Abstract
Leishmania RNA virus (LRV) has been shown to be a symbiotic component of Leishmania parasites in South America. Nested retro-transcription polymerase chain reaction was employed to investigate LRV1 presence in leishmaniasis lesions from Brazil. In endemic areas of Rio de Janeiro (RJ), no LRV1 infection was observed even with mucosal involvement. LRV1 was only detected in Leishmania (V.) guyanensis cutaneous lesions from the northern region, which were obtained from patients presenting with disease reactivation after clinical cure of their primary lesions. Our results indicated that the severity of leishmaniasis in some areas of RJ, where Leishmania (V.) brazi-liensis is the primary etiological agent, was not associated with Leishmania LRV1 infection.
Keywords: Leishmania, LRV, tegumentary leishmaniasis, nested retro-transcription PCR
Tegumentary leishmaniasis (TL) comprises a spectrum of clinical manifestations caused by a group of protozoa, Leishmania spp, that are transmitted to humans and other mammals by phlebotomine sandflies. Clinical manifestations of TL include severe diffuse cutaneous leishmaniasis, localised cutaneous leishmaniasis (LCL), metastastatic and destructive mucosal leishmaniasis (ML) and mucocutaneous leishmaniasis (MCL). More than 10 species of Leishmania from the subgeneras Leishmania (Viannia) and L. ( Leishmania) cause TL in South America (Reithinger et al. 2007). In Brazil, Leishmania (V.) guyanensis and Leishmania (V.) braziliensis are the primary etiological agents of the disease. Approximately 1-10% of TL infections may evolve to mucosal manifestation months or years after the primary cutaneous lesion is cured, primarily in cases caused by L. (V.) braziliensis (Marsden 1986, Reithinger et al. 2007). Recently, cases of ML progression have also been associated with L. (V.) guyanensis infection (Guerra et al. 2011).
Leishmania RNA virus (LRV) and other protozoal and fungal viruses are members of the family Totiviridae. LRV is a dsRNA virus that persistently infects different Leishmania lineages. These viruses have been categorised as LRV1 and LRV2. LRV2 was found in Leishmania (L.) major , an Old World species (Scheffter et al. 1995) and LRV1 was identified in some strains of L. (V.) braziliensis and L. (V.) guyanensis, which circulate in specific regions of South America (Salinas et al. 1996, Saiz et al. 1998). High loads of LRV1 have been related to parasite persistence and metastasis and evasion of the host immune response (Ives et al. 2011). Several parasite strains must be examined to support the hypothesis that the virus presence favours the development of MCL or increases the rate of disease relapse (Scott 2011).
This study used nested retro-transcription polymerase chain reaction (PCR) targeting conserved viral sequences to investigate the presence of LRV1 in leishmaniasis lesions from patients with different clinical presentations diagnosed with L. (Viannia) infections.
Our cohort was selected from the southeastern (n = 40) and northern/northeastern (n = 8) regions of Brazil. All patients were diagnosed through clinical and epidemiological data, Montenegro skin testing, direct observation for the presence of the parasite using imprint or histopathology and/or kDNA PCR. This study was conducted with the approval of the Ethical Committee of the Oswaldo Cruz Institute (IOC), Oswaldo Cruz Foundation (Fiocruz) [Rio de Janeiro (RJ), Brazil] (protocol 695/11) and signed consent forms were obtained from all participants. Biopsy fragments were collected and kept frozen in liquid nitrogen until processing. This study included 27 LCL, 9 ML, 2 MCL, 5 recidivant LCL and five scars. Tissue samples from patients with sporothricosis and skin tumors were included as negative controls. All samples were retrospectively characterised as Leishmania (Viannia ) by kDNA hybridisation (Pirmez et al. 1999 ).
RNA was isolated from frozen sections of biopsy fragments using a commercial extraction kit (Real Biotech Corporation, Banqiao, Taiwan) according to the manufacturer's recommendations. Total RNA was reverse transcribed with Superscript II RT (Invitrogen Carlsbad, USA) using oligodT (Invitrogen). An LRV1-positive L. (V.) guyanensis isolate (MHOM/BR/1989/IM3597) obtained from the Leishmania Collection at the IOC was used as the positive control and processed using the same procedures as those used for the clinical samples. Amplifications were carried out in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, USA). Ethidium bromide-stained agarose gel electrophoresis was used to visualise the amplification products. Human β-actin was used as an endogenous control gene for the reactions before performing LRV1 RNA detection. The nested PCR detection limit was archived with serial dilutions of spectrophotometrically quantified purified PCR product. Nested PCR was able to detect 10 copies of the LRV1 amplicon. The 5' to 3' sequences of the primer sets used were as follows: β-actin: TAATGTCACGCACGATTTCCC and TCACCGAGCGCGGCT, LRV1 pair 1: CTGACTGGACGGGGGGTAAT and CAAAACACTCCCTTACGC, and LRV1 pair 2: GGTAATCGAGTGGGAGTCC and GCGGCAGTAACCTGG. PCR was performed using 35 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec.
The hsp70 gene was used to identify Leishmania species in LRV1 positive samples. A 234 bp fragment was amplified as previously described (da Graça et al. 2012), purified using the HiYieldTMGel/PCR DNA Mini kit (Real Biotech Corporation, Banqiao, Taiwan) and sequenced. The sequencing was performed by an automated sequencer (ABI PRISM(r) BigDye™Terminator Cycle Sequencing) at the Fiocruz facilities (Genomic Platform - DNA sequencing, Program for Technological Development of Health Products, Fiocruz).
From the 48 cases evaluated for the presence of LRV1, two were positive for the virus (Figure and Table). These two cases were from the Amazon Region, representing 25% of the cases analysed (n = 2/8) in the northern/northeastern area. Both LRV1 cases showed disease reactivation after clinical cure of the primary lesion. Sequencing of the Leishmania hsp70 gene was performed to determine which Leishmania species caused the TL. The two samples with LRV1 infection were determined to be caused by L. (V.) guyanensis . These results corroborated other studies demonstrating the presence of LRV1 infection in L. (V.) guyanensis strains circulating in the Amazon River Basin (Salinas et al. 1996) and Peru (Saiz et al. 1998). No LRV1 infection was observed in samples from the endemic regions of RJ (n = 0/40), regardless of the severity of the disease. The samples from patients infected in the northeastern states, where L. (V.) braziliensis is the predominant species of TL, were negative for LRV1.
Presence of Leishmania RNA virus 1 in clinical samples. Representative nested retro-transcription polymerase chain reaction (PCR) ethidium bromide stained 2.5% agarose gel. Ladders are 50 bp (left) and 100 bp (right). A: first round PCR using pair 1 primers; B: second and nested round using pair 2 primers; -: negative control; +: positive control with Leishmania (V.) guyanensis (MHOM/BR/1989/IM3597) cDNA; -*: double negative control using first round negative control as template; Lanes 1-3: state of Rio de Janeiro (RJ)-recidivant localised cutaneous leishmaniasis; 4, 8, 9: state of Amazonas-localised cutaneous leishmaniasis (LCL); 5-7: RJ-LCL; 10: RJ-scar; 11: RJ-mucosal leishmaniasis.
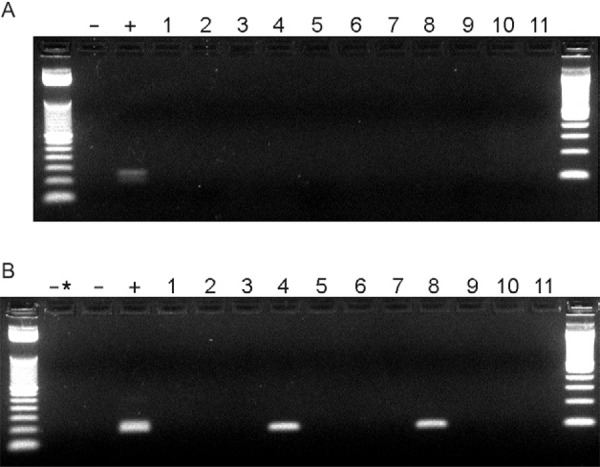
TABLE. Clinical and epidemiological characteristics of patients tested for the presence of Leishmania RNA virus 1 (LRV1).
| Brazilian Regions LRV1+ | |||||
|---|---|---|---|---|---|
| Southeast | North | Northeast | |||
| Clinical form | RJ | AM | BA | PB | Total (n) |
| LCL | 23 (0) | 4 (2) | - | - | 27 |
| ML | 7 (0) | - | 2 (0) | - | 9 |
| MCL | 2 (0) | - | - | - | 2 |
| REC | 4 (0) | 1 (0) | - | - | 5 |
| Scar | 4 (0) | - | - | 1 (0) | 5 |
| Sub-total (n) | 40 | 8 | 48 | ||
| LRV1+ (%) | 0 | 25 | |||
number of LRV1 positive cases are indicated in parenthesis for each clinical form and state. AM: state of Amazonas; BA: state of Bahia; LCL: localised cutaneous leishmaniasis; MCL: mucocutaneous leishmaniasis; ML: mucosal leishmaniasis; PB: state of Paraíba; REC: recidivant localised cutaneous leishmaniasis; RJ: state of Rio de Janeiro.
Presence of Leishmania RNA virus 1 in clinical samples. Representative nested retro-transcription polymerase chain reaction (PCR) ethidium bromide stained 2.5% agarose gel. Ladders are 50 bp (left) and 100 bp (right). A: first round PCR using pair 1 primers; B: second and nested round using pair 2 primers; -: negative control; +: positive control with Leishmania (V.) guyanensis (MHOM/BR/1989/IM3597) cDNA; -*: double negative control using first round negative control as template; Lanes 1-3: state of Rio de Janeiro (RJ)-recidivant localised cutaneous leishmaniasis; 4, 8, 9: state of Amazonas-localised cutaneous leishmaniasis (LCL); 5-7: RJ-LCL; 10: RJ-scar; 11: RJ-mucosal leishmaniasis.
It has been demonstrated that LRV types can be clustered according to the geographical origin of the parasite strains (Widmer & Dooley 1995). A molecular epidemiological study of L. (V.) braziliensis isolates from Brazil demonstrated variation in strain prevalence by region. The Amazonian Region group presented the highest level of genetic diversity, while isolates from RJ were more homogeneous (Cupolillo et al. 2003). Wide vectors diversity, sympatric Leishmania species and multiple host infections could be important factors for maintaining LRV prevalence in Amazon Basin Region. Our findings suggest that the strain of L. (V.) braziliensis that disseminated in the RJ region was not infected with LRV1 or had undetectable LRV1 titres and/or had developed differential RNAi pathway functionality. Lye et al. (2010) recently demonstrated that even if active, the RNAi pathway is less efficient in L. (V.) guyanensis M4147 (with or without LRV1) than in the virus-free L. (V.) braziliensis M2903. It is not only the parasite features, but also the host factors, such as age, gender and immune status, that are able to affect disease progression (Reithinger et al. 2007). Although metastasis can occur in the absence of LRV, as observed with Leishmania (V.) panamensis (reviewed by Hartley et al. 2012), LRV1-mediated hyper-inflammatory immune responses through toll-like receptor 3 may explain the differences in the clinical outcomes observed in different Leishmania species and/or strains. Our study showed that LRV1 infection was not correlated with severe and chronic TL cases caused by L. (V.) braziliensis in RJ and other endemic areas in Brazil.
Footnotes
Financial support: PROEP/CNPq, PAPES V-FIOCRUZ, POM-FIOCRUZ
REFERENCES
- Cupolillo E, Brahim LR, Toaldo CB, Oliveira-Neto MP de, Brito ME de, Falqueto A, Naiff M de Farias, Grimaldi G., Jr Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J Clin Microbiol. 2003;41:3126–3132. doi: 10.1128/JCM.41.7.3126-3132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça GC da, Volpini AC, Romero GAS, Oliveira MP de, Neto, Hueb M, Porrozzi R, Boité MC, Cupolillo E. Development and validation of PCR-based assays for diagnosis of American cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz. 2012;107:664–674. doi: 10.1590/s0074-02762012000500014. [DOI] [PubMed] [Google Scholar]
- Guerra JA, Prestes SR, Silveira H, Coelho LI, Gama P, Moura A, Amato V, Barbosa MG, Ferreira LC. Mucosal leishmaniasis caused by Leishmania (Viannia) braziliensis and Leishmania (Viannia) guyanensis in the Brazilian Amazon. PLoS Negl Trop Dis. 2011;5: doi: 10.1371/journal.pntd.0000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley MA, Ronet C, Zangger H, Beverley SM, Fasel N. Leishmania RNA virus: when the host pays the toll. Front Cell Infect Microbiol. 2012;2:99–99. doi: 10.3389/fcimb.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives A, Ronet C, Prevel F, Ruzzante G, Fuertes-Marraco S, Schutz F, Zangger H, Revaz-Breton M, Lye LF, Hickerson SM, Beverley SM, Acha-Orbea H, Launois P, Fasel N, Masina S. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science. 2011;331:775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6: doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden PD. Mucosal leishmaniasis ("espundia" Escomel, 1911) Trans R Soc Trop Med Hyg. 1986;80:859–876. doi: 10.1016/0035-9203(86)90243-9. [DOI] [PubMed] [Google Scholar]
- Pirmez C, Trajano VS, Paes-Oliveira M, Neto, da-Cruz AM, Gonçalves-da-Costa SC, Catanho M, Degrave W, Fernandes O. Use of PCR in diagnosis of human american tegumentary leishmaniasis in Rio de Janeiro, Brazil. J Clin Microbiol. 1999;37:1819–1823. doi: 10.1128/jcm.37.6.1819-1823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- Saiz M, Llanos-Cuentas A, Echevarria J, Roncal N, Cruz M, Muniz MT, Lucas C, Wirth DF, Scheffter S, Magill AJ, Patterson JL. Short report: detection of leishmania virus in human biopsy samples of leishmaniasis from Peru. Am J Trop Med Hyg. 1998;58:192–194. doi: 10.4269/ajtmh.1998.58.192. [DOI] [PubMed] [Google Scholar]
- Salinas G, Zamora M, Stuart K, Saravia N. Leishmania RNA viruses in Leishmania of the Viannia subgenus. Am J Trop Med Hyg. 1996;54:425–429. doi: 10.4269/ajtmh.1996.54.425. [DOI] [PubMed] [Google Scholar]
- Scheffter SM, Ro YT, Chung IK, Patterson JL. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an Old World parasite strain. Virology. 1995;212:84–90. doi: 10.1006/viro.1995.1456. [DOI] [PubMed] [Google Scholar]
- Scott P. Leishmania-a parasitized parasite. N Engl J Med. 2011;364:1773–1774. doi: 10.1056/NEJMcibr1101694. [DOI] [PubMed] [Google Scholar]
- Widmer G, Dooley S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 1995;23:2300–2304. doi: 10.1093/nar/23.12.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]


