Abstract
Currently, several assays can confirm acute dengue infection at the point-of-care. However, none of these assays can predict the severity of the disease symptoms. A prognosis test that predicts the likelihood of a dengue patient to develop a severe form of the disease could permit more efficient patient triage and treatment. We hypothesise that mRNA expression of apoptosis and innate immune response-related genes will be differentially regulated during the early stages of dengue and might predict the clinical outcome. Aiming to identify biomarkers for dengue prognosis, we extracted mRNA from the peripheral blood mononuclear cells of mild and severe dengue patients during the febrile stage of the disease to measure the expression levels of selected genes by quantitative polymerase chain reaction. The selected candidate biomarkers were previously identified by our group as differentially expressed in microarray studies. We verified that the mRNA coding for CFD, MAGED1, PSMB9, PRDX4 and FCGR3B were differentially expressed between patients who developed clinical symptoms associated with the mild type of dengue and patients who showed clinical symptoms associated with severe dengue. We suggest that this gene expression panel could putatively serve as biomarkers for the clinical prognosis of dengue haemorrhagic fever.
Keywords: dengue, biomarkers, apoptosis, innate immunity, quantitative real-time PCR
Dengue fever (DF) is a viral illness that is transmitted by the Aedes genus of mosquitoes (Tsai & Halstead 2002). Annually, the disease affects several million people in tropical regions of the world (Gibbons & Vaughn 2009). Dengue virus (DENV) infection has a large spectrum of clinical manifestations, ranging from asymptomatic to a mild DF to the severe symptoms of haemorrhage and shock named dengue haemorrhagic fever (DHF) or severe dengue. DF is usually a self-limiting condition (Cordeiro et al. 2009); however, the main concern is the development of the severe form, which occurs in up to 5-10% of cases and is lethal if not treated properly (Senanayake 2006, Huerta-Zepeda et al. 2008). The World Health Organization (WHO) defines DHF as an illness with similar symptoms to DF with the addition of circulatory failure manifested by a rapid and weak pulse with the narrowing of the pulse pressure or hypotension, resulting in cold, clammy skin and restlessness (WHO 2012). The outcome of DHF depends largely on early diagnosis and the immediate replacement of fluid (Mairuhu et al. 2004). The main problem facing clinicians when treating dengue-infected patients is deciding which ones can be sent home and which ones need to be hospitalised. Although intensive efforts have been made to study the early clinical pathophysiology of dengue infection with the objective of identifying a potential cause of DHF, we still do not have a reliable early marker that can predict which patients will develop DHF (Noisakran & Perng 2008).
The coincidence of severe disease manifestations simultaneously with defervescence and viral control was highlighted by Screaton and Mongkolsapaya (2006), who suggested that the symptoms observed in severe dengue could be a consequence of the exaggerated and misdirected cellular immune response to the virus rather than viral-induced cytopathology. Mathew and Rothman (2008) postulated that a skewed T-cell cytokine response leads to plasma leakage in DHF. Furthermore, several studies have shown that host genetic factors can be correlated with severe dengue susceptibility or protection (Chaturvedi et al. 2006, Coffey et al. 2009). Many of these genes, such as CD209 (Kwan et al. 2005, Sakuntabhai et al. 2005, Durbin et al. 2008), tumour necrosis factor-alpha (Espina et al. 2003) and FC gamma receptor IIA (Loke et al. 2002), highlight the role of innate immunity in dengue pathogenesis. Moreover, our group has found significant correlations between wild-type AA MBL2 genotype and age as associated risk factors for the development of dengue-related thrombocytopaenia (Acioli-Santos et al. 2008).
The search for the molecular events involved in DHF development and for host genes related to the protection/infection of the DENV has instigated several high-throughput studies that have elucidated the role of various molecules and pathways during a DENV infection event (Fink et al. 2007, de Kruif et al. 2008, Higa et al. 2008, Chang et al. 2009, Long et al. 2009, Nascimento et al. 2009a, b). Despite controversial and inconclusive results, there is a consensus that the innate immune and apoptotic processes trigger haemorrhagic symptoms depending on the magnitude of the response/expression (Espina et al. 2003, Ubol et al. 2008, Brown et al. 2009, Cheng et al. 2009, Gil et al. 2009, Jaiyen et al. 2009, Nasirudeen & Liu 2009, Nascimento et al. 2009a, Conceição et al. 2010).
In previous work (Calzavara-Silva et al. 2009), we suggested that it is possible to define genes that are differentially expressed between DF and DHF patients during the onset of symptoms and, consequently, predict the propensity of a dengue patient to progress to the severe form of the disease. Here, we propose the use of five genes that may predict a propensity of a DENV-infected patient to develop the severe symptoms of dengue as a molecular tool to aid DHF clinical diagnosis.
SUBJECTS, MATERIALS AND METHODS
Patients and samples - Patients were classified following the WHO criteria. We used peripheral blood mononuclear cells (PBMCs) from 15 patients (5 DF and 10 DHF) presenting different clinical forms of the disease during the acute phase (up to 7 days of fever) or convalescent phase (more than 21 days post onset of symptoms) and five samples obtained from febrile non-dengue (ND) individuals. Table shows a summary of patient data. Acute phase samples were subjected to polymerase chain reaction (PCR) (Lanciotti et al. 1992) to indicate the absence or presence of viral RNA and serotyping and anti-dengue IgM-capture ELISAs (PanBio) and anti-dengue IgG indirect ELISAs (PanBio) to determine the presence of anti-DENV IgM and IgG antibodies, respectively. Primary infection was characterised by the absence of dengue-specific IgG antibodies in the acute serum sample and the presence of anti-dengue IgM and/or viral RNA detection, followed by the presence of anti-dengue IgG in convalescent serum samples. Sequential infection was characterised by detection of specific anti-dengue IgG in the acute sample and the absence of anti-dengue IgM, associated with a positive reverse transcription-PCR, followed by the presence of anti-dengue IgM in convalescent serum samples (Cordeiro et al. 2007, 2009). Samples characterised as "never infected" were obtained from febrile volunteers and were characterised by all negative results. Blood samples from patients enrolled in this study were collected in heparin Vacutainer tubes (BD Vacutainer) and within 2 h of collection, PBMC samples were separated by gradient density using Ficoll-Paque (GE Healthcare) and cryopreserved in 10% (v/v) dimethyl sulfoxide (Sigma-Aldrich) in inactivated foetal bovine sera (Thermo Scientific Hyclone).
TABLE. Samples used in the quantitative real-time polymerase chain reaction assays.
| Patient ID | Pathology | Serotype | IgG/IgM | Infection | Days of fever |
|---|---|---|---|---|---|
| 305 | DHF | 3 | -/+ | Primary | 4 |
| 428 | DHF | 3 | +/- | Sequential | 3 |
| 564 | DHF | - | -/+ | Primary | 2 |
| 586 | DHF | 3 | +/+ | Sequential | 0 |
| 629 | DHF | 3 | +/- | Sequential | 4 |
| 400 | DHF | 3 | -/- | Primary | 2 |
| 265 | DHF | 3 | -/+ | Primary | 7 |
| 379 | DHF | 3 | +/- | Sequential | 4 |
| 429 | DHF | 3 | +/- | Sequential | 3 |
| 248 | DHF | 3 | +/- | Sequential | 5 |
| 650 | DF | 3 | +/- | Sequential | 1 |
| 593 | DF | 3 | -/- | Primary | 6 |
| 582 | DF | - | -/+ | Sequential | 2 |
| 506 | DF | 3 | +/- | Sequential | 2 |
| 424 | DF | 3 | +/- | Sequential | 4 |
| Col1 | ND | - | -/- | Never infected | 3 |
| Col2 | ND | - | -/- | Never infected | 6 |
| Col3 | ND | - | -/- | Never infected | 5 |
| Col4 | ND | - | -/- | Never infected | 5 |
| Col5 | ND | - | -/- | Never infected | 2 |
DF: dengue fever; DHF: dengue haemorrhagic fever; ND: non-dengue; -: negative; +: positive.
Quantitative real-time PCR (qPCR) - Genes were amplified and detected using TaqMan (r) gene expression assays (Applied Biosystems, cat. 4331182 - gene id: CFD - Hs00157263_m1, MT2A - Hs02379661_g1, MYD88 - Hs01573837_g1, PDCD4 - Hs00377253_m1, MAGED1 - Hs00986269_m1, PSMB9 - Hs00160610_m1, FCGR3B - Hs00275547_m1, PRDX4 - Hs01056076_m1, PYCARD - Hs01547324_g1). Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and treated with DNAse (Qiagen) following the manufacturer's protocols. Total RNA (1 µg) was reverse transcribed to cDNA using a SuperScript III First-Strand Synthesis System (Invitrogen) and Random Hexamer Primers (Invitrogen) under the following the reaction conditions: 50ºC for 30 min, 85ºC for 5 min and then incubation on ice. RNase H (2 U) (Invitrogen) was added and samples were incubated at 37ºC for 20 min. qPCR was performed using the ABI PRISM 7500 (Applied Biosystems). cDNA obtained from the total RNA of the patients described above was used. A mix of five ND cDNA samples was used as a reference for the DF and DHF results. β-actin gene expression was used to normalise the gene expression data due to its constitutive expression. Reactions were performed in triplicate and included 2 µL of cDNA, 6.25 µM of each specific assay or human Beta-Actin (Applied Biosystems), TaqMan Universal PCR Master Mix (Applied Biosystems) and water added to a final volume of 25 µL. Triplicates of non-template controls were included for each qPCR experiment. Cycle conditions were as follows: after initial holds for 2 min at 50ºC and 10 min at 95ºC, the samples were cycled 40 times at 95ºC for 15 s and 60ºC for 1 min. The baseline and threshold for cycle threshold (Ct) calculations were set automatically using Sequence Detection Software, version 1.4 (Applied Biosystems). The efficiency of amplification (E) of each target molecule was calculated from the slope of the standard curve (plot of Ct vs. the negative log10 concentration of the target) derived from the slopes {E={10(-1/Slope)}-1}. For relative calculations, the 2 -∆∆Ct method was used (Livak & Schmittgen 2001, Applied Biosystems 2012a, b) once all assays met the amplification efficiency criteria of 100% ± 10% (Livak & Schmittgen 2001).
Statistical analyses - Data were analysed and plotted using an unpaired two-tailed t-test (95% confidence interval) with Welch's correction using GraphPad Prism version 4.0a for Macintosh OS X (GraphPad Software) and p values < 0.05 were considered significant.
Ethics - The study was performed using samples from patients enrolled in a dengue cohort study performed in Recife, state of Pernambuco, Brazil (Cordeiro et al. 2007). This study was reviewed, approved by and in accordance with the Ethical Committee of the Brazilian Ministry of Health (CONEP 4909, process 25000.119007/2002-03, CEP 68/02) and with Helsinki Declarations of 1975, as revised in 1983.
RESULTS
In our previous study using the cDNA microarray approach (Nascimento et al. 2009b), we identified nearly 2,000 genes that were differentially expressed between DF, DHF and ND during the acute phase of dengue and most of the genes were involved in the innate immune response. In that study, we suggested that some of these genes contribute to the development of the severe symptoms observed in DHF patients. Based on those results, we selected nine genes (CFD, MT2A, MYD88, PDCD4, MAGED1, PSMB9, FCGR3B, PRDX4 and PYCARD) {named as cited in Gray et al. (2013)} that are directly involved in innate immunity and apoptotic processes to further explore for use as potential candidates as molecular markers of DHF development.
For the qPCR analysis, we extracted total RNA obtained either from PBMCs collected from DF or DHF patients who were in an acute or convalescent phase of the disease, from samples stored in our dengue cohort (Cordeiro et al. 2007) or from healthy volunteer to obtain the cDNA samples used for qPCR. We compared the gene expression levels between DENV-infected vs. non-infected subjects and between DF and DHF patients, during both phases of the disease.
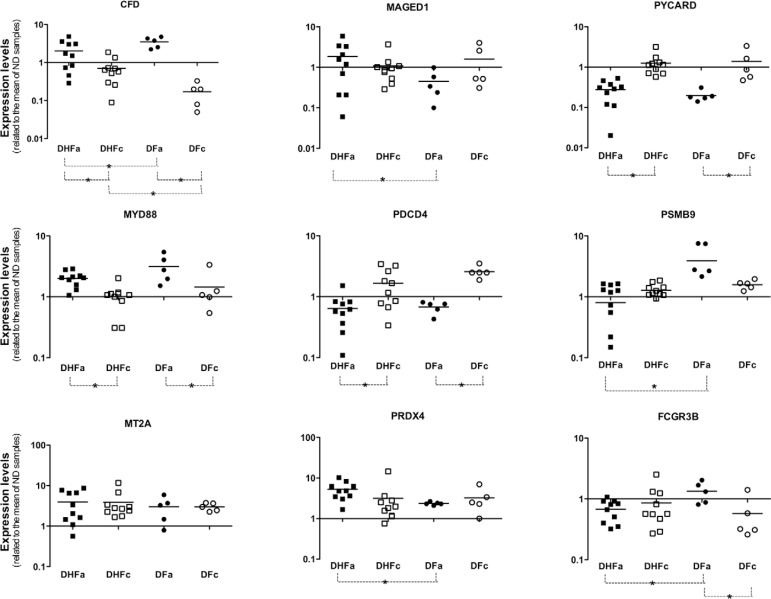
The panel presented in Fig. 1 shows the mRNA levels of all tested genes. We verified that MYD88 mRNA levels were increased in DENV-infected patients during the acute phase of dengue. The opposite was observed for the expression levels of the PYCARD and PDCD4 genes. Thus, MYD88, PYCARD and PDCD4 had characteristics of good acute dengue markers because their mRNA levels were consistently increased or reduced in DENV-infected patients, independently of the severity of symptoms. We also observed that fever, by itself, decreased the expression levels of genes PDCD4, MT2A and PRDX4. There was a considerable difference in the expression of CFD between DHF and DF patients during the convalescent phase of dengue. The expression levels of CFD and PSMB9 were significantly reduced in patients with acute DHF in comparison with patients with acute DF. Furthermore, PSMB9 expression was significantly higher in acute DF patients in comparison with acute DHF patients, while the opposite behaviour was observed for MAGED1 and PRDX4, whose expression levels were increased in DHF patients.
Fig. 1. quantification of mRNA coding for innate immunity or apoptosis genes. Peripheral blood mononuclear cells samples of 10 dengue haemorrhagic fever (DHF) and five dengue fever (DF) patients in acute (DHFa or DFa) or convalescent (DHFc or DFc) phase of disease were used to extract total RNA and produce the cDNA used on quantitative real-time polymerase chain reaction assays. Beta-actin was used as normalisation gene. A mix of cDNA obtained from five non-dengue (ND) volunteers was used as calibrator sample. Asterisk means p < 0.05.
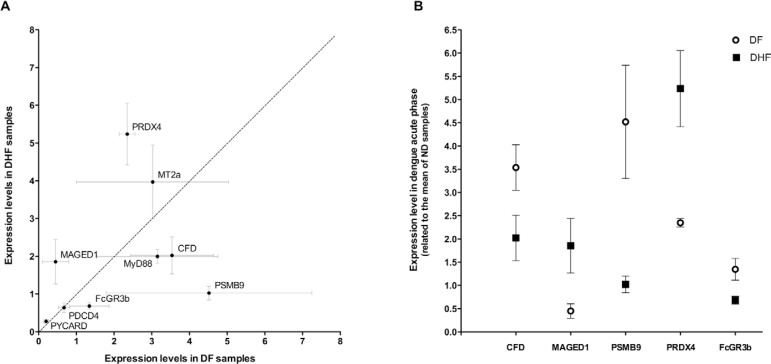
CFD, FCGR3B, PSMB9, MAGED1 and PRDX4 showed significant differences in expression when the acute phases of DHF and DF are compared (Fig. 2A). MAGED1 and PRDX4 expression levels were greater in DHF patients than in DF patients during the acute phase of dengue, in contrast with the expression levels observed for CFD, FCGR3B and PSMB9 (Fig. 2B).
Fig. 2. potential biomarkers for prognosis of dengue haemorrhagic fever (DHF). A: genes that showed significant differences in its mRNA expression levels between DHF and dengue fever (DF) patients during the acute phase of dengue were plotted according its DF x DHF expression levels. Each circle represents mean values of 15 patients (10 DHF and 5 DF); B: comparison of the expression levels of the five potential biomarkers between acute DF and DHF samples. Each point represents mean values from 10 DHF or five DF patients. A mix of cDNA obtained from five non-dengue (ND) volunteers was used as calibrator sample.
DISCUSSION
The primary controversial aspect concerning dengue prognosis resides in the fact that a substantial number of deaths caused by this disease could be avoided through simple and inexpensive fluid reposition if severe dengue could be detected earlier (Mairuhu et al. 2004). However, it is difficult to identify which DENV-infected patients will require observation for intravenous fluid treatment at early stages of infection. It is common for patients to come to the hospital, but subsequently be dismissed because they appear to be in good clinical condition. Nevertheless, within two or three days, they rapidly present dengue shock syndrome (DSS), often too late for treatment. Therefore, it is critical to complement the available clinical diagnosis techniques for dengue with more sensitive laboratory approaches. Several accurate diagnostic methods can identify a positive infection by DENV, determine with which serotype of the virus the patient is infected, measure the viral load and distinguish between primary and sequential dengue infection. However, none of the available tests can accurately indicate the propensity for dengue-infected patients to develop the haemorrhagic symptoms of the disease.
As described by Mairuhu et al. (2004), the simultaneous occurrence of several risk factors related to individual, epidemiological and viral aspects could determine whether a person in a given population may develop DHF. However, most researchers who are trying to identify parameters to predict the evolution of DF to its severe form agree that the DHF diagnosis is far from simple for several reasons. The clinical manifestation of dengue is dependent on (i) individual risk factors such as gender, body size, age and blood type, nutritional and immunological state, (ii) primary or secondary/sequential infections, viral burden, serotype and charge of infection and (iii) genetic predisposition, which has also been correlated to the development of severe disease (Screaton & Mongkolsapaya 2006).
Recently, Hoang et al. (2010) conducted a nested, case-control comparison of the early host transcriptional features in DSS patients within sex, age, viral serotype-matched and uncomplicated dengue patients. Their data suggest an association between neutrophil activation, pathogenesis and the development of severe symptoms of dengue and point to future strategies for guiding prognosis. Coffey et al. (2009) tested the extremely valuable identification of markers and candidate genes of dengue severity. Previously, we suggested that the measurement of several genes by qPCR in PBMC samples collected from dengue-infected patients during the acute phase of the disease could be useful for prognosis of DHF development (Calzavara-Silva et al. 2009). Here, we reinforce the viability of using a panel of carefully selected genes to assess the likelihood of developing DHF.
Infection by DENV induces endothelial cell production of reactive nitrogen/oxygen species and apoptotic cell death (Yen et al. 2008, Levy et al. 2010). Haemorrhage and severe haemorrhage are greatly reduced in mice lacking iNOS or p47 (phox) and in mice treated with an oxidase inhibitor, which highlights the critical roles of reactive nitrogen and oxygen species in dengue haemorrhage (Yen et al. 2008). PBMCs from DHF patients underwent apoptosis at a significantly higher rate than PBMCs from patients suffering from DF alone, suggesting that the severity of natural DENV infection correlates with PBMC apoptosis (Jaiyen et al. 2009). The low expression of the PYCARD gene, which encodes a protein that mediates assembly of large signalling complexes in the inflammatory and apoptotic-signalling pathways via the activation of caspase, in dengue patients suggests an attempt by the organism to balance inflammatory and apoptotic processes during DENV infection. Interestingly, over-expression of the pro-apoptotic PDRX4 and MAGED1 genes was observed in patients who developed the severe symptoms of dengue, the acute DHF patients. MAGED1 has been associated with the p75 neurotrophin receptor-mediated programmed cell death pathway (Põld et al. 1999). Indeed, the increased expression of pro-apoptotic genes, such as MAGED1, observed in DHF patients, could be because DENV infection augments apoptosis in patients with the severe symptoms of the disease.
PRDX4 is a member of the peroxiredoxins family that may exert a protective function against oxidative damage by scavenging reactive oxygen species in the extracellular space (Okado-Matsumoto et al. 2000). In addition, PRDX4 also plays a regulatory role in the activation of the transcription nuclear factor (NF)-kappa B (Jin et al. 1997), which is activated concomitantly with viral protein synthesis before the appearance of apoptotic cells. Apoptosis is inhibited when DENV-infected cells are treated with NF-kappa B decoys, indicating that this transcription factor is involved in the induction of cell death (Marianneau et al. 1997). The apoptosis process itself generates reactive oxygen species and higher PDRX4 expression could be interpreted as an attempt to protect the endothelium against tissue damage induced by free radicals.
Regulation of the complement pathways is an important biological process involved in dengue infection (Avirutnan et al. 2008, Basu & Chaturvedi 2008). Previous findings from our group (Nascimento et al. 2009a) highlighted the relationship between the balance of complement pathway activation and the pathogenesis of or protection against dengue infection. The authors reported that, during acute dengue infection, the levels of CFD protein in DF patients are lower than in either DHF patients or healthy controls. In contrast, during the convalescent phase, levels of CFD protein in DF patients were as high as those found in DHF patients, suggesting that the patients who are more capable of modulating their levels of factor D during acute dengue infection will exhibit the mild disease phenotype. These data are in contrast with the results observed in this work because we detected lower levels of mRNA coding for CFD in acute phase DHF patients and a lower variation of CFD mRNA levels in patients who developed DHF between the acute and convalescent phases as compared with the variation observed in DF patients. However, our group (Nascimento et al. 2009a) measured total levels of protein CFD in the serum of dengue patients and showed that fat cells are the main CFD producers, whereas antigen-presenting cells have several reduced functions when infected. In fact, using a cDNA microarray approach, we also reported that DHF patients had lower levels of mRNA coding for CFD compared with DF patients (Nascimento et al. 2009b), corroborating our results presented here. Furthermore, the low levels of CFD during the convalescent phase of dengue in DF patients reinforce the relationship between CFD modulation and severity of dengue.
FCGR3B, or CD16b, is a glycosyl-phosphatidylinositol-linked receptor found on human neutrophils. Its soluble form, sCD16b, plays a regulatory role in inflammatory processes and provides a molecular basis for the interaction between Fc-gamma RIII-B and the CR3 human complement receptor on the cell membrane (Galon et al. 1996), favouring adhesion to and phagocytosis of viral-infected cells. Kindzelskii et al. (2000) verified that Ebola virus' secretory glycoprotein binds to CD16b and inhibits L-selectin shedding, diminishing the physical linkage between CR3-CD16b on neutrophils. In addition, Mandelboim et al. (1999) showed that CD16b is directly involved in the lysis of some viral-infected and tumour cells, independent of antibody binding. Moreover, soluble CD16 also induces the maturation of dendritic cells (DCs) and the production of several cytokines, including interleukin (IL)-12, which consequently initiates T-helper 1 type immune response (de la Salle et al. 1997). Thus, the diminished expression of FCGR3B in acute DHF patients could be silencing the first cellular immune responses against DENV infection.
We observed that, during the acute phase of the disease, DF patients showed augmented expression levels of PSMB9, whereas DHF patients did not have any alterations in their PSMB9 expression levels. The PSMB9 gene encodes a member of the proteasome B-type family. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an adenosine triphosphate/ubiquitin-dependent process through a non-lysosomal pathway. An essential function of the immunoproteasome, which is a modified proteasome, is the processing of class I major histocompatibility complex peptides. Drugs that block proteasome activity are potent inhibitors of West Nile virus (WNV) genome amplification and activity even if cells are treated 12 h after infection, indicating that the proteasome is required at a post-entry stage(s) of the WNV infection cycle (Gilfoy et al. 2009). This requirement is in contrast with our observations that DHF patients exhibit higher levels of viral-induced changes; thus, if the proteasome is important to intracellular flavivirus replication, PSMB9 should be augmented in DHF individuals.
We also verified that MYD88 expression levels were upregulated in DENV infected patients. This gene codes for a pan-adapter protein in the signal transduction pathway that is mediated by IL-1 and Toll-like receptors (TLR). MYD88 activation induces type I interferon (IFN) responses, especially in plasmacytoid DCs, as an attempt of the organism to control the DENV infection. Japanese encephalitis virus can induce a functional impairment of DCs through MYD88-dependent pathways, which subsequently leads to poor CD4 (+) and CD8 (+) T cell responses and results in increased viral survival and dissemination in the body (Aleyas et al. 2009). Conceição et al. (2010) measured gene expression changes in HepG2 cells after 6 h, 24 h and 48 h of infection with DENV. They showed that MYD88 was upregulated at 6 h of infection, corroborating our data. However, MYD88 levels diminished after 24 h of infection. Moreover, using a microarray assay, Nascimento et al. (2009b) also verified that MYD88 expression levels were decreased in acute DHF patients in comparison with DF patients. In our results, we did not observe a significant difference of MYD88 expression levels between DHF and DF patients even though the MYD88 expression level tended to be increased in DF patients, most likely due to the low number of samples used in our study.
We suggest here that the quantification of mRNA coding for CFD, MAGED1, FCGR3B, PSMB9 and PRDX4 could predict the propensity of a DENV-infected patient to develop the severe symptoms of the disease. A limitation of this study is the low number of tested samples, which may affect the exclusion and more importantly, the inclusion of new genes in the panel of DHF biomarkers, as noted in the case of MYD88. In a previous study (Gomes et al. 2010), we presented a new use of the Support Vector Machine algorithm to classify DF and DHF patients using the expression data of seven putative critical genes related to dengue symptoms: TLR3, MDA5, IRF3, IFN-alpha, CLEC5A and, most importantly, MYD88 and TLR7. Our in silico results reinforce the search for new biomarker to dengue prognosis. We are currently working on selecting new candidate genes for inclusion in our present panel and obtaining a larger panel of patients, especially those presenting acute DENV infections.
In this work, we verified that the mRNA levels for CFD, MAGED1, PSMB9, PRDX4 and FCGR3B are differentially expressed between patients who developed the mild type of dengue and patients who showed severe clinical symptoms. We now suggest the use of a panel of these genes as a useful tool to improve clinical prognosis for dengue.
Acknowledgments
To the NPT/CPqAM, for the support, and to Marli T Cordeiro, Ana Maria Silva and Carlos A Brito, for cohort study and their significant contributions.
Funding Statement
Financial support: FAPEMIG (CBB-APQ-00741-11), CNPq (550140/2010-7, 477569/2011-0)
REFERENCES
- Acioli-Santos B, Segat L, Dhalia R, Brito CA, Braga-Neto UM, Marques ET, Crovella S. MBL2 gene polymorphisms protect against development of thrombocytopenia associated with severe dengue phenotype. Hum Immunol. 2008;69:122–128. doi: 10.1016/j.humimm.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Aleyas AG, George JA, Han YW, Rahman MM, Kim SJ, Han SB, Kim BS, Kim K, Eo SK. Functional modulation of dendritic cells and macrophages by Japanese encephalitis virus through MyD88 adaptor molecule-dependent and independent pathways. J Immunol. 2009;183:2462–2474. doi: 10.4049/jimmunol.0801952. [DOI] [PubMed] [Google Scholar]
- Applied Biosystems Amplification efficiency of TaqManR Gene Expression Assays. Assays tested extensively for qPCR efficiency. 2012 3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_040377.pdf
- Applied Biosystems User Bulletin. Applied Biosystems 7500/7500 Fast Real-Time PCR Systems. 2012 3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_050637.pdf
- Avirutnan P, Mehlhop E, Diamond MS. Complement and its role in protection and pathogenesis of flavivirus infections. Vaccine. 2008;26(Suppl. 8):100–107. doi: 10.1016/j.vaccine.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Chaturvedi UC. Vascular endothelium: the battlefield of dengue viruses. FEMS Immunol Med Microbiol. 2008;53:287–299. doi: 10.1111/j.1574-695X.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MG, Huang YY, Marshall JS, King CA, Hoskin DW, Anderson R. Dramatic caspase-dependent apoptosis in antibody-enhanced dengue virus infection of human mast cells. J Leukoc Biol. 2009;85:71–80. doi: 10.1189/jlb.0308167. [DOI] [PubMed] [Google Scholar]
- Calzavara-Silva CE, Gomes AL, Maia RC, Acioli-Santos B, Gil LH, Marques ET., Jr Early molecular markers predictive of dengue hemorrhagic fever. An Acad Bras Cienc. 2009;81:671–677. doi: 10.1590/s0001-37652009000400006. [DOI] [PubMed] [Google Scholar]
- Chang K, Lu PL, Ko WC, Tsai JJ, Tsai WH, Chen CD, Chen YH, Chen TC, Hsieh HC, Pan CY, Harn MR. Dengue fever scoring system: new strategy for the early detection of acute dengue virus infection in Taiwan. J Formos Med Assoc. 2009;108:879–885. doi: 10.1016/S0929-6646(09)60420-4. [DOI] [PubMed] [Google Scholar]
- Chaturvedi U, Nagar R, Shrivastava R. Dengue and dengue haemorrhagic fever: implications of host genetics. FEMS Immunol Med Microbiol. 2006;47:155–166. doi: 10.1111/j.1574-695X.2006.00058.x. [DOI] [PubMed] [Google Scholar]
- Cheng HJ, Lin CF, Lei HY, Liu HS, Yeh TM, Luo YH, Lin YS. Proteomic analysis of endothelial cell auto antigens recognized by anti-dengue virus nonstructural protein 1 antibodies. Exp Biol Med (Maywood) 2009;234:63–73. doi: 10.3181/0805-RM-147. [DOI] [PubMed] [Google Scholar]
- Coffey LL, Mertens E, Brehin AC, Fernandez-Garcia MD, Amara A, Després P, Sakuntabhai A. Human genetic determinants of dengue virus susceptibility. Microbes Infect. 2009;11:143–156. doi: 10.1016/j.micinf.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Conceição TM, El-Bacha T, Villas-Bôas CS, Coello G, Ramírez J, Montero-Lomeli M, da Poian AT. Gene expression analysis during dengue virus infection in HepG2 cells reveals virus control of innate immune response. J Infect. 2010;60:65–75. doi: 10.1016/j.jinf.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Cordeiro MT, Braga-Neto U, Nogueira RM, Marques ET., Jr Reliable classifier to differentiate primary and secondary acute dengue infection based on IgG ELISA. PLoS ONE. 2009;4: doi: 10.1371/journal.pone.0004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro MT, Silva AM, Brito CA, Nascimento EJ, Magalhães MC, Guimarães GF, Lucena-Silva N, Carvalho EM de, Marques ET., Jr Characterization of a dengue patient cohort in Recife, Brazil. Am J Trop Med Hyg. 2007;77:1128–1134. [PubMed] [Google Scholar]
- Salle H de la, Galon J, Bausinger H, Spehner D, Bohbot A, Cohen J, Cazenave JP, Fridman WH, Sautès C, Hanau D. Soluble CD16/Fc gamma RIII induces maturation of dendritic cells and production of several cytokines including IL-12. Adv Exp Med Biol. 1997;417:345–352. doi: 10.1007/978-1-4757-9966-8_56. [DOI] [PubMed] [Google Scholar]
- Kruif MD de, Setiati TE, Mairuhu AT, Koraka P, Aberson HA, Spek CA, Osterhaus AD, Reitsma PH, Brandjes DP, Soemantri A, Gorp EC van. Differential gene expression changes in children with severe dengue virus infections. PLoS Negl Trop Dis. 2008;2: doi: 10.1371/journal.pntd.0000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376:429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina LM, Valero NJ, Hernández JM, Mosquera JA. Increased apoptosis and expression of tumor necrosis factor-alpha caused by infection of cultured human monocytes with dengue virus. Am J Trop Med Hyg. 2003;68:48–53. [PubMed] [Google Scholar]
- Fink J, Gu F, Ling L, Tolfvenstam T, Olfat F, Chin KC, Aw P, George J, Kuznetsov VA, Schreiber M, Vasudevan SG, Hibberd ML. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl Trop Dis. 2007;1: doi: 10.1371/journal.pntd.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Gauchat JF, Mazières N, Spagnoli R, Storkus W, Lötze M, Bonnefoy JY, Fridman WH, Sautès C. Soluble Fc-gamma receptor type III (Fc-gammaRIII, CD16) triggers cell activation through interaction with complement receptors. J Immunol. 1996;157:1184–1192. [PubMed] [Google Scholar]
- Gibbons RV, Vaughn DW. Dengue: an escalating problem. BMJ. 2009;324:1563–1566. doi: 10.1136/bmj.324.7353.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil L, López C, Blanco A, Lazo L, Martín J, Valdés I, Romero Y, Figueroa Y, Guillén G, Hermida L. The cellular immune response plays an important role in protecting against dengue virus in the mouse encephalitis model. Viral Immunol. 2009;22:23–30. doi: 10.1089/vim.2008.0063. [DOI] [PubMed] [Google Scholar]
- Gilfoy F, Fayzulin R, Mason PW. West Nile virus genome amplification requires the functional activities of the proteasome. Virology. 2009;385:74–84. doi: 10.1016/j.virol.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AL, Wee LJ, Khan AM, Gil LH, Marques ET, Jr, Calzavara-Silva CE, Tan TW. Classification of dengue fever patients based on gene expression data using support vector machines. PLoS ONE. 2010;5: doi: 10.1371/journal.pone.0011267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–D552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LM, Caruso MB, Canellas F, Soares MR, Oliveira-Carvalho AL, Chapeaurouge DA, Almeida PM, Perales J, Zingali RB, da Poian AT. Secretome of HepG2 cells infected with dengue virus: implications for pathogenesis. Biochim Biophys Acta. 2008;1784:1607–1616. doi: 10.1016/j.bbapap.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Hoang LT, Lynn DJ, Henn M, Birren BW, Lennon NJ, Le PT, Duong KT, Nguyen TT, Mai LN, Farrar JJ, Hibberd ML, Simmons CP. The early whole-blood transcriptional signature of dengue virus and features associated with progression to dengue shock syndrome in Vietnamese children and young adults. J Virol. 2010;84:12982–12994. doi: 10.1128/JVI.01224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Zepeda A, Cabello-Gutiérrez C, Cime-Castillo J, Monroy-Martínez V, Manjarrez-Zavala ME, Gutiérrez-Rodríguez M, Izaguirre R, Ruiz-Ordaz BH. Crosstalk between coagulation and inflammation during dengue virus infection. Thromb Haemost. 2008;99:936–943. doi: 10.1160/TH07-08-0438. [DOI] [PubMed] [Google Scholar]
- Jaiyen Y, Masrinoul P, Kalayanarooj S, Pulmanausahakul R, Ubol S. Characteristics of dengue virus-infected peripheral blood mononuclear cell death that correlates with the severity of illness. Microbiol Immunol. 2009;53:442–450. doi: 10.1111/j.1348-0421.2009.00148.x. [DOI] [PubMed] [Google Scholar]
- Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappa B activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- Kindzelskii AL, Yang Z, Nabel GJ, Todd RF 3rd, Petty HR. Ebola virus secretory glycoprotein (sGP) diminishes Fc gamma RIIIB-to-CR3 proximity on neutrophils. J Immunol. 2000;164:953–958. doi: 10.4049/jimmunol.164.2.953. [DOI] [PubMed] [Google Scholar]
- Kwan WH, Helt AM, Marañón C, Barbaroux JB, Hosmalin A, Harris E, Fridman WH, Mueller CG. Dendritic cell precursors are permissive to dengue virus and human immunodeficiency virus infection. J Virol. 2005;79:7291–7299. doi: 10.1128/JVI.79.12.7291-7299.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Valero N, Espina LM, Añez G, Arias J, Mosquera J. Increment of interleukin 6, tumor necrosis factor alpha, nitric oxide, C-reactive protein and apoptosis in dengue. Trans R Soc Trop Med Hyg. 2010;104:16–23. doi: 10.1016/j.trstmh.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loke H, Bethell D, Phuong CX, Day N, White N, Farrar J, Hill A. Susceptibility to dengue hemorrhagic fever in Vietnam: evidence of an association with variation in the vitamin D receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67:102–106. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- Long HT, Hibberd ML, Hien TT, Dung NM, van Ngoc T, Farrar J, Wills B, Simmons CP. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J Infect Dis. 2009;199:537–546. doi: 10.1086/596507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairuhu AT, Wagenaar J, Brandjes DP, Gorp EC van. Dengue: an arthropod-borne disease of global importance. Eur J Clin Microbiol Infect Dis. 2004;23:425–433. doi: 10.1007/s10096-004-1145-1. [DOI] [PubMed] [Google Scholar]
- Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL. Human CD16 as a lyses receptor mediating direct natural killer cell cytotoxicity. Proc Natl Acad Sci USA. 1999;96:5640–5644. doi: 10.1073/pnas.96.10.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianneau P, Cardona A, Edelman L, Deubel V, Desprès P. Dengue virus replication in human hepatoma cells activates NF-kappa B which in turn induces apoptotic cell death. J Virol. 1997;71:3244–3249. doi: 10.1128/jvi.71.4.3244-3249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev. 2008;225:300–313. doi: 10.1111/j.1600-065X.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- Nascimento EJ, Braga-Neto U, Calzavara-Silva CE, Gomes AL, Abath FG, Brito CA, Cordeiro MT, Silva AM, Magalhães C, Andrade R, Gil LH, Marques ET., Jr Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS ONE. 2009;4: doi: 10.1371/journal.pone.0007892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento EJ, Silva AM, Cordeiro MT, Brito CA, Gil LH, Braga-Neto U, Marques ET. Alternative complement pathway deregulation is correlated with dengue severity. PLoS ONE. 2009;4: doi: 10.1371/journal.pone.0006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirudeen AM, Liu DX. Gene expression profiling by microarray analysis reveals an important role for caspase-1 in dengue virus-induced p53-mediated apoptosis. J Med Virol. 2009;81:1069–1081. doi: 10.1002/jmv.21486. [DOI] [PubMed] [Google Scholar]
- Noisakran S, Perng GC. Alternate hypothesis on the pathogenesis of dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS) in dengue virus infection. Exp Biol Med (Maywood) 2008;233:401–408. doi: 10.3181/0707-MR-198. [DOI] [PubMed] [Google Scholar]
- Okado-Matsumoto A, Matsumoto A, Fujii J, Taniguchi N. Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J Biochem. 2000;127:493–501. doi: 10.1093/oxfordjournals.jbchem.a022632. [DOI] [PubMed] [Google Scholar]
- Põld M, Zhou J, Chen GL, Hall JM, Vescio RA, Berenson JR. Identification of a new unorthodox member of the MAGE gene family. Genomics. 1999;59:161–167. doi: 10.1006/geno.1999.5870. [DOI] [PubMed] [Google Scholar]
- Sakuntabhai A, Turbpaiboon C, Casadémont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM, Tangnararatchakit K, Tangthawornchaikul N, Vasanawathana S, Chaiyaratana W, Yenchitsomanus PT, Suriyaphol P, Avirutnan P, Chokephaibulkit K, Matsuda F, Yoksan S, Jacob Y, Lathrop GM, Malasit P, Desprès P, Julier C. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton G, Mongkolsapaya J. T cell responses and dengue hemorrhagic fever. Novartis Found Symp. 2006;277:164–171. [PubMed] [Google Scholar]
- Senanayake S. Dengue fever and dengue hemorrhagic fever-a diagnostic challenge. Aust Fam Physician. 2006;35:609–612. [PubMed] [Google Scholar]
- Tsai TF, Halstead SB. Tropical viral infections. Curr Opin Infect Dis. 2002;11:547–553. doi: 10.1097/00001432-199810000-00006. [DOI] [PubMed] [Google Scholar]
- Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, Kasisith J. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever, but not dengue hemorrhagic fever. J Infect Dis. 2008;197:1459–1467. doi: 10.1086/587699. [DOI] [PubMed] [Google Scholar]
- WHO - World Health Organization . Dengue/dengue haemorrhagic fever. Global Alert and Response (GAR); 2012. who.int/csr/disease/dengue/en/index.html [Google Scholar]
- Yen YT, Chen HC, Lin YD, Shieh CC, Wu-Hsieh BA. Enhancement by tumor necrosis factor alpha of dengue virus-induced endothelial cell production of reactive nitrogen and oxygen species is key to hemorrhage development. J Virol. 2008;82:12312–12324. doi: 10.1128/JVI.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]