gli2Δ852-1183–GFP compared to the percentage that displayed Gli2–GFP (Fig. 5A,B). Similarly, a smaller percentage of Gli2Δ852-1183–GFP localized to cilia in response to SAG, as compared to Gli2–GFP (Fig. 1E; Fig. 5B). The fold increase in ciliary localization upon SAG treatment was not significant, indicating that the steady-state levels of ciliary Gli2Δ852-1183–GFP are decreased. To determine whether a smaller domain mediates ciliary localization, we created Floxin alleles Gli2Δ852-940–GFP, Gli2Δ941-1044–GFP and Gli2Δ1045-1183–GFP (supplementary material Fig. S4A). However, these sub-deletions did not exhibit defective ciliary localization. Therefore, we renamed Gli2Δ852-1183–GFP as Gli2-CLD–GFP, for ‘ciliary localization defective’.
Fig. 5.
The central region of Gli2 is necessary for ciliary localization, stabilization and transcriptional activity. (A) Gli2Gli2–GFP/+, Gli2Gli2Δ562-707–GFP/+ and Gli2Gli2-CLD–GFP/+ (lacking amino acids 852–1183) ESCs were stained for cilia (AcTub, red) and GFP (green); white arrowheads depict ciliary localization, whereas red arrowheads depict lack of ciliary localization. Scale bar: 5 µm. (B) The percentage of ESCs or differentiated cells (diff) in the presence of SAG or vehicle control that display ciliary localization of full-length Gli2–GFP (WT) or Gli2-CLD–GFP (CLD). *P<0.05 (Student's t-test). (C) Gli2–GFP, Gli2-CLD–GFP, Ci–Myc and Ci–Gli2 (a fusion of the N-terminal half of Ci and the C-terminal half of Gli2) were transiently transfected into NIH3T3 cells and stained for cilia (AcTub, red) and either GFP or Myc (green). Insets depict Gli2–GFP or homologs at the individual cilia indicated by arrowheads. Scale bar: 5 µm. (D) Gli2Rev/+ (Rev), Gli2Gli2–GFP/+ (WT) and Gli2Gli2-CLD–GFP/+ (CLD) ESCs were assessed for Gli2 and GFP expression, normalized to β-actin. Whole cell lysate of Gli2Gli2–GFP/+ and Gli2Gli2-CLD–GFP/+ ESCs was probed for GFP and tubulin. The absence of the ciliary localization region does not reduce Gli2 mRNA or protein levels. (E) Gli2Gli2–GFP/+ and Gli2Gli2-CLD–GFP/+ ESC lysates were blotted for GFP and tubulin, or were immunoprecipitated with anti-GFP or anti-Sufu and blotted for GFP and Sufu. Gli2-CLD–GFP is stabilized compared to wild-type Gli2–GFP. Gli2-CLD–GFP interacts with Sufu. (F) Left and middle: differentiated Gli2Rev/+(Rev), Gli2Gli2–GFP/+ and Gli2Gli2-CLD–GFP/+ ESCs were assayed for Gli1 and Ptch1 expression levels following 18 hours of SAG treatment. Expression was normalized to that of β-actin. Right: the effect of Gli2 and Gli2-mutant protein expression in Smo−/− MEFs on Gli–luciferase reporter activity. The expression of Gli2-CLD–GFP in either ESCs or MEFs interferes with Gli-dependent transcription. *P<0.05 (Student's t-test as compared to WT).
To assess whether the ciliary localization region participates in the ciliary localization of other Gli proteins, we deleted the homologous region of human Gli3. Similar to Gli2-CLD–GFP, Gli3Δ912-1223–GFP failed to reach the cilium (supplementary material Fig. S4B). To assess whether the Gli2 ciliary localization region is sufficient to direct ciliary localization, we expressed this domain fused to GFP or fused to the N-terminal half of Ci. Ci, the Drosophila homolog of the Gli proteins, does not localize to the cilium when expressed in mammalian cells (Zeng et al., 2010). Neither the Gli2 ciliary localization region (amino acids 852–1183) nor Ci fused to the Gli2 ciliary localization region localized to cilia, indicating that this region is not sufficient for ciliary targeting (Fig. 5C).
To ensure that the failure of Gli2-CLD–GFP to efficiently localize to cilia was not due to diminished expression, we assessed Gli2 and GFP expression, as well as protein levels by immunoblot. Gli2-CLD–GFP mRNA levels were comparable to those of wild-type Gli2, and the protein levels of Gli2-CLD–GFP were greater than those of wild-type Gli2 (Fig. 5D,E). The regions of Gli1 and Gli3 that are homologous to the Gli2 ciliary localization region have been shown to contain binding motifs for the E3 ubiquitin ligases Itch and SPOP, respectively, suggesting this region might promote Gli protein turnover (Di Marcotullio et al., 2006; Wang et al., 2010; Zhang et al., 2009). Consistent with this possibility, we found that deletion of the ciliary localization region in Gli3 similarly abrogated its turnover (supplementary material Fig. S4C).
To assess whether Gli2-CLD–GFP was stabilized by its failure to reach cilia, we expressed Gli2–GFP and Gli2-CLD–GFP in MEFs lacking Kif3a, a protein that is required for ciliogenesis (Corbit et al., 2008). In the absence of Kif3a, Gli2-CLD–GFP protein was stabilized compared to wild-type Gli2–GFP, similar to ciliated cells (supplementary material Fig. S4D). Levels of Gli2–GFP were also equivalent in cells with and without Kif3a (supplementary material Fig. S4D). Thus, the ciliary localization region of Gli2 promotes protein turnover independently of its role in promoting ciliary localization.
The hypothesis that Gli2 localization to cilia is crucial for its transcriptional activation predicts that Gli2-CLD–GFP will not activate target gene expression. Therefore, we tested the transcriptional activity of Gli2-CLD–GFP using two different cell models. In differentiated ESCs, Gli2-CLD–GFP partially suppressed the expression of the Hh target genes Gli1 and Ptch1, indicating that it might act as a dominant negative (Fig. 5F). In MEFs, co-expression of Gli2-CLD–GFP with wild-type Gli2 confirmed that Gli2-CLD–GFP inhibits Gli2 transcriptional activity, similar to the dominant-negative activity of Gli2ΔAct, which lacks the C-terminal activator domain of Gli2 and functions as a constitutive repressor (Fig. 5F). The finding that the ciliary localization domain is also crucial for Gli2 transcriptional activity suggests that ciliary localization is essential for the ability of Gli transcription factors to induce target genes in response to Hh signals.
DISCUSSION
Gli2 represses or activates target genes in a Hh-responsive manner. In our study, we used the Floxin system to express GFP-tagged forms of Gli2 from the Gli2 locus in ESCs, creating a genetically tractable means of studying Gli2 subcellular dynamics. Floxin allowed the systematic genetic tailoring of domains and residues, alone and in combination, to precisely investigate the function of Gli2 under endogenous regulatory control (supplementary material Fig. S2D). Using this approach, we identified a domain within Gli2 that is required for ciliary localization and transcriptional activity. These findings support a model in which the regulation of Gli2 at the primary cilium is an important step in Gli2 activation.
A central region of Gli2 (amino acids 852–1183) is necessary, but not sufficient, for ciliary localization. Zeng et al. identified a partially overlapping central region (amino acids 570–967) that is required for overexpressed GFP–Gli2 to localize to cilia (Zeng et al., 2010). However, we tested the localization of Gli2Δ562-707–GFP, Gli2ΔPC–GFP (lacking amino acids 767–852) and Gli2Δ852-940–GFP using the Floxin system and observed intact ciliary localization with all three individual deletions. Differences in our observations could be due to the different locations of the GFP tag or differences in protein levels, as our studies examined Gli2–GFP expressed under the control of its endogenous regulatory elements, whereas Zeng et al. (Zeng et al., 2010) examined overexpressed GFP–Gli2. Alternatively, because Gli2-CLD–GFP could be detected at a very small percentage of ciliary tips, additional domains might facilitate the localization of Gli2 to cilia. Gli2ΔPC–GFP and Gli2ΔRep–GFP both exhibited diminished ciliary localization, suggesting that the phosphorylation cluster and the N-terminal repressor domains might cooperate with the ciliary localization domain to promote Gli2 entry or retention to cilia.
In addition to failing reach the cilium, Gli2-CLD–GFP was more stable than wild-type Gli2–GFP. To ascertain whether the stabilization of Gli2 was due to its failure to reach cilia, we assessed Gli2-CLD–GFP levels in MEFs lacking Kif3a, a protein required for ciliogenesis. Because Gli2–GFP and Gli2-CLD–GFP levels were unaffected by the absence of cilia, the central domain of Gli2 reduces protein stability independently of its role in ciliary localization. Two mechanisms by which this domain could destabilize Gli2 are through interaction with the E3 ubiquitin ligases Itch and SPOP. Itch recognizes proline-rich motifs within Gli1 to promote its turnover (Di Marcotullio et al., 2006). The homologous proline-rich motifs found in Gli2 are within the ciliary localization region, and might function as Itch-dependent degrons. Similarly, SPOP, a nuclear-speckle-associated protein that recruits Gli proteins to Cul3-based ubiquitin ligases, interacts with portions of the C-termini of Gli2 and Gli3 and this interaction might also require the central ciliary localization region (Bhatia et al., 2006; Chen et al., 2009; Pan et al., 2006; Zhang et al., 2009).
While investigating whether ciliary localization is required for transcriptional activity, we found that Gli2-CLD–GFP dominantly inhibited the activation of Gli target genes in ESCs and MEFs. Unlike Smo, the requirement for cilia in the activation of Gli2 can be circumvented by overexpressing wild-type Gli2 in both ciliated and non-ciliated cells. Similarly, an oncogenic version of Gli2 can activate Hh targets even in the absence of cilia (Han et al., 2009; Sheng et al., 2002; Wong et al., 2009). Thus, our finding that Gli2-CLD–GFP inhibits the expression of Hh target genes is consistent with the idea that ciliary localization is required for Gli2 activation. However, the ability to circumvent the ciliary requirement through overexpression or through deletion of the N-terminal repressor domain indicates that this requirement is not absolute. It will be interesting to determine whether Hh signals induce the cilium to activate Gli2 through inhibition of its repressor domain or other mechanisms.
Although the ciliary localization region of Gli2 plays a crucial role in the ciliary localization and activity of the protein, other regions control other important aspects of its behavior. For example, the C-terminal activator domain, the N-terminal repressor domain and a Sufu-binding site each inhibit nuclear accumulation. Notably, the Gli2 protein that lacked an important Sufu-binding site, Gli2ΔSYGH, was enriched in the nucleus as compared to full-length Gli2 and was destabilized, consistent with findings that the loss of Sufu leads to decreased Gli2 protein levels (Chen et al., 2009; Wang et al., 2010). Because SPOP and Sufu have opposing effects on Gli2 stability (Wang et al., 2010), SPOP-mediated degradation might account for the decreased stability of Gli2ΔSYGH. Despite its destabilization, Gli2ΔSYGH retained its ability to interact with Sufu both in the cytoplasm and in the nucleus. Therefore, the SYGH motif is important for Sufu-mediated nuclear exclusion and stabilization of Gli2 but other sites also contribute to the Sufu–Gli interaction, either directly or indirectly.
Multiple sites mediating the interaction of Gli2 with Sufu might correlate with the multiple roles that Sufu plays in Gli2 regulation. In addition to stabilizing Gli proteins, Sufu inhibits their nuclear localization and suppresses their transcriptional activity (Cheng and Bishop, 2002; Merchant et al., 2004). Because Gli2ΔSYGH–GFP shows increased nuclear localization without misactivating transcription, Sufu binding through a site distinct from the SYGH motif might restrict Gli2 transcriptional activity.
Somewhat surprisingly, mutating Gli2 phosphorylation and sumoylation sites did not affect ciliary localization. Because a non-sumoylatable form of Gli2 still localizes to cilia, sumoylation is dispensable for ciliary localization. Because the non-sumoylatable mutant displays increased transcriptional activity and the E2 Sumo-conjugating enzyme Ubc-9 can localize to cilia in C. elegans, it is possible that sumoylation occurs at the cilium to constrain Gli2 function in the absence of Hh pathway activation (Li et al., 2012). It will be interesting to determine whether Hh signals activate Gli2 by inducing de-sumoylation of Gli2 at cilia.
We also explored the intriguing possibility that mechanisms similar to nuclear trafficking control the ciliary localization of Gli2. The deletion of three putative nuclear export signals led to a detectable increase in the intensity of ciliary GFP. Both leptomycin B and siRNA-mediated knockdown of Exportin1 recapitulated this phenotype, but we did not detect an interaction between Gli2 and Exportin1. Thus, further work is needed to elucidate whether the inhibition of Exportin1 directly or indirectly regulates the ciliary exit of Gli2.
Although it has previously been evident that cilia are required for Hh signals to modulate the activator and repressor functions of Gli transcription factors, it has not been clear whether the localization of Gli to cilia is essential for this regulation. The ciliary localization region that we have identified is necessary for the ciliary localization of both Gli2 and Gli3 and is also essential for Gli2 transcriptional activity. These findings are consistent with ciliary localization being a crucial and essential step in the Hh-induced activation of Gli2.
MATERIALS AND METHODS
Cell lines and cell culture
Gli2gt/+ (XG045) E14 ESC lines were obtained from BayGenomics. Cells were cultured on 0.1% gelatin in knockout DMEM (Invitrogen) supplemented with 10% FBS, 1 mM glutamine, 1 mM pyruvate, 100 µM non-essential amino acids, 100 µM β-mercaptoethanol (BME) and 1000 U/ml leukemia inhibitory factor (LIF). Floxin cell lines were grown in 150 µg/ml G418. Differentiated cells were grown as a monolayer or as embryoid bodies in the absence of LIF for eight days. Smo−/− MEFs (a gift from J. Taipale) and NIH3T3 cells were cultured in DMEM H21 supplemented with 10% FBS and 50 µg/ml penicillin/streptomycin.
SAG (Enzo) was added to fresh medium at 1 µM in DMSO for 18 hours for transcriptional assays or 1 hour for localization assays. Leptomycin B (Sigma L2913) was used at 20 nM for 90 minutes.
cDNA constructs and cloning
Gli2 exons 2–14 were amplified and cloned into the pFloxin shuttle vector, in frame with the C-terminal GFP tag. QuikChange II XL-site directed mutagenesis (Stratagene) was used to introduce specific deletions and substitutions within Gli2. All products were sequenced before electroporation or transfection.
Electroporation, selection and screening
Electroporation for reversion and Floxin were performed as described previously (Singla et al., 2010). Floxin clones were selected with 150 µg/ml G418 and were assessed by genomic PCR and β-galactosidase activity using the Galacto-Light Plus System (Applied Biosystems).
Immunofluorescence and microscopy
ESCs were plated on coverslips coated with poly D-lysine and 1% Matrigel. The cells were fixed for 8 minutes in 4% paraformaldehyde (PFA), washed in PBS and incubated in PBST (PBS with 0.1% Triton X-100) for 20 minutes. The cells were blocked in 2% bovine serum albumin (BSA) in PBS for 1 hour at 4°C and were incubated overnight with primary antibody at 4°C. The cells were then washed in PBS and incubated in Alexa Fluor secondary antibodies (Invitrogen) for 1 hour at room temperature. The cells were mounted in Prolong Gold with DAPI. Images were acquired on one of the following microscopes: Zeiss Axio Observer D1, Nikon C1 confocal, Leica TCS SP5 or Leica TCS SPE.
Antibodies used for immunofluorescence were against: GFP (Aves Lab GFP-1020), 1∶1000; acetylated-tubulin (Sigma, T6793), 1∶1000; Ninein (a gift from J. Sillibourne), 1∶20,000; Gli2 (a gift from J. Eggenshwiler), 1∶2000; Gli3 (a gift from S. Scales), 1∶500; Sufu (Santa Cruz Biotechnology), 1∶200; and Xpo1 (Santa Cruz Biotechnology), 1∶200. Alexa Fluor secondaries (Invitrogen) were used at 1∶1000.
Integrated density quantification
Images were acquired by confocal microscopy using identical settings within experiments, and experiments were repeated three times. ImageJ or Fiji (NIH) was used to measure an 8×8 circular region of interest encompassing Gli2–GFP (ROIGFP). An identical ROI was taken in an adjacent region to account for background (ROIBACK). Integrated density readings, or pixel intensities, were measured as ROIGFP − ROIBACK and were binned into four equal data clusters. + represents the lowest pixel intensity, whereas ++++ represents the highest pixel intensity. Statistical significance was assessed using the Chi-squared goodness-of-fit test.
Immunoblots and immunoprecipitation
Cells were lysed in buffer containing 50 mM Tris-HCl pH 7.4, 0.5% NP-40, 0.25% sodium deoxycholate (NaDOC), 150 mM NaCl and 1× protease (Calbiochem) and 1× phosphatase (Pierce) inhibitors. The lysates were cleared at 16,000 g for 10 minutes and the protein concentration was quantified by Bradford assay. NE-PER Nuclear Cytoplasmic Reagent (Pierce) was used for cell-fractionation assays. Detection was performed using SuperSignal West Pico Chemiluminescent Substrate (Pierce). For Li-Cor Odyssey analysis, cells were lysed as above and subjected to SDS-PAGE and detection as per the manufacturer's instructions.
Antibodies used for western detection were against: GFP (Aves Lab GFP-1020), 1∶1000; Sufu (Santa Cruz Biotechnology), 1∶2000; α-tubulin (Sigma), 1∶5000; Xpo1 (Santa Cruz Biotechnology), 1∶1000; and Gli2 (a gift from J. Eggenshwiler), 1∶2000. Secondary antibodies (Jackson Immuno, Li-Cor IRDye) were used at 1∶10,000. Antibodies used for immunoprecipitation from 1 mg of cleared lysate include: anti-Sufu (Santa Cruz Biotechnology), 2 µg; GFP-Trap Resin (Allele), 15 µl of a 50% slurry.
Gli–luciferase reporter assays
Smo−/− MEFs were transfected with JetPrime as per the manufacturer's instructions (0.5 µg total DNA per well of a 24-well plate). DNA comprised 50% construct of interest, 40% Gli-luc and 10% pRLTK control. At 24 hours post-transfection, the cells were starved in Opti-MEM for 18 hours with or without SAG. The cells were assayed using the Dual Luciferase Reporter Kit (Promega). Statistical significance was assessed using the unpaired Student's t-test.
Quantitative PCR
RNA was extracted from ESCs or differentiated cells using RNeasy Plus (Qiagen). cDNA synthesis was performed using First Strand cDNA Synthesis (Fermentas). Transcript levels were measured in quadruplicate using a 7300 Real-time PCR machine (Applied Biosystems) and were normalized to β-actin. Primer sequences are: βactinF, 5′-CACAGCTTCTTTGCAGCTCCTT-3′ and βactinR, 5′-CGTCATCCATGGCGAACTG-3′; Gli2F, 5′-GCTGCACCAAGAGGTACACA-3′ and Gli2R, 5′-GGACATGCACATCATTACGC-3′; Gli1F, 5′-GGTGCTGCCTATAGCCAGTGTCCTC-3′ and Gli1R, 5′-GTGCCAATCCGGTGGAGTCAGACCC-3′; Ptch1F, 5′-CTCTGGAGCAGATTTCCAAGG-3′ and Ptch1R, 5′-TGCCGCAGTTCTTTTGAATG-3′; and gfpF, 5′-CGACCACTACCAGCAGAACA-3′ and gfpR, 5′-GAACTCCAGCAGGACCATGT-3′. Statistical significance was assessed using the unpaired Student's t-test or ANOVA.
In vitro sumoylation assays
In vitro sumoylation of immunoprecipitated Gli2–GFP was performed with 0.12 µM E1, 4 µM Ubc9 and 10 µM Sumo1 in a sumoylation buffer containing 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 10 mM MgCl2, 2 mM ATP and 2 µM fresh dithiothreitol (DTT) at 37°C for 1.5 hours. Reactions were then resolved by 7.5% SDS-PAGE and detected by immunoblot.
siRNA transfections
NIH3T3 and IMCD3 cells were plated at a density of 20,000 cells per 24-well coverslip 24 hours before transfection. Xpo1 siRNA (Invitrogen, 20 nM) was transfected by RNAiMax (Invitrogen) as per the manufacturer's instructions. At 36 hours post-transfection, cells were starved in Opti-MEM supplemented with 1% FBS for an additional 36 hours to promote ciliation. After 72 hours, cells were fixed and stained as described above.
Supplementary Material
Acknowledgments
We thank the members of the Reiter Lab for helpful discussions. We thank Jussi Taipale (University of Helsinki), James Sillibourne (Institut Curie), Jonathan Eggenshwiler (Univ. of Georgia), Suzie Scales (Genentech), Holly Ingraham and Tom Kornberg (UC San Francisco) for reagents.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
N.S. performed the experiments and data analysis. N.S. and J.R. wrote the manuscript.
Funding
This work was supported by the National Science Foundation to N.S., and the National Institutes of Health [grant numbers R01AR054396, R01GM095941]; the Burroughs Wellcome Fund; the Packard Foundation; and the Sandler Family Supporting Foundation to J.F.R. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.139253/-/DC1
References
- Aanstad P., Santos N., Corbit K. C., Scherz P. J., Trinh A., Salvenmoser W., Huisken J., Reiter J. F., Stainier D. Y. (2009). The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol. 19, 1034–1039 10.1016/j.cub.2009.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramírez-Weber F. A., Laget M. P., Schwartz C., Kornberg T. B. (1997). Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 10.1016/S0092--8674(00)80292--5 [DOI] [PubMed] [Google Scholar]
- Barnfield P. C., Zhang X., Thanabalasingham V., Yoshida M., Hui C. C. (2005). Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation 73, 397–405 10.1111/j.1432--0436.2005.00042.x [DOI] [PubMed] [Google Scholar]
- Barzi M., Berenguer J., Menendez A., Alvarez-Rodriguez R., Pons S. (2010). Sonic-hedgehog-mediated proliferation requires the localization of PKA to the cilium base. J. Cell Sci. 123, 62–69 10.1242/jcs.060020 [DOI] [PubMed] [Google Scholar]
- Bhatia N., Thiyagarajan S., Elcheva I., Saleem M., Dlugosz A., Mukhtar H., Spiegelman V. S. (2006). Gli2 is targeted for ubiquitination and degradation by beta-TrCP ubiquitin ligase. J. Biol. Chem. 281, 19320–19326 10.1074/jbc.M513203200 [DOI] [PubMed] [Google Scholar]
- Chen J. K., Taipale J., Young K. E., Maiti T., Beachy P. A. (2002). Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 99, 14071–14076 10.1073/pnas.182542899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. H., Wilson C. W., Li Y. J., Law K. K., Lu C. S., Gacayan R., Zhang X., Hui C. C., Chuang P. T. (2009). Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910–1928 10.1101/gad.1794109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. Y., Bishop J. M. (2002). Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl. Acad. Sci. USA 99, 5442–5447 10.1073/pnas.082096999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005). Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., Reiter J. F. (2008). Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 10, 70–76 10.1038/ncb1670 [DOI] [PubMed] [Google Scholar]
- Cox B., Briscoe J., Ulloa F. (2010). SUMOylation by Pias1 regulates the activity of the Hedgehog dependent Gli transcription factors. PLoS ONE 5, e11996 10.1371/journal.pone.0011996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. (1999). Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274, 8143–8152 10.1074/jbc.274.12.8143 [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Po A., Sico M. A., Alimandi M., Giannini G., Maroder M., Screpanti I. et al. (2006). Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 8, 1415–1423 10.1038/ncb1510 [DOI] [PubMed] [Google Scholar]
- Ding Q., Motoyama J., Gasca S., Mo R., Sasaki H., Rossant J., Hui C. C. (1998). Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125, 2533–2543 [DOI] [PubMed] [Google Scholar]
- Ding Q., Fukami S., Meng X., Nishizaki Y., Zhang X., Sasaki H., Dlugosz A., Nakafuku M., Hui C. (1999). Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 9, 1119–1122 10.1016/S0960--9822(99)80482--5 [DOI] [PubMed] [Google Scholar]
- Dishinger J. F., Kee H. L., Jenkins P. M., Fan S., Hurd T. W., Hammond J. W., Truong Y. N., Margolis B., Martens J. R., Verhey K. J. (2010). Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat. Cell Biol. 12, 703–710 10.1038/ncb2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva M., Michelson P., Kogerman P., Toftgard R. (2003). Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 278, 5116–5122 10.1074/jbc.M209492200 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J. T., Anderson K. V. (2007). Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345–373 10.1146/annurev.cellbio.23.090506.123249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S. C., Anderson K. V. (2010). The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. G., Kim H. J., Dlugosz A. A., Ellison D. W., Gilbertson R. J., Alvarez-Buylla A. (2009). Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 15, 1062–1065 10.1038/nm.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Pan Y., Wang B. (2012). Small ubiquitin-like Modifier (SUMO) modification inhibits GLI2 protein transcriptional activity in vitro and in vivo. J. Biol. Chem. 287, 20483–20489 10.1074/jbc.M112.359299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K. (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1, e53 10.1371/journal.pgen.0010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Anderson K. V. (2005). Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA 102, 11325–11330 10.1073/pnas.0505328102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Hui C. C., Angers S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 10.1146/annurev--cellbio--092910--154048 [DOI] [PubMed] [Google Scholar]
- Humke E. W., Dorn K. V., Milenkovic L., Scott M. P., Rohatgi R. (2010). The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 10.1101/gad.1902910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd T. W., Fan S., Margolis B. L. (2011). Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. J. Cell Sci. 124, 718–726 10.1242/jcs.070839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., Nakano Y., Seger C. (2011). Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 10.1038/nrg2984 [DOI] [PubMed] [Google Scholar]
- Kim J., Kato M., Beachy P. A. (2009). Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 106, 21666–21671 10.1073/pnas.0912180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K. W., Ruppert J. M., Bigner S. H., Vogelstein B. (1988). The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature 332, 371–374 10.1038/332371a0 [DOI] [PubMed] [Google Scholar]
- Kiprilov E. N., Awan A., Desprat R., Velho M., Clement C. A., Byskov A. G., Andersen C. Y., Satir P., Bouhassira E. E., Christensen S. T. et al. (2008). Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J. Cell Biol. 180, 897–904 10.1083/jcb.200706028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R., Zaphiropoulos P. G. (1999). Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312–319 10.1038/13031 [DOI] [PubMed] [Google Scholar]
- Lee F. Y., Faivre E. J., Suzawa M., Lontok E., Ebert D., Cai F., Belsham D. D., Ingraham H. A. (2011). Eliminating SF-1 (NR5A1) sumoylation in vivo results in ectopic hedgehog signaling and disruption of endocrine development. Dev. Cell 21, 315–327 10.1016/j.devcel.2011.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Wei Q., Zhang Y., Ling K., Hu J. (2012). SUMOylation of the small GTPase ARL-13 promotes ciliary targeting of sensory receptors. J. Cell Biol. 199, 589–598 10.1083/jcb.201203150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Wang B., Niswander L. A. (2005). Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 132, 3103–3111 10.1242/dev.01894 [DOI] [PubMed] [Google Scholar]
- McMahon A. P., Ingham P. W., Tabin C. J. (2003). Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1–114 10.1016/S0070--2153(03)53002--2 [DOI] [PubMed] [Google Scholar]
- Merchant M., Vajdos F. F., Ultsch M., Maun H. R., Wendt U., Cannon J., Desmarais W., Lazarus R. A., de Vos A. M., de Sauvage F. J. (2004). Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol. Cell. Biol. 24, 8627–8641 10.1128/MCB.24.19.8627--8641.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R., Freer A. M., Zinyk D. L., Crackower M. A., Michaud J., Heng H. H., Chik K. W., Shi X. M., Tsui L. C., Cheng S. H. et al. (1997). Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124, 113–123 [DOI] [PubMed] [Google Scholar]
- Motoyama J., Liu J., Mo R., Ding Q., Post M., Hui C. C. (1998). Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat. Genet. 20, 54–57 10.1038/1711 [DOI] [PubMed] [Google Scholar]
- Pan Y., Wang B. (2007). A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 282, 10846–10852 10.1074/jbc.M608599200 [DOI] [PubMed] [Google Scholar]
- Pan Y., Bai C. B., Joyner A. L., Wang B. (2006). Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 26, 3365–3377 10.1128/MCB.26.9.3365--3377.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Wang C., Wang B. (2009). Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev. Biol. 326, 177–189 10.1016/j.ydbio.2008.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca di Magliano M., Hebrok M. (2003). Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3, 903–911 10.1038/nrc1229 [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. (1993). Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science 261, 1701–1707 10.1126/science.8378770 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Scott M. P. (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Sasaki H., Nishizaki Y., Hui C., Nakafuku M., Kondoh H. (1999). Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126, 3915–3924 [DOI] [PubMed] [Google Scholar]
- Sheng H., Goich S., Wang A., Grachtchouk M., Lowe L., Mo R., Lin K., de Sauvage F. J., Sasaki H., Hui C. C. et al. (2002). Dissecting the oncogenic potential of Gli2: deletion of an NH(2)-terminal fragment alters skin tumor phenotype. Cancer Res. 62, 5308–5316 [PubMed] [Google Scholar]
- Singla V., Hunkapiller J., Santos N., Seol A. D., Norman A. R., Wakenight P., Skarnes W. C., Reiter J. F. (2010). Floxin, a resource for genetically engineering mouse ESCs. Nat. Methods 7, 50–52 10.1038/nmeth.1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempé D., Casas M., Karaz S., Blanchet-Tournier M. F., Concordet J. P. (2006). Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol. Cell. Biol. 26, 4316–4326 10.1128/MCB.02183--05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H., Lopez L. V., Salic A. (2010). A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415–428 10.1083/jcb.201004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M., He M., Anderson K. V. (2011). Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development 138, 4921–4930 10.1242/dev.070805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Li Y. (2006). Evidence for the direct involvement of betaTrCP in Gli3 protein processing. Proc. Natl. Acad. Sci. USA 103, 33–38 10.1073/pnas.0509927103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Pan Y., Wang B. (2010). Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137, 2001–2009 10.1242/dev.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Lai C. K., Evangelista M., Hongo J. A., de Sauvage F. J., Scales S. J. (2010). Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 30, 1910–1922 10.1128/MCB.01089--09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. Y., Seol A. D., So P. L., Ermilov A. N., Bichakjian C. K., Epstein E. H., Jr, Dlugosz A. A., Reiter J. F. (2009). Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat. Med. 15, 1055–1061 10.1038/nm.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Jia J., Liu A. (2010). Coordinated translocation of mammalian Gli proteins and suppressor of fused to the primary cilium. PLoS ONE 5, e15900 10.1371/journal.pone.0015900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B., Jiang J. (2009). Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 106, 21191–21196 10.1073/pnas.0912008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.