Abstract
A single polymerase chain reaction (PCR) reaction targeting the spliced-leader intergenic region of Trypanosoma cruzi I was standardised by amplifying a 231 bp fragment in domestic (TcIDOM) strains or clones and 450 and 550 bp fragments in sylvatic strains or clones. This reaction was validated using 44 blind coded samples and 184 non-coded T. cruzi I clones isolated from sylvatic triatomines and the correspondence between the amplified fragments and their domestic or sylvatic origin was determined. Six of the nine strains isolated from acute cases suspected of oral infection had the sylvatic T. cruzi I profile. These results confirmed that the sylvatic T. cruzi I genotype is linked to cases of oral Chagas disease in Colombia. We therefore propose the use of this novel PCR reaction in strains or clones previously characterised as T. cruzi I to distinguish TcIDOMfrom sylvatic genotypes in studies of transmission dynamics, including the verification of population selection within hosts or detection of the frequency of mixed infections by both T. cruzi I genotypes in Colombia.
Keywords: spliced-leader intergenic region, Trypanosoma cruzi I, PCR, Chagas disease, domestic cycle, sylvatic cycle
Trypanosoma cruzi, the causative agent of Chagas disease, has been divided into six discrete taxonomic units (DTUs) named T. cruzi I-VI (Zingales et al. 2009). Recently, many authors have attempted to determine the geographical distribution of T. cruzi DTUs and their relationships with transmission dynamics and clinical outcomes. As summarised by Vallejo et al. (2009a), studies using TCC/TC1/TC2 primers designed for the spliced-leader intergenic region (SL-IR) (Souto et al. 1996) have demonstrated that T. cruzi I predominates in Colombian domestic and sylvatic vectors, with 2.1-78.6% having been detected in naturally-infected Rhodnius prolixus, Rhodnius colombiensis and Rhodnius pallescens mid-gut specimens. Studies of Colombian Chagas disease patients have also revealed the predominance of T. cruzi I over other DTUs in blood and tissue explants (Zafra et al. 2008, 2011, Mantilla et al. 2010, Ramírez et al. 2010).
Although there is a current consensus regarding the predominance of T. cruzi I in Colombian, Venezuelan and Central American countries, T. cruzi I has tremendous genetic variability, as demonstrated by several molecular markers (Guhl & Ramírez 2011). Based on the SL-IR region, four T. cruzi I genotypes associated with Chagas disease transmission cycles have been described in Colombia (Ia, Ib, Ic, Id) and corroborated through the American continent by the detection of a novel genotype, named T. cruzi Ie, associated with Mepraia species in Bolivia and Chile (Herrera et al. 2007, 2009, Cura et al. 2010). Specific primers have also been developed to identify three of these four genotypes (Ia, Ib and Id) (Falla et al. 2009). Other molecular markers have confirmed the intraspecific variability within T. cruzi I, suggesting the emergence of novel genotypes such as a TcI clade, which has been named TcIDOM (formerly TcIa⁄VENDOM) and is associated with the domestic transmission cycle throughout the American continent (Ramírez et al. 2012c). Several authors have thus proposed the separation of the two main T. cruzi I groups based on associations with domestic and sylvatic cycles (Llewellyn et al. 2009, Ocaña-Mayorga et al. 2010, Ramírez et al. 2012a, b).
The domestic and sylvatic cycles of T. cruzi are not strictly separated. Several reports have shown that sylvatic triatomines can invade housing units and contaminate food with T. cruzi, causing outbreaks of "oral Chagas disease". For example, the application of high-resolution molecular markers to biological clones from strains isolated during such outbreaks have incriminated T. cruzi I sylvatic strains and clones as causal agents of oral Chagas disease outbreaks in Colombia (Ramírez et al. 2013). This association between domestic and sylvatic cycles in T. cruzi I highlights the potential for further research in this field. The objective of this present study was to develop a method for single polymerase chain reaction (PCR) amplification of the SL-IR to discriminate between domestic and sylvatic genotypes and to obtain a better understanding of domestic and sylvatic cycles as well as the transmission dynamics of this DTU.
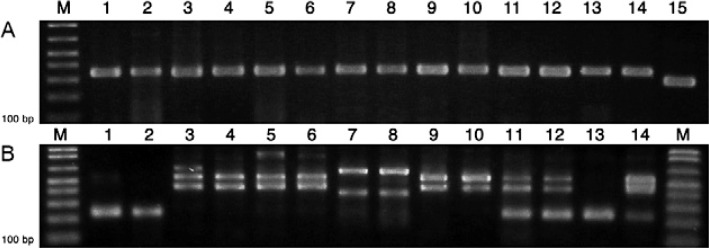
SL-IR sequences (accessions AM259467-AM259477 and EU626722-EU626738) were retrieved from GenBank and subsequently aligned. The 1Am primer (5'-TGTGTGTGTATGTATGTG-3') was designed and used with primer 1B (5'-CGGAGCGGTGTGTGCAG-3') (Falla et al. 2009) to perform PCR reactions in a total volume of 20 µL containing 2 µL of 10X reaction buffer (Invitrogen), 200 µM of a mixture of deoxynucleotide triphosphates, 1.5 mM MgCl2 , 25 µM each primer, 0.5 units of Taq DNA polymerase (Invitrogen). The template (20 ng) included DNA from a strain or clone previously characterised as T. cruzi I using different molecular markers, including multilocus enzyme electrophoresis or PCR of large subunit rDNA (Souto et al. 1996), PCR-restriction fragment length polymorphism (RFLP) of heat shock protein 60-EcoRV or glycosylphosphatidylinisotol-HhaI (Westenberger et al. 2005), PCR-RFLP of COll-Alul (Freitas et al. 2006) and PCR with TCC/TC1/TC2 primers designed in the SL-IR (Souto et al. 1996), as shown in Fig. 1A.
Fig. 1: Lanes 1-14 in A and B contain the same Trypanosoma cruzi I strains or clones. A: differentiation between Trypanosoma cruzi I and T. cruzi II using TCC/TC1/TC2 primers (Souto et al. 1996). 1-14: T. cruzi I (350 bp); 15: T. cruzi II (300 bp); M: 100 bp ladder; 1: MHOM/C0/04/MG strain; 2: CG clone 1; 3: ORT15 Rcol strain; 4: Rcol clone 03; 5: Rpal clone 139-2; 6: Rpal clone 89-2; 7: Rpro (sylvatic) clone 8; 8: Rpro (sylvatic) clone 10; 9: D1 ( Didelphis ) strain; 10: D2 ( Didelphis ) strain; 11: X1081 Rpro (domestic) strain; 12: X1082 Rpro (domestic) strain; 13: CG clone 13; 14: EH strain; 15: T. cruzi II (300 bp); B: genotyping T. cruzi I strains and clones using a single pair of primers (1Am/1B) designed in the spliced-leader intergenic region. Domestic strains were identified by amplifying a 231 bp fragment and sylvatic strains were identified by amplifying a 450-550 bp fragment. M: 100 bp ladder; 1: MHOM/C0/04/MG strain; 2: CG clone 1; 3: ORT15 Rcol strain; 4: Rcol clone 03; 5: Rpal clone 139-2; 6: Rpal clone 89-2; 7: Rpro (sylvatic) clone 8; 8: Rpro (sylvatic) clone 10; 9: D1 ( Didelphis ) strain; 10: D2 ( Didelphis ) strain; 11: X1081 Rpro (domestic) strain; 12: X1082 Rpro (domestic) strain; 13: CG clone 13; 14: EH strain; M: 100 bp ladder.
All PCR reactions were conducted for 35 amplification cycles using a thermal minicycler (MJ Research PTC-150-16). Cycling include a denaturing step at 94ºC for 30 s (4 min for initial denaturing), annealing at 51ºC for 30 s, extension at 72ºC for 30 s and a final extension step at 72ºC for 10 min.
After standardising the reaction conditions using the previously described primers, the TcIDOM and clones produced the expected 231 bp amplification product; however, the sylvatic clones and strains produced 450-550 bp amplification products when using the same primers (Fig. 1B). Several T. cruzi I samples and clones isolated from sylvatic triatomines and reservoirs produced the 450-550 bp profile, as shown in Fig. 1B. The ORT15 strain, isolated by xenoculture from a rectal ampulla from R. colombiensis in the department of Tolima, was correctly typed after its isolation in February 2013 (Fig. 1B, Lane 3). T. cruzi I clones from the intestinal contents of R. colombiensis, R. pallescens and sylvatic R. prolixus, obtained by direct plating in a sensitive solid medium (Yeo et al. 2007), produced the 450-550 bp profile (Fig. 1B, Lanes 4-8). The D1 and D2 strains, isolated from Didelphis marsupialis and maintained in culture in biphasic medium, also produced the sylvatic profile (Fig. 1B, Lanes 9, 10). The existence of mixed profiles in T. cruzi I strains is possible, as 175 of 182 (96.2%) clones obtained from sylvatic R. colombiensis, R. pallescens and R. prolixus produced the 450-550 bp profile and seven of these 182 clones (3.8%) produced a 231 bp profile (data not shown). These results support the assertion that the 450-550 bp profile is predominant in sylvatic cycles.
Alternately, MHOM/CO/04/MG, a human strain that is maintained by periodic passages in mice, produced the 231 bp profile (Fig. 1B, Lane 1) as did two human CG strain clones (Fig. 1B, Lanes 2, 13). Three domestic samples had mixed profiles. The X-1081 and X-1082 strains, which were isolated by xenoculture from the intestinal content of domiciliated R. prolixus in the department of Boyaca and kept in culture for several years, displayed the 231 and 450-550 bp profiles (Fig. 1B, Lanes 11, 12). A similar mixed profile was observed in the human EH strain (Lane 14), suggesting a mixed infection of both genotypes, as previously reported by Ramírez et al. (2013).
Domestic or sylvatic reservoirs may also select for subpopulations within T. cruzi I. The possibility that domestic or sylvatic vectors may selectively transmit T. cruzi I genotypes cannot be excluded, as the selective transmission of both Trypanosoma rangeli genotypes has been demonstrated for various Rhodnius species (Pulido et al. 2008, Vallejo et al. 2009b, Urrea et al. 2011).
When diagnostic or molecular characterisation techniques are developed, it is necessary to verify that the technique is specific to T. cruzi and demonstrate a lack of cross-reaction with T. rangeli, as T. rangeli frequently occurs in mixed infections with T. cruzi in triatomines and in vertebrate reservoirs in Colombia. We emphasise that the purpose of this work was to use a PCR reaction with 1Am/1B primers in strains that had previously been typed as T. cruzi I sensu lato and thus we initially used strains or clones that were previously characterised as T. cruzi I. Nevertheless, because mixed T. rangeli infections in vectors and vertebrates can be found in nearly all T. cruzi-endemic areas in Colombia, strains can therefore be isolated together with T. cruzi I and T. rangeli. T. rangeli DNA and 1Am/1B primers, which did not produce any amplification products, were used as controls to rule out cross-reactions (data not shown).
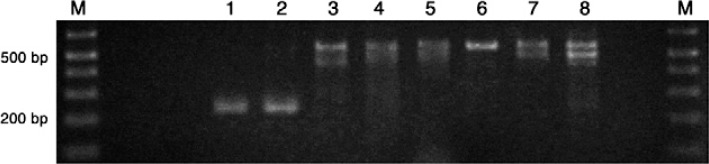
Our results demonstrated that Colombian domestic and sylvatic T. cruzi I strains can be discriminated using a single PCR amplification of the SL-IR from TcIDOM (formerly TcIa/VENDOM) and sylvatic strains. Nine isolates from patients suffering acute Chagas disease involving different transmission routes and different clinically diagnosed symptomatology were used to verify whether this single PCR reaction could be used to discriminate between T. cruzi I infections of TcIDOM and sylvatic genotype origin. Six of the nine isolates had the sylvatic profile, corresponding to three cases of acute myocarditis in outbreaks of oral transmission (Santander, Colombia), a congenital case (Santander, Colombia) and two cases of sylvatic vector transmission (Putumayo, Colombia) (Fig. 2). The remaining three cases were identified as T. cruzi II when using D71/D72, V1/V2 primers (Brisse et al. 2001). T. cruzi II was found in two congenital cases (Boyaca, Colombia), as previously described (Pavia et al. 2009), and in a case of reactivation caused by human immunodeficiency virus/acquired immune deficiency syndrome (Caquetá, Colombia). Human T. cruzi strains were isolated by haemoculture during the acute phase of infection and were typed long after isolation and thus there was no patient follow-up to obtain new isolates for further characterisation. Selection among T. cruzi genotype mixtures when maintained in culture or in animal models could not be ruled out, as selection towards TcII in mixed infections with TcI strains (with subtle TcII parasites or clones) has been reported (Pena et al. 2011). Our single PCR reaction for discriminating T. cruzi I genotypes could be of great use for determining the genotype frequency as well as follow-up analyses comparing the profiles of initially isolated genotypes with their later stability or selection after maintenance in culture or biological models for varied lengths of time.
Fig. 2. characterisation of human isolates using 1Am/1B primers. Lanes 1-2 contain the same samples of MHOM/CO/03/CG C1 clone of human origin and Lanes 3-8 contain the strains SMA, XCH, JEM, YLY, EH, HMriv isolated from acute cases of Chagas disease in Colombia.
The results of this present study corroborated previous reports by Ramírez et al. (2013) highlighting the fact that the sylvatic T. cruzi I genotype is implicated in oral and sylvatic vector transmission in Colombia. New studies involving a greater number of human isolates should further support these observations.
In summary, after T. cruzi I has been diagnosed in any isolate, we propose that an additional PCR reaction be used with a single pair of primers for discriminating between TcIDOM and sylvatic T. cruzi I. This methodology has led to the identification of the genotypes involved in outbreaks of oral Chagas disease as well as supporting studies of the transmission dynamics of T. cruzi as the causal agent of Chagas disease in Colombia. Our results agree with those of other investigators reporting that T. cruzi I encompasses two main groups that, to date, are not known to be stable in time and space. Further investigation will be useful for verifying population selection within hosts and detecting the frequency of mixed genotypes of T. cruzi I in Colombia.
ACKNOWLEDGEMENTS
To Javier Andrés Trejo, Uriel Alvarado and Diana Catalina Serrato, from the Tropical Parasitology Research Laboratory, for providing technical assistance.
Funding Statement
Financial support: COLCIENCIAS (110551929038), UT Research Fund, Chagas EpiNet/FP7 (223034)
Footnotes
Financial support: COLCIENCIAS (110551929038), UT Research Fund, Chagas EpiNet/FP7 (223034)
REFERENCES
- Brisse S, Verhoef J, Tibayrenc M. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol. 2001;36:337–346. doi: 10.1016/s0020-7519(01)00238-7. [DOI] [PubMed] [Google Scholar]
- Cura CI, Mejía-Jaramillo AM, Duffy T, Burgos JM, Rodriguero M, Cardinal MV, Kjos S, Gurgel-Gonçalves R, Blanchet D, de Pablos LM, Tomasini N, da Silva A, Russomando G, Cuba CA, Aznar C, Abate T, Levin MJ, Osuna A, Gürtler RE, Diosque P, Solari A, Triana-Chávez O, Schijman AG. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int J Parasitol. 2010;40:1599–1607. doi: 10.1016/j.ijpara.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Trop. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Freitas JM, Pinto LA, Pimenta JR, Rodrigues LB, Goncalves VF, Teixeira SM. Ancestral genomes, sex and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2: doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhl F, Ramírez JD. Trypanosoma cruzi I diversity: towards the need of genetic subdivision? Acta Trop. 2011;119:1–4. doi: 10.1016/j.actatropica.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Herrera C, Bargues MD, Fajardo A, Montilla M, Triana O, Vallejo GA, Guhl F. Identifying four Trypanosoma cruzi I isolate haplotypes from different geographic regions of Colombia. Infect Genet Evol. 2007;7:535–539. doi: 10.1016/j.meegid.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Herrera C, Bargues MD, Falla A, Fajardo A, Montilla M, Vallejo GA, Guhl F. Genetic variability and phylogenetic relationships within Trypanosoma cruzi I isolates in Colombia based on minexon gene sequences. J Parasitol Res. 2009;897364 doi: 10.1155/2009/897364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Vargas J, Torrico F, Diosque P, Valente V, Valente SA, Gaunt MW. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5: doi: 10.1371/journal.ppat.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantilla JC, Zafra GA, Macedo AM, González CI. Mixed infection of Trypanosoma cruzi I and II in a Colombian cardiomyopathic patient. Hum Pathol. 2010;41:610–613. doi: 10.1016/j.humpath.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Ocaña-Mayorga S, Llewellyn M, Costales J, Miles MA, Grijalva MJ. Sex, subdivision and domestic dispersal of Trypanosoma cruzi lineage I in southern Ecuador. PLoS Negl Trop Dis. 2010;4: doi: 10.1371/journal.pntd.0000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavia PX, Montilla M, Flórez C, Herrera G, Ospina JM, Manrique F, Nicholls RS, Puerta C. The first case of congenital Chagas disease analyzed by AP-PCR in Colombia. Biomedica. 2009;29:513–522. [PubMed] [Google Scholar]
- Pena DA, Eger I, Nogueira L, Heck N, Menin Á, Báfica A, Steindel M. Selection of TcII Trypanosoma cruzi population following macrophage infection. J Infect Dis. 2011;204:478–486. doi: 10.1093/infdis/jir292. [DOI] [PubMed] [Google Scholar]
- Pulido XC, Pérez G, Vallejo GA. Preliminary characterization of a Rhodnius prolixus hemolymph trypanolytic protein, this being a determinant of Trypanosoma rangeli KP1(+) and KP1(-) subpopulations' vectorial ability. Mem Inst Oswaldo Cruz. 2008;103:172–179. doi: 10.1590/s0074-02762008000200008. [DOI] [PubMed] [Google Scholar]
- Ramírez JD, Duque MC, Montilla M, Cucunubá Z, Guhl F. Natural and emergent Trypanosoma cruzi I genotypes revealed by mitochondrial (Cytb) and nuclear (SSU rDNA) genetic markers. Exp Parasitol. 2012;132:487–494. doi: 10.1016/j.exppara.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Ramírez JD, Duque MC, Montilla M, Cucunubá ZM, Guhl F. Multilocus PCR-RFLP profiling in Trypanosoma cruzi I highlights an intraspecific genetic variation pattern. Infect Genet Evol. 2012;12:1743–1750. doi: 10.1016/j.meegid.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Ramírez JD, Guhl F, Messenger LA, Lewis MD, Montilla M, Cucunuba Z, Miles MA, Llewellyn MS. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol Ecol. 2012;21:4216–4226. doi: 10.1111/j.1365-294X.2012.05699.x. [DOI] [PubMed] [Google Scholar]
- Ramírez JD, Guhl F, Rendón LM, Rosas F, Marin-Neto JA, Morillo CA. Chagas cardiomyopathy manifestations and Trypanosoma cruzi genotypes circulating in chronic chagasic patients. PLoS Negl Trop Dis. 2010;4: doi: 10.1371/journal.pntd.0000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez JD, Montilla M, Cucunubá ZM, Flórez AC, Zambrano P, Guhl F. Molecular epidemiology of human oral Chagas disease outbreaks in Colombia. PLoS Negl Trop Dis. 2013;7: doi: 10.1371/journal.pntd.0002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- Urrea DA, Herrera CP, Falla A, Carranza JC, Cuba-Cuba C, Triana-Chávez O, Grisard EC, Guhl F, Vallejo GA. Sequence ana-lysis of the spliced-leader intergenic region (SL-IR) and random amplified polymorphic DNA (RAPD) of Trypanosoma rangeli strains isolated from Rhodnius ecuadoriensis, R. colombiensis, R. pallescens and R. prolixus suggests a degree of co-evolution between parasites and vectors. Acta Trop. 2011;120:59–66. doi: 10.1016/j.actatropica.2011.05.016. [DOI] [PubMed] [Google Scholar]
- Vallejo GA, Guhl F, Carranza JC, Herrera C, Urrea DA, Falla A, Zabala D, Villa LM. Trypanosoma cruzi population variability in Colombia: possible co-evolution in different vector species. Rev Soc Bras Med Trop. 2009;42(2):27–34. [Google Scholar]
- Vallejo GA, Guhl F, Schaub GA. Triatominae-Trypanosoma cruzi/T. rangeli: vector-parasite interactions. Acta Trop. 2009;110:137–147. doi: 10.1016/j.actatropica.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Westenberger SJ, Barnabe C, Campbell DA, Sturm NR. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171:527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lewis MD, Carrasco HJ, Acosta N, Llewellyn M, Valente SAS, Valente VC, de Arias AR, Miles MA. Resolution of multiclonal infections of Trypanosoma cruzi from naturally infected triatomine bugs and from experimentally infected mice by direct plating on a sensitive solid medium. Int J Parasitol. 2007;37:111–120. doi: 10.1016/j.ijpara.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Zafra G, Mantilla JC, Jácome J, Macedo AM, González CI. Direct analysis of genetic variability in Trypanosoma cruzi populations from tissues of Colombian chagasic patients. Hum Pathol. 2011;42:1159–1168. doi: 10.1016/j.humpath.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Zafra G, Mantilla JC, Valadares HM, Macedo AM, González CI. Evidence of Trypanosoma cruzi II infection in Colombian chagasic patients. Parasitol Res. 2008;103:731–734. doi: 10.1007/s00436-008-1034-0. [DOI] [PubMed] [Google Scholar]
- Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]