INTRODUCTION
Xenopus laevis is an ideal model system for investigating the mechanisms of pattern formation. The ability to express endogenous mRNA or introduce morpholinos into cleavage-stage Xenopus embryos has allowed gain- and loss-of-function experiments that reveal molecular-genetic control of development and regeneration. However, injection of mRNAs into cleavage-stage embryos provides limited spatio-temporal control: it is difficult to limit targeting to small regions (e.g., inducing foci of expression) and the fate-map does not facilitate targeting some tissues, such as those of the tail. Likewise, early injection can result in unwanted developmental defects, as mRNA can be translated long before the desired timepoint. These are especially important limitations when studying developmental and regenerative processes during gastrula – tailbud stages. While transgenic techniques allow precise control over spatio-temporal expression of genes when the appropriate promoter is available, the process of creating stable transgenic animals is time-consuming. Electroporation provides an alternative method for delivering mRNA and other nucleic acids, enabling the targeting of single cells or groups of cells at any stage of development. This protocol describes detailed electroporation parameters for the transfection of mRNA into a wide range of tissues in embryos at gastrula–tailbud stages, with high efficiency, low-toxicity, and expression as early as four hours post-electroporation. We present several sets of parameter values that take into account the resistance of electroporating solution (thus allowing this procedure to be performed in concert with physiology or drug screening experiments in which the medium composition can be varied), and allow control over the size of the expressing region, facilitating studies of signaling by specific regions or cell groups.
RELATED INFORMATION
Protocols are also available for Improved mRNA Electroporation Method for Xenopus Neurula Embryos (Sasagawa et al. 2002), Electroporation of CDNA/Morpholinos to targeted areas of embryonic CNS in Xenopus (Falk et al. 2007), Single-Cell Electroporation of Xenopus Tadpole Tectal Neurons (Liu and Haas 2011).
MATERIALS
Recipes and Caution
Please see appendix 1 for recipes of reagents marked with <R> and appendix 2 for cautions marked with <!>.
Be sure to read the safety caution and instructions contained in the operating manual of your electroporation machine.
Reagents
Agarose solution (1% prepared in 0.1x MMR)
<R>Marc's modified Ringer's (MMR) (1X)
mRNA encoding protein of interest
Tricaine Methanesulfonate (MS222) (0.03% prepared in 0.1x MMR)
Xenopus embryos/tadpoles, stage of interest
In this protocol, Xenopus embryo stages are given according to Nieuwkoop and Faber (Nieuwkoop and Faber 1967).
Equipment
Electroporation chamber
See Step 1
Forceps, fine (for breaking needles)
Injection needles (pulled capillary glasses)
Injector (e.g., Medical Systems PLI-100)
Micromanipulator, for injection (e.g., M3301R; World Precision Instruments)
Micropipettor and tips
Microscope, dissection
Microscope, equipped with the necessary filter sets to visualize fluorescent or fluorescently-tagged proteins.
In figure 2, an Olympus BX-61 Spinning Disc confocal microscope equipped with a Hamamatsu ORCA AG CCD camera, controlled by MetaMorph software, was used to image the expression of green-to-red photonvertible protein (EosFP) (Wacker et al. 2007) using the filters, EX 470/20; BS 485; EM 517/23 (Chroma filter set 41001), and the red fluorescent protein (tdTomato) (Shaner et al. 2004) using the filters EX 545/20; BS 565; EM 595/50.
FIGURE 2.
Generation of tDTomato- and EosFP-expressing animals via electroporation at stages 28 and 35. Panel (A) images were taken from Nieuwkoop and Faber (Nieuwkoop and Faber 1967) and retrieved from the Xenbase database (Bowes et al. 2009). Panel (A) shows regions of EosFP (I and IV) and Tdtomato (II and III) electroporations. The corresponding expressions in myotomes (I), hindgut (II), eye (III), and tail muscles are shown in panel (B).
Parafilm
Petri dishes (e.g., Fischerbrand 100mm × 15mm or 60mm × 15mm dishes)
A 60mm × 15mm dish with a hole 20mm in diameter at the center will be used in Step 1.
Pipettes, disposable transfer
Pulse generator (e.g., CUY21EDIT (square wave electroporator) by Nepa Gene Co., LTD)
Additional equipment required with CUY21EDIT include: Petridish Round Platinum Plate Electrode (anode) 20 mm diameter (CUY700P20E), Needle Cover Platinum Electrode (cathode) 0.3mm, 20mm-Tip (CUY195P0.3) and two cables connecting each electrode to the voltage plug terminal of CUY21EDIT.
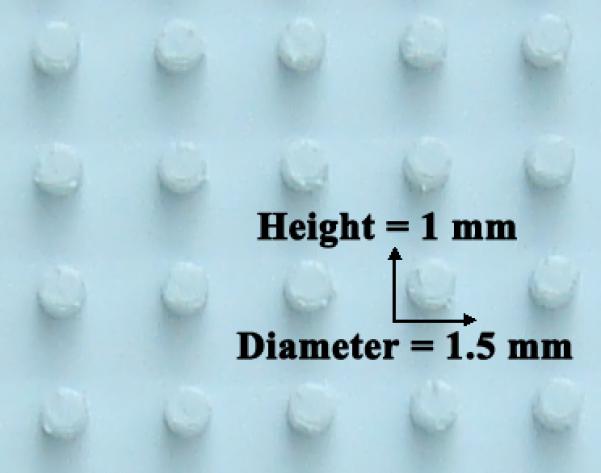
Rubber dimples
METHOD
Preparing electroporation chamber (40 min)
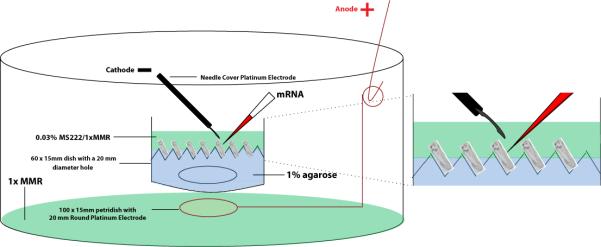
Cover the bottom of a 60mm × 15mm dish with a hole 20mm in diameter at the center with parafilm.
Pour 1% agarose (in 0.1x MMR) onto the dish to a thickness of 2mm – 3mm.
Immediately insert rubber dimples (for up to 20 pre-tailbud embryos) or 5mm micropipette tip cuts (for up to 7 tailbud/tadpole stage embryos) into the 1% agarose to make agarose bed pockets that keep animals in place during injection and electroporation.
See Fig. 1.
FIGURE 1.
Schematic for injection/electroporation set up. See steps 1-7 for detailed instructions.
Pre-injection/electroporation set-up
The set-up process can be completed while the agarose cools and solidifies (~30min) in Step 2. Ideally, set up micromanipulator and electroporator on the opposite side (right/left) of the dissection microscope.
-
4
Adjust micromanipulator (with needle) position so that it is ~ 450 with respect to the base of the dissection scope.
-
5
Calibrate needle (e.g., using fine forceps, break needle so that it outputs 10-20 nl solution per injection).
-
6
Fill needle with the appropriate mRNA (e.g., 0.5 μl of 1-2 μg/μl concentration) solution using the “fill” button on the micromanipulator.
Keep needle tip in water to prevent clogging.
-
7
Perform electroporation set up.
See Fig. 1. Remove rubber dimples or pipette tips before adding the 0.03% MS222 (in 1x MMR) solution to the injection/electroporation dish. Hold on to adding animals until after completing Step 10.
Determining electroporation parameters (~5 min)
The parameters that need to be determined are poring pulse (to form pores in the plasma membrane) and driving pulse (to drive mRNA into the cytosol) (Akaneya et al. 2005).
-
8
Placing the cathode into the 1x MMR solution of the injection / electroporation dish, perform a resistance (ohm) measurement by pressing the “ohm check” button.
-
9
Depending on the resistance value measured in Step 8, set up the poring and driving pulse parameters according to Table 1.
Table 1.
Resistance-dependent poring and driving pulse parameters for mRNA electroporation
| Resistance (Ω) | Parameters | |||
|---|---|---|---|---|
| Poring Pulse / Driving Pulse | ||||
| Volt (V) | Length (ms) | Interval (ms) | Repeat count | |
| 200 | 10 / 3 | 50 / 50 | 50 / 50 | 3 / 10 |
| 200 - 400 | 20 / 5 | 50 / 50 | 50 / 50 | 3 / 10 |
| 400 - 600 | 35 / 7 | 50 / 50 | 50 / 50 | 3 / 10 |
| 600 - 1000 | 50 / 10 | 50 / 50 | 50 / 50 | 3 / 10 |
In CUY21EDIT, poring and driving parameters can be stored in “Memory 1” and “Memory 2” and easily retrieved by simply pressing M1 and M2 buttons respectively.
See troubleshooting for out of range resistance values.
Injecting/Electroporating (~1 min per embryo or tadpole)
-
10
Anesthetize tailbud/tadpole stage embryos in 0.03% MS222 (~5min).
Skip this step for embryos younger than the neurula stage.
-
11
Transfer anaesthetized animals into injection/electroporation dish, carefully orienting each of them in the agarose bed pockets.
-
12
Inject 10-20 nl of mRNA into the desired location and immediately proceed to Step 13.
The injection volume can be varied to target fewer or more cells.
Injection/electroporation can be performed on early stage embryos without removing the vitelline membrane
See troubleshooting for issues concerning targeted expression.
-
13
Position the cathode about 1mm away from the surface of injection and apply poring and driving pulses respectively.
-
14
Repeat Steps 12 and 13 until all animals in agarose bed pockets have been electroporated.
-
15
Transfer to and keep animals in fresh 1x MMR for 1 hour.
-
16
Transfer animals to 0.1X MMR and rear them overnight at 14 or 180C.
Scoring for protein expression and Imaging
-
17
Anesthetize tailbud/tadpole stage animals in 0.03% MS222 (~5min).
-
18
Mount anesthetized animals onto the stage of a dissection scope (to view and score developmental phenotypes) or a microscope, equipped with the necessary filtersets to visualize fluorescently-tagged proteins encoded by the electroporated mRNA.
TROUBLESHOOTING
Problem: Measured resistance values are out of range.
[Step 9]
Solution: 1) If resistance values are greater than 18 KΩ, check to make sure that the electrodes are properly connected to the pulse generator.
2) If resistance measurements range between 1 – 18 KΩ, consider lowering these values using an injection/electroporation media with more concentrated MMR (higher ionic strength).
Problem: Protein expression site does not match that of injection/electroporation site.
[Step 12]
Solution: 1) Electroporate immediately following injection (since pressure-injected mRNA diffuses).
2) Use ~450 – angled needle for accurate delivery of mRNA to target site
Problem: No protein expression is observed.
Solution: Increase volume and/or concentration of injected mRNA.
DISCUSSION
Under the above electroporating conditions, we achieve 86–93% transfection efficiency with 100% viability using stage 15-35 animals. In addition to the observed high rates of transfection and viability, our techniques allows for targeted expression in tissues such as the flank and tail, both of which are hard to get by cleavage stage injection (Fig. 2). The high voltage / short duration poring pulse and the low voltage / longer duration driving pulse parameters can also be used successfully for targeted endotoxin-free DNA and morpholino delivery (data not shown).
FIGURE 3.
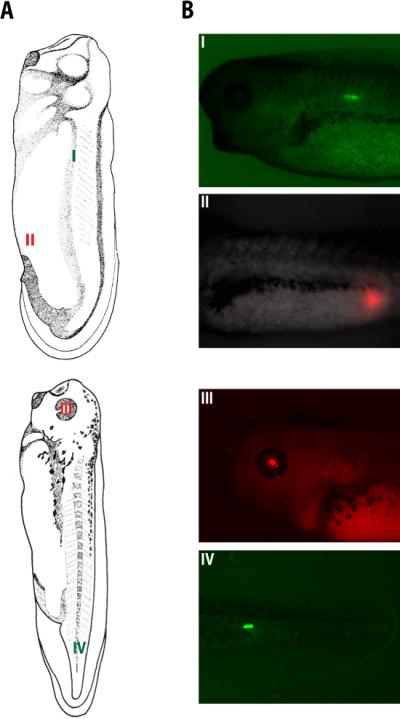
Generation of tdTomato- and EosFP-expressing Xenopus embryos via electroporation at stages 28 and 35. Illustrations in Panel A were retrieved from the Xenbase database (Bowes et al. 2009). (Reproduced with permission from Normal Table of Xenopus laevis (Daudin) by Nieuwkoop and Faber [1994]; ©1994 Garland Science/Taylor & Francis Books, LLC.) (A) Illustrations showing regions of EosFP (I and IV) and tdTomato (II and III) expression following electroporation. (B) Images showing expression in myotomes (I), hindgut (II), eye (III), and tail muscles (IV). These images were obtained with an Olympus BX-61 spinning disk confocal microscope equipped with a Hamamatsu ORCA AG CCD camera, controlled by MetaMorph software. Expression of EosFP (a greento- red photoconvertible protein; Wacker et al. 2007) was imaged using the filters EX 470/20, BS 485, and EM 517/23 (Chroma filter set 41001). Expression of tdTomato (a red fluorescent protein; Shaner et al. 2004) was imaged using the filters EX 545/20, BS 565, and EM 595/50.
ACKNOWLEDGEMENTS
We thank Junji Morokuma and Kelly McLaughlin for helpful discussions, and Franz Oswald for the kind gift of EosFP construct. M.L. is grateful for support by the NIH (EY018168, AR061988, GM078484, AR055993), the G. Harold and Leila Y. Mathers Charitable Foundation, and the Telemedicine and Advanced Technology Research Center (TATRC) at the U.S. Army Medical Research and Materiel Command (USAMRMC) through award W81XWH-10-2-0058.
Footnotes
RECIPE
Marc's modified Ringer's (MMR) (1X)
0.1 M NaCl
<!>2.0 mM KCl
<!>1.0 mM MgSO4
<!>2.0 mM CaCl2
5.0 mM HEPES (pH 7.8)
0.1 mM EDTA
CAUTIONS
Calcium chloride
is harmful by inhalation, ingestion, or skin absorption. Wear appropriate gloves and safety glasses. Use in a chemical fume hood.
Magnesium Sulfate
may be harmful by inhalation, ingestion, or skin absorption. Wear appropriate gloves and safety glasses.
Potassium Chloride
may be harmful by inhalation, ingestion, or skin absorption. Wear appropriate gloves and safety glasses.
REFERENCES
- Falk J, Drinjakovic J, Leung KM, Dwivedy A, Regan AG, Piper M, Holt CE. Electroporation of cDNA/Morpholinos to targeted areas of embryonic CNS in Xenopus. BMC Dev Biol. 2007;7:107. doi: 10.1186/1471-213X-7-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Haas K. Single-cell electroporation of Xenopus tadpole tectal neurons. Cold Spring Harb Protoc. 2011;2011(9) doi: 10.1101/pdb.prot065615. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin) North-Holland Publishing Company; Amesterdam: 1967. [Google Scholar]
- Sasagawa S, Takabatake T, Takabatake Y, Muramatsu T, Takeshima K. Improved mRNA electroporation method for Xenopus neurula embryos. Genesis. 2002;33(2):81–5. doi: 10.1002/gene.10094. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Wacker SA, Oswald F, Wiedenmann J, Knochel W. A green to red photoconvertible protein as an analyzing tool for early vertebrate development. Dev Dyn. 2007;236(2):473–80. doi: 10.1002/dvdy.20955. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2009;38(Database issue):D607–12. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]


