Abstract
Background:
Synaptic connections are disrupted in patients with Huntington’s disease (HD). Synaptosomes from postmortem brain are ideal for synaptic function studies because they are enriched in pre- and post-synaptic proteins important in vesicle fusion, vesicle release, and neurotransmitter receptor activation.
Objective:
To examine striatal synaptosomes from 3, 6 and 12 month old WT and Hdh140Q/140Q knock-in mice for levels of synaptic proteins, methionine oxidation, and glutamate release.
Methods:
We used Western blot analysis, glutamate release assays, and liquid chromatography tandem mass spectrometry (LC-MS/MS).
Results:
Striatal synaptosomes of 6 month old Hdh140Q/140Q mice had less DARPP32, syntaxin 1 and calmodulin compared to WT. Striatal synaptosomes of 12 month old Hdh140Q/140Q mice had lower levels of DARPP32, alpha actinin, HAP40, Na+/K+-ATPase, PSD95, SNAP-25, TrkA and VAMP1, VGlut1 and VGlut2, increased levels of VAMP2, and modifications in actin and calmodulin compared to WT. More glutamate released from vesicles of depolarized striatal synaptosomes of 6 month old Hdh140Q/140Q than from age matched WT mice but there was no difference in glutamate release in synaptosomes of 3 and 12 month old WT and Hdh140Q/140Q mice. LC-MS/MS of 6 month old Hdh140Q/140Q mice striatal synaptosomes revealed that about 4% of total proteins detected (>600 detected) had novel sites of methionine oxidation including proteins involved with vesicle fusion, trafficking, and neurotransmitter function (synaptophysin, synapsin 2, syntaxin 1, calmodulin, cytoplasmic actin 2, neurofilament, and tubulin). Altered protein levels and novel methionine oxidations were also seen in cortical synaptosomes of 12 month old Hdh140Q/140Q mice.
Conclusions:
Findings provide support for early synaptic dysfunction in Hdh140Q/140Q knock-in mice arising from altered protein levels, oxidative damage, and impaired glutamate neurotransmission and suggest that study of synaptosomes could be of value for evaluating HD therapies.
Keywords: Mutant huntingtin, HD, synaptosomes, oxidative damage, methionine oxidation, glutamate release, synaptic proteins, mass spectrometry
INTRODUCTION
Early cognitive decline occurs in patients with Huntington’s disease (HD) and is attributed in part to the loss of neuronal connections, especially those of the corticostriatal pathway. Huntingtin (htt), the protein affected in HD, may have multiple roles at the synapse through interactions with proteins and lipids [1-4]. Htt moves in axons on vesicular membranes along with other vesicle-associated proteins including dynein, HAP1 and the vesicle docking protein SNAP-25 [5]. Htt co-exists in subcellular fractions with transferrin receptor, synaptophysin, vesicle membrane-associated protein (SV2) and clathrin and has been localized at the electron microscopic level to pre- and post-synaptic regions of synapses [1]. Mutant htt (mhtt) interferes with the phosphorylation of synapsin 1, which is important for neurotransmitter release [6]. Mutant htt also abnormally associates with phospholipids that function in signaling at membranes including lipid rafts and can alter the membrane distribution of NMDA and AMPA receptor subunits [7-9]. Htt also interacts with syntaxin 1 which mediates vesicle fusion at membranes, and levels of syntaxin 1 modulate mhtt toxicity [10]. Full length mhtt expressed in Drosophila increased excitatory potentials arising from an increase in Ca2+ dependent neurotransmitter release [11]. Transgenic mice that selectively express mutant htt fragment of exon 1 in presynaptic terminals have reduced survival and altered neurostransmitter function [6]. These studies suggest that mhtt may be in position in pre and post synaptic regions to affect neuronal connections, synaptic strength and stability.
The Hdh140Q/140Q mouse model has been used to study changes in the HD brain. In this model, human HD exon 1 inserted into full-length htt is expressed under the control of the endogenous mouse htt promoter. Mhtt localizes in the nucleus diffusely and in aggregates by 4 months of age. Climbing a pole is impaired at 1.5 months and the deficit worsens at 6.5 months. A transient rearing phenotype appears at two months [12, 13]. The levels of striatal immunoreactive DARPP32, which is a selective marker of striatal medium spiny neurons, were normal in 4 month old Hdh140Q/140Q mice but significantly depleted by 41% compared to WT at 12 months [13]. Lipid rafts are cholesterol and sphingolipid enriched domains in membranes that partition proteins and lipids involved in mediating signaling by growth factors, neurotransmitters and other stimuli [14-18]. We found full length mhtt and GSK3-beta accumulated in lipid raft fractions from the cortex of one month old Hdh140Q/140Q mice [9]. We also observed abnormal distribution and activity of Rab11 on striatal endosomal membranes of 2 month old Hdh140Q/140Q mice and impaired recycling of Rab11 dependent cargoes back to the plasma membrane in HD primary cortical neurons derived from embryonic brain [19]. These findings suggest that there are changes in the brain of young Hdh140Q/140Q mice that may affect synaptic function before frank motor disease.
Synaptosomal fractions prepared from brain are enriched in pre-synaptic and postsynaptic membranes [20, 21]. Striatal synaptosomes have been used in biochemical analysis and functional assays to study connectivity related to acetylcholine, dopamine, glutamate and GABA neurotransmission [22]. For example, depolarization of striatal synaptosomes with potassium chloride releases glutamate which induces Ca2+ entry through different types of calcium channels, including N, P and Q [23]. Recently we found that synaptosomal fractions of the striatum and cortex of Hdh140Q/140Q mice have increased levels of NADPH oxidase (NOX) activity, which produces the free radical anion superoxide and additional reactive oxygen species (ROS) as secondary products. We proposed that increased NOX2 activity is a source of oxidative stress in HD at the level of synapses since inhibitors of NOX2 enzymatic activity or deleting NOX2 in Hdh140Q/140Q neurons reduces ROS levels in Hdh140Q/140Q primary neurons [21]. Our analysis of NOX activity in synaptosomes suggested that synaptosomal fractions could be useful to identify other potential sources of early synaptic dysfunction in HD.
In this study we show that striatal synaptosomes of Hdh140Q/140Q mice differ from WT in levels of synaptic proteins involved with vesicle fusion and release, protein oxidation, and release of glutamate. We suggest that striatal synaptosomes may be a useful and efficient way to evaluate disease progression related to synaptic function in the Hdh140Q/140Q model and to monitor the effectiveness of treatments for HD.
METHODS
Animals
All mice used in this study were obtained from a colony maintained at the MGH animal facility. The MGH Subcommittee on Research Animal Care (SRAC)-OLAW Assurance # A3596-01 has reviewed and approved our animal protocol which conforms to the USD Animal Welfare Act, PHS Policy on Humane Care and Use of Laboratory Animals, the ‘ILAR Guide for the Care and Use of Laboratory Animals’ and other relevant laws and regulations [9, 21, 24]. The WT and Hdh140Q/140Q mice had a background strain of C57BL/6. The Hdh140Q/140Q knock-in mice were first described by Dr. Scott Zeitlin and colleagues and has exon 1 of the human htt gene with 140 CAG (glutamine) repeats in place of exon 1 of the htt mouse gene [12]. The number of CAGs in exon 1 of the Hdh140Q/140Q mice in this study is about 128. The genotypes of the mice were verified by PCR using DNA from the tail of every mouse. All mice used in this study were males.
Preparation of synaptosomes
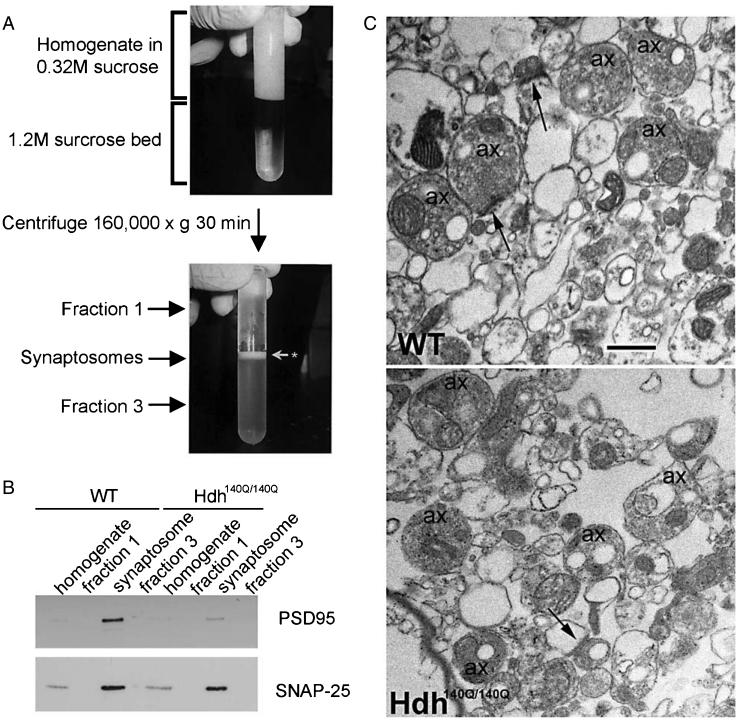
Synaptosomes from fresh WT and Hdh140Q/140Q mouse striata were isolated from brain tissue by a sucrose gradient centrifugation using a method described before [20, 21]. WT and Hdh140Q/140Q mice of 3, 6 and 12 months of age were overdosed with Avertin (250 mg/kg of weight). The brain was removed and kept in ice-cold PBS. The cortex and striatum were dissected, homogenized on ice in 7 ml of 0.32 M sucrose plus protease inhibitors. Ten mM dithiothreitol (DTT) was added to prevent ex vivo oxidation. Samples were processed with a Dounce homogenizer (tight piston B, 8 strokes per sample). Homogenates were centrifuged at 4°C for 10 min at 1,000× g. Then, 6.5 ml of the supernatant was layered onto×6.5 ml of 1.2 M sucrose in 14 × 89 mm Ultra-Clear centrifuge tubes (Beckman, Palo Alto CA). Samples were centrifuged at 4°C for 35 min at 160,000× g using a SW41 rotor in a Beckman L8-80 M Ultracentrifuge with low acceleration and no brakes. A distinguishable cloudy band at the interface of 0.32 M and 1.2 M sucrose was recovered (see Fig. 1A). A total volume of 500 μl aliquots from the supernatant (0.32 M sucrose layer, fraction 1) and the lower face (1.2 M sucrose layer, fraction 3) were collected and used to confirm by biochemical assay the purity of the fractionation based on the enrichment of PSD95 and SNAP25. Protein levels were measured using the Bradford protocol. Most samples were then frozen at −80°C in aliquots containing small volumes for future use. Some samples were processed for electron microscopy to evaluate the content of the preparation.
Fig. 1.
Preparation of synaptosomal fractions. A. Centrifugation tube shows homogenate before (top image) and after (lower image) fractionation by sucrose gradient as explained in Methods. Synaptosome band is indicated at white arrow in lower image between fractions 1 and 3. B. Western blot analysis shows enrichment of the presynaptic marker SNAP25 and the postsynaptic marker PSD95 in the synaptosomes compared to fractions 1 and 3. The blot corresponds to striatal synaptosomes prepared from 6 month old WT and Hdh140Q/140Q mice. C. Electron micrographs of WT and Hdh140Q/140Q striatal synaptosomes show the presence of membrane profiles that are empty and vesicle filled. Some axon terminals (ax) form synaptic contacts (arrows). There are few mitochondria and myelin figures present in this preparation. Scale bar = 500 nm.
SDS-PAGE, western blot and densitometry
Synaptosomal proteins (5–20 μg/lane) were separated in 3–8% Tris-acetate or 4–12% Bis-Tris gels (Life Technologies). Proteins were transferred to nitrocellulose using the iBlot system (Life Technologies). Nitrocellulose blots were blocked in 5% milk in TBS + 0.1% Tween-20 (TBST) and incubated overnight in primary antibody diluted in blocking solution (see next section for sources and dilutions of antisera). After blots were washed in TBST and incubated in peroxidase labeled secondary antibody for 1 hour in blocking solution, bands were detected using the West-pico SuperSignal substrate (Pierce) and Hyperfilm ECL (GE Healthcare). Blots were re-probed with anti-actin, anti-spectrin or anti-GAPDH. Actin signal was used as loading control for the study of synaptosomes from 3 and 6 month old mice and spectrin or GAPDH signals were used for the 12 month old groups since actin signal at 12 months had an additional band of higher molecular mass that was not seen in WT mice or in 3 and 6 month old Hdh140Q/140Q synaptosomes. Signal intensity was measured using ImageJ software (NIH). Signal intensity was normalized to the signal for spectrin, actin or GAPDH.
Sources and dilutions of antisera used for western blots
Actin (Sigma, 1:400), DARPP32 (Chemicon, 1:500), Huntingtin 3B5H10 (Sigma, 1:2000), Huntingtin S830 (gift from Dr. Gillian Bates, 1:750), Huntingtin Ab1 ([1], 1:3000), calmodulin (Abcam, 1:1000), syntaxin-1 (Millipore, 1:1000), GAPDH (Millipore, 1:6000), spectrin (Chemicon, 1:15000), TrkA (Abcam, 1:1000), alpha-actinin (Abcam, 2μg/ml), Na+/K+-ATPase (Affinity Bioreagents, 1:10000), PSD95 (Cell Signalling, 1:500), synaptophysin (Boehringer Mannheim Biochemicals, 1:5000), HAP40 (Chemicon, 1:500), SNAP-25 (BD Transduction Laboratories, 1:5000), VAMP1 (Abcam, 1:3000), VGlut1 (Synaptic Systems, 1:10000), VGlut2 (Synaptic Systems, 1:5000), VAMP2 (Abcam, 1:1000). Peroxidase-labeled secondary antibodies were purchased from Jackson Immunoresearch and were used at a 1:5000 dilution.
Electron microscopy analysis of synaptosomal fraction
A 200μl aliquot of the striatal synaptosome fraction was pelleted at 16,000× g for 5 minutes then re-suspended in 100 μl 2.5%×glutaraldehyde/PBS and fixed for 30 minutes. Synaptosomes were washed in PBS, centrifuged again at 16,000× g for 5 minutes then processed as a pellet. Synaptosome × pellets were incubated in 1% osmium tetroxide/PBS for 1 hour, dehydrated, and then incubated in 1% uranyl acetate/70% ethanol for 45 minutes. Pellets were dehydrated and embedded in EPON 812. Utlrathin sections were cut on a Reichert-Jung Ultracut E. Synaptosomes were imaged on a JEM1011 electron microscope (JEOL) and images were acquired with a digital camera from Advanced Microscopy Techniques, Inc (Danvers, MA) and processed using AMT V601 software.
Glutamate release assay after depolarization
Glutamate levels in synaptosomes were measured using Glutamate Assay Kit (BioVision Cat# K629-100). The assay is based on the activity of glutamate oxidase in the presence of glutamate as a substrate. Glutamate enzyme mix was reconstituted with 220 μl Locke’s solution pH 7.4 (1.8 mM CaCl2, 0.8 mM, MgCl2, 0.8 mM Na2HPO4, 5.5 mM glucose, 30 mM HEPES, 3 mM NaHCO3, 120 mM NaCl and 5 mM KCl). Absorbance was read at 450 nm on a Wallac Victor2 1420 plate reader (Perkin Elmer). Hdh140Q/140Q and WT synaptosomes were treated with 125 mM KCl to release glutamate and the absorbance at 450 nm (levels of glutamate) was monitored every 20 seconds for 7 minutes until a plateau was reached. The amount of glutamate released in each sample was calculated according to the standard curve (see below) and normalized to the protein content in the sample. All samples were measured in duplicate (3 month old mice: WT N = 6 and Hdh140Q/140Q N = 6; 6 month old mice WT N = 7 and Hdh140Q/140Q N = 7 mice; 12 month old mice, WT N = 4 and Hdh140Q/140Q N = 4). The level of KCl-dependent glutamate release was determined by subtracting readings of synaptosomes without KCl from readings of synaptosomes with KCl. The glutamate standard curve was established by measuring absorbance using fixed concentrations of glutamate (0, 2, 4, 6, 8, 10, 20, 30 nmol). Values of glutamate release were normalized to protein content and expressed as nanomoles of glutamate per 1 μg protein for each genotype and age group.
Data plotting and statistical analysis
For the Western blot studies mean signal intensity ± SD was determined for each protein analyzed in the WT and Hdh140Q/140Q groups. Data were plotted using the mean percent signal intensity of the analyzed protein in the Hdh140Q/140Q synaptosomes relative to the average WT signal with SDs indicated on the bar graph. To determine if the variance of the means differed, an unpaired Student’s t-test was performed using Microsoft Excel software. For the glutamate release assays the mean levels of glutamate increase after KCl stimulation expressed in nmol per μg protein were plotted for WT and Hdh140Q/140Q on one graph per age group. Variance of the means for WT and Hdh140Q/140Q was compared using a paired two-tailed Student’s t-test. One synaptosome preparation was represented for each mouse and for all analysis N is the number of mice per group. Differences were considered significant if there was a P value of <0.05.
LC-MS/MS and data analysis
Synaptosome lysates from striatum of 6 month old WT and Hdh140Q/140Q mice and cortex of 12 month old WT and Hdh140Q/140Q mice corresponding to 20 μg total protein were reduced in 15 μL of a 15mM DTT solution for 30 minutes at 60°C followed by alkylation of cysteines in 20 μL of a 60 mM iodoacetamide solution for one hour in the dark. The proteins were digested overnight at 30°C with 100 ng trypsin (Promega) in 0.1% Rapigest (Waters) reagent. Rapigest was cleaved by the addition of 1.1 μL of a 10% TFA solution followed by incubation at 37°C for 45 minutes. Following centrifugation, the peptides were recovered from the supernatant.
Triplicates of each brain sample were processed and analyzed. For each digest, a 0.5 μg aliquot was spiked with 200 fmol alcohol dehydrogenase (Saccharomyces cerevisiae) and injected onto a custom packed trap column (100 μm I.D. fused silica with Kasil frit) containing 2.0 cm of 200A,Å 5 μm Magic C18AQ particles (Bruker-Michrom) configured to a custom packed analytical column (75 μm I.D. fused silica) with gravity-pulled tip and packed with 25 cm 100A,Å 5 μm Magic C18AQ particles. Peptides were separated at 300 nL/min using a Proxeon Easy-nLC (Thermo) system using a linear gradient from 100% solvent A (5% acetonitrile with 0.1% (v/v) formic acid) to 35% solvent B (acetonitrile with 0.1% formic acid) in 90 minutes and eluted directly into an LTQ Orbitrap Velos hybrid mass spectrometer (Thermo Scientific) [25]. Data were acquired using a data-dependent acquisition routine of acquiring one mass spectrum over m/z 350–2000 in the Orbitrap (resolution 60,000) followed by tandem mass spectrometry scans in the linear ion trap of the 10 most abundant precursor ions found in the mass spectrum. To minimize data redundancy and maximize peptide identification charge state rejection of singly-charged ions and dynamic exclusion was utilized [26].
Raw data files were processed with ExtractMSN (Thermo Scientific) into peak lists and then searched against the murine UniProt database (03/29/11) using the Mascot (Version 2.3.02; Matrix Science) search engine. Tolerances for parent and fragment masses were set to 15 ppm and 0.5 Da, respectively. The software was set to identify peptides with up to 2 missed cleavages. Variable modifications included acetylation (protein N-term), pyroglutamation (N-term glutamine), and oxidation (methionine) while carbamidomethylation (cysteine) was considered as fixed. Mascot search results were loaded into Scaffold (Version 3.3.1; Proteome Software) and proteins were scored and filtered at the 95% or greater probability level before comparative analyses using spectral counting of tandem mass spectra [27]. Filters were applied to find unique peptide sequences with oxidized methionines that were present in at least two of three Hdh140Q/140Q mice that were not present in any of the WT animals. Peptide extracted ion chromatograms (XIC) were then used for label-free quantitation based on the replicate method [28] and was implemented by both Mascot Distiller (Version 2.4.2; Matrix Science) and ProteoIQ (Version 2.3.08; NuSep) quantitative analysis software packages.
RESULTS
Characterization of synaptosome preparations
Synaptosomes were isolated from striatum of WT and Hdh140Q/140Q mouse brain as described in Methods [20, 21]. Figure 1A shows images of the synaptosomal gradient preparation before and after the ultracentrifugation process. The synaptosomal fraction is indicated with an arrow between the two sucrose layers (Fig. 1A, fraction 1 and fraction 3). The quality of the synaptosomal preparation was evaluated by Western blot analysis. The synaptosomal fraction had an enrichment of the pre-synaptic protein SNAP-25 and the post-synaptic marker PSD95 compared to fractions 1 and 3 (Fig. 1B). Electron microscopic analysis of the synaptosome preparation (Fig. 1C) revealed a mix of profiles that were vesicle-filled or devoid of organelles representing presynaptic and postsynaptic structures, respectively. There were relatively few mitochondria present in the preparation. Some of the vesiclefilled profiles formed synaptic contacts (Fig. 1C, arrows).
Biochemical analysis of striatal synaptosomes from WT and Hdh 140Q/140Q mice
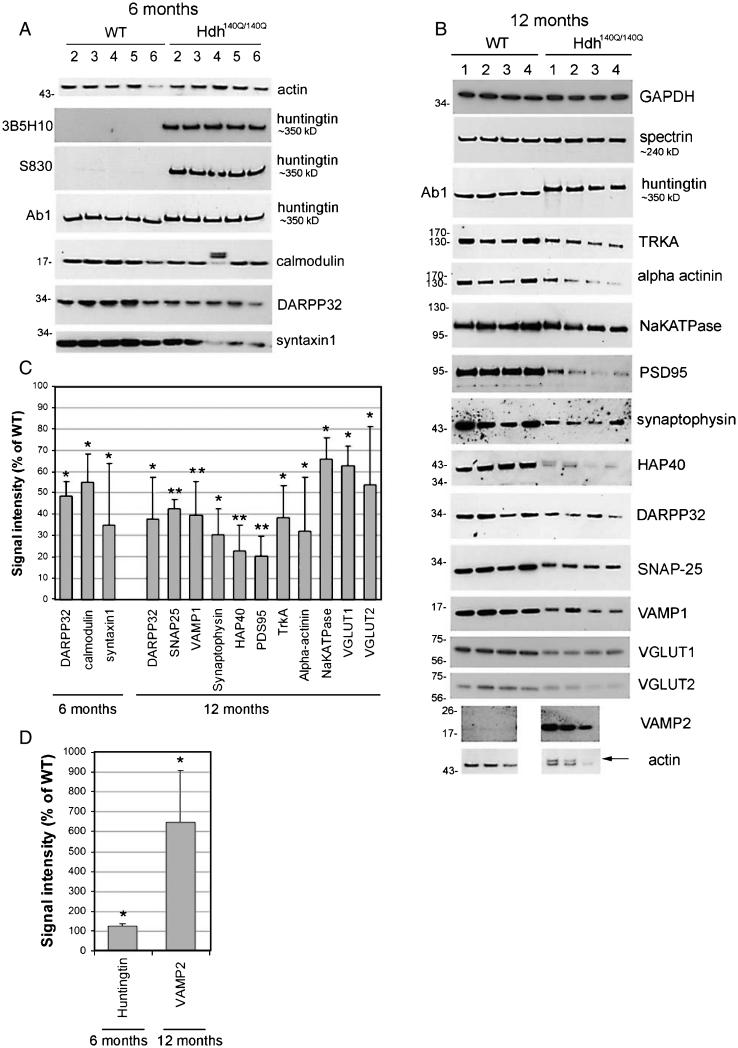
Striatal synaptosomal fractions were prepared from brains of 3, 6 and 12 month old WT and Hdh140Q/140Q mice as described. Four to six synaptosome preparations of each genotype per age group were examined. SDS-PAGE and Western blot assays were performed as described in Methods. Actin signal was used as loading control in 3 and 6 month analysis and spectrin or GAPDH for 12 month old groups because of an additional actin band in the 12 month Hdh140Q/140Q synaptosomes (Fig. 2B, arrow).
Fig. 2.
Biochemical assay and quantitative analysis of proteins detected in striatal synaptosomes of 6 and 12 month old WT and Hdh140Q/140Q mice. A and B. Western blots show the results for proteins that were changed in Hdh140Q/140Q synaptosomes in this study. Huntingtin and loading controls (actin, GAPDH, or spectrin) are also shown. As explained in methods because of additional band in actin signal at 12 months in Hdh140Q/140Q mice, the actin signal was only used as loading control for results in 3 and 6 month old mice and spectrin or GAPDH was used for the 12 month old groups. Location of molecular weight markers are shown on the left (in kD). Huntingtin and spectrin proteins are too high on the gel to include molecular weight markers so approximate protein size is listed on the right. The different huntingtin antibodies used are shown on the left. C and D. Bar graphs show Mean percent ± SD signal intensity in synaptosomes from 6 and 12 month old Hdh140Q/140Q mice relative to WT mice. All proteins indicated were significantly different from WT (Student’s t-Test, 6 month, N = 5 WT, 5 Hdh140Q/140Q; 12 month N = 4 WT, 4 Hdh140Q/140Q, *p < 0.05 **p < 0.005). Note that in C the levels of proteins shown in Hdh140Q/140Q are less than in WT and in D the levels of proteins shown in Hdh140Q/140Q synaptosomes are greater than in WT.
We evaluated the levels of htt and 51 other proteins, most of them with known roles in synapses for localization by Western blot assay using previously characterized antisera. Proteins that were found to be at similar levels in WT and Hdh140Q/140Q striatal and cortical synaptosomes included AP2, beta catenin, DNP, dynamin, gp91-phox, GSK3beta, Kalirin, LAMP1, LC3, LSAMP, Myosin10, p22.phox, PI3K P85, PI3K P110, pascin1, profilin1, profilin2, RabGDPalpha, Rac1, synaptotagmin, syntaxin6, and TRKB. We could not detect a specific signal using commercially available antibodies against CB1, cofilin, D1, D2, ezrin, p47.phox, phospho-cofilin and Rab11Fip5. Shown in Fig. 2AB are the Western blots for those proteins that were found to be affected by the presence of the HD gene and the loading controls (actin, spectrin, GAPDH). All signals on Western blot films were quantified as a total density normalized to the standard protein and mean signals for Hdh140Q/140Q samples are expressed as signal intensity (% of WT) for the average age-matched WT (Fig. 2C, D). Also shown are levels of htt. Four antibodies that detect full length mhtt were tested, Ab1 which recognizes htt1-17, 1C2 (MAB1574, Millipore, not shown) and 3B5H10 which recognize the expanded polyglutamine tract, and S830, which reacts against htt-Exon1-Q53 (generous gift of Dr. Gillian Bates); the results obtained with these antisera are shown for the 6 month old mice and confirmed that endogenous full length WT and mhtt are robustly expressed in the Hdh140Q/140Q synaptosomes (Fig. 2A). Since the lower part of the nitrocellulose membrane was usually separated for probing with other antisera, the presence of htt fragments was not routinely evaluated in our study. However in some blots where the membrane remained intact no N-terminal htt fragments were observed in the WT or Hdh140Q/140Q synaptosomes. Interestingly in synaptosomes from 6 month old Hdh140Q/140Q mice, the levels of full length mhtt were significantly increased by ~20% compared to WT htt when detected with antibody Ab1 (anti-htt1-17) (Fig. 2A, D).
In striatal synaptosomes of 3-month old Hdh140Q/140Q mice no significant changes were detected compared to WT for the proteins we tested. Only DARPP32 showed a modest decline in some of the Hdh140Q/140Q mice compared to the WT (results not shown). In striatal synaptosomes obtained from 6-month old Hdh140Q/140Q mice, the levels of three proteins were significantly lower than in WT synaptosomes, namely DARPP32 (~55% calmodulin (~45% decrease), and syntaxin-1 (~65% decrease), (Fig. 2A, C). Eleven proteins were affected in the striatal synaptosomes of 12 month old Hdh140Q/140Q mice (Fig. 2B–D). There was a significant decline in the levels of SNAP25 (~55% decrease), VAMP1 (~60% decrease), synaptophysin (~70% decrease), HAP40 (~75% decrease), PSD95 (~80% decrease), TrkA (~60% decrease), alpha actinin (~65% decrease), Na+/K+-ATPase (~35% decrease), VGlut1 (~35% decrease), VGlut2 (~45% decrease), and DARPP32 (~70% decrease) (Fig. 2B, C). In contrast, VAMP2 showed increased levels (~6-fold) in the striatal synaptosomes of the 12 month old Hdh140Q/140Q mice compared to WT (Fig. 2B, D).
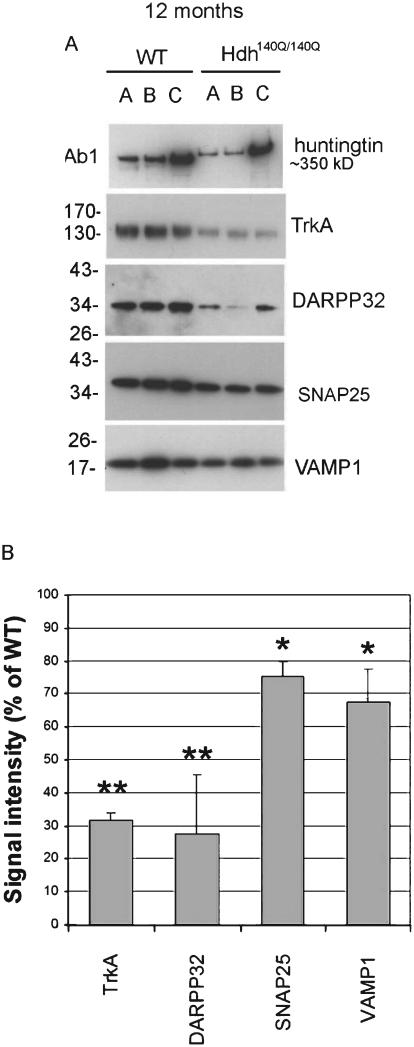
We also studied synaptosomes from cortex of 12 month old WT and Hdh140Q/140Q mice (N = 3 per genotype) and found a decline in the levels of 4 proteins that were also changed in the striatum of 12 month old Hdh140Q/140Q mice namely TrkA (~65% decrease), DARPP32 (~70% decrease), SNAP25 (~25% decrease), and VAMP1 (~30% decrease) (Fig. 3).
Fig. 3.
Biochemical assay and quantitative analysis of proteins reduced in cortical synaptosomes of 12 month old WT and Hdh140Q/140Q mice. A. Western blots show the results for proteins that were changed in Hdh140Q/140Q synaptosomes compared to WT. B. Bar graphs show Mean percent±SD signal intensity in Hdh140Q/140Q synaptosomes compared to WT mice. All proteins indicated were significantly different from WT (Student’s t-Test, N = 4 WT, 4 Hdh140Q/140Q evels of proteins show, *p < 0.05 **p < 0.005).
Analysis of glutamate release from stimulated striatal synaptosomes of Hdh 140Q/140Q and WT mice
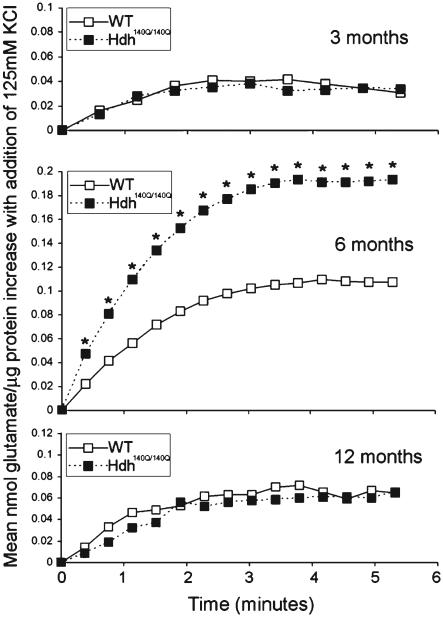
The release of glutamate was evaluated in freshly prepared synaptosomes of WT and Hdh140Q/140Q mice. Basal glutamate levels were monitored for 7 minutes before stimulation with KCl. Synaptosomes were treated with 125 mM KCl and readings of glutamate levels were taken every 20 seconds up to 7 minutes after stimulation. In both WT and Hdh140Q/140Q samples, glutamate levels in the presence of KCl rose rapidly in the first 1–3 minutes following stimulation, and then reached a plateau (Fig. 4). WT and Hdh140Q/140Q synaptosomes from the 3 and 12 month old mice did not differ in the levels of glutamate measured up to 7 minutes following KCl stimulation. However at each time point synaptosomes from the 6 month old Hdh140Q/140Q mice had a significant increase in the levels of glutamate detected compared to WT (Mean ± SD with paired t-test). The rate of release was determined by calculating the slope in the linear range of the curves in the first 2.5 minutes after stimulation. The rate of glutamate release was significantly higher at 6 months for striatal synaptosomes from Hdh140Q/140Q animals compared to WT (0.072 nmol/min±0.032 for Hdh140Q/140Q compared to 0.04 nmol/min±0.025 for WT, mean±SD; p < 0.05, n = 7, paired t-test). ± No differences in release rates were found between WT and Hdh140Q/140Q synaptosomes at 3 months or 12 months.
Fig. 4.
Stimulated release of glutamate in WT and Hdh140Q/140Q evels of proteins show striatal synaptosomes. Glutamate levels were measured using a commercial assay kit as described in Methods and measurements were taken using Wallac Victor2 1420 plate reader (Perkin Elmer). Shown are changes in mean glutamate levels detected every 20 seconds following stimulation with 125 mM KCl. Glutamate levels are significantly higher in the Hdh140Q/140Q synaptosomes of 6 month old mice compared to those of WT (Student’s paired t-test p < 0.05). Standard deviation bars were large and are omitted from the graph.
LC-MS/MS analysis of oxidized methionines in Hdh140Q/140Q and WT synaptosomes
Previously we found evidence for potential imbalances in ROS in Hdh140Q/140Q mice that might contribute to oxidative stress. Primary cortical neurons derived from embryos of Hdh140Q/140Q mice have lower levels of glutathione caused by reduced uptake of cysteine at the plasma membrane [19]. We also found that the activity of the ROS generator NOX2 was significantly elevated in Hdh140Q/140Q striatal synaptosomes at 3, 6 and 12 months of age with highest levels occurring in striatal synaptosomes of 6 month old Hdh140Q/140Q mice [21]. To look for signs of unusual protein oxidation in the Hdh140Q/140Q synaptosomes, we analyzed WT and Hdh140Q/140Q samples from 6 month old mice by mass spectrometry (LC/MS/MS) as described in Methods. Unique peptide sequences containing oxidized methionines were identified. Oxidized methionines in Hdh140Q/140Q proteins were considered novel if methionine oxidation did not occur in the corresponding WT protein or if the specific oxidized residue was unique to those found in WT synaptosomes. As a positive control, we used WT synaptosomes treated with 100 μM of H2O2 for 30 minutes. The data from these samples were used to identify those methionine sites with potential for ex vivo oxidation (see below for comparison to in vivo oxidation in Hdh140Q/140Q samples).
A total of 622 and 623 proteins were identified in the WT and Hdh140Q/140Q striatal synaptosomes respectively by LC-MS/MS. The vast majority –610–were the same proteins in the WT and Hdh140Q/140Q samples. Although readily detected by Western blot, htt proteins (wild type htt and mtt) were not among the proteins detected in the WT or Hdh140Q/140Q striatal and cortical synaptosomes by LC-MS/MS nor were the majority of other proteins that were found by Western blot changed in Hdh140Q/140Q samples compared to the WT samples. Only VGlut1 and VAMP2, which were altered in Hdh140Q/140Q striatum based on Western blot data, were among the proteins detected by mass spectrometry. In view of these results we did not perform a quantitative analysis of protein levels in WT and Hdh140Q/140Q samples using the LC-MS/MS results.
Based only on the presence of novel methionine oxidation in Hdh140Q/140Q proteins compared to WT, we found 25 proteins in the 6 month old Hdh140Q/140Q samples with oxidized methionines (40 oxidized methionines in 25 proteins) that were not present in the corresponding proteins of the WT synaptosomes (Table 1, proteins 1–25). Fourteen of the 25 proteins with novel oxidized methionines in the Hdh140Q/140Q synaptosomes are involved in vesicle transport, synaptic function, cytoskeleton organization, and neurotransmitter uptake, and included synapsin 2, synaptophysin, syntaxin B1, EAAC2, calcium/calmodulin dependent proteinase kinase K type II subunit alpha, and 14-3-3 protein gamma (Table 1). Four other proteins –tubulin beta2A, actin cytoplasmic 2, dihydropyrimidase-related protein 2 and 14-3-3 protein epsilon–had a total of 14 oxidized methionines in Hdh140Q/140Q synaptosomes that differed from the oxidized methionines that were detected in the WT proteins (Table 1, proteins 26–29 show methionines oxidized in Hdh140Q/140Q proteins and not WT proteins). Table 1 columns 6–8 indicate the positions of the novel oxidized methionines in proteins of each Hdh140Q/140Q mouse. Most proteins have one oxidized methionine in at least two mice. Nine Hdh140Q/140Q proteins had 2–4 oxidized methionines that were not seen in corresponding WT proteins (spectrin alpha chain, EAAC2, calcium/calmodulin-dependent proteinase K type II subunit alpha, neurofilament light polypeptide, tubulin beta 1B, neuronal cell adhesion molecule 1, creatine kinase B, ATPase subunit beta-mitochondrial, and dihydropyrimidinase-related protein). Results suggest that there is methionine oxidation occurring in proteins at striatal synapses of 6 month old Hdh140Q/140Q mice.
Table 1.
Distribution of novel oxidized proteins in striatal synaptosomes of 6 month Hdh140Q/140Q mice
| # | Protein* | Function | Gene code accession# |
# mice affected |
# of peptides with oxidized methionines |
Sites of oxidized methionines HD1 |
Sites of oxidized methionines HD2 |
Sites of oxidized methionines HD3 |
|---|---|---|---|---|---|---|---|---|
| 1 | Synapsin2 | Synaptic vesicle protein |
SYN2_MOUSE | 3 | 1-1-2 | 66 | 66 | 66 |
| 2 | Synaptophysin | Synaptic vesicle protein |
SYPH_MOUSE | 3 | 1-1-1 | 170 | 170 | 170 |
| 3 | Syntaxin B1 | SNARE complex | STXB1_MOUSE | 2 | 1-0-1 | 355 | 0 | 355 |
| 4 | Vesicle-fusing ATPase | Vesicle transport and fusion |
NSF_MOUSE | 2 | 0-1-1 | 0 | 504 | 504 |
| 5 | Spectrin alpha chain | Membrane associate protein (axons and dendrite) |
SPTN1_MOUSE | 3 | 1-1-1 | 162, 561 | 561 | 561 |
| 6 | Excitatory amino acid transporter 2 |
Uptake of excitatory amino acids |
EAAC2_MOUSE | 2 | 0-1-1 | 0 | 18 | 10 |
| 7 | V-type proton ATPase subunit G |
Regulates ion balance in vesicles (synaptic) |
VATG_MOUSE | 3 | 1-1-1 | 63 | 63 | 63 |
| 8 | V-type proton ATPase subunit B, brain isoform |
Regulates ion balance in vesicles (synaptic) |
VATB_MOUSE | 2 | 1-0-1 | 410 | 0 | 410 |
| 9 | Calcium/ Calmodulin- dependent proteinase K type II subunit alpha |
Signaling pathways regulator |
KCC2A_MOUSE | 3 | 1-2-3 | 281 | 281, 307 | 281, 307, 372 |
| 10 | Neurofilament light polypeptide |
Assembly of cytoskeleton structures |
NFL_MOUSE | 3 | 3-3-2 | 55, 326, 435 | 326, 348, 435 | 326, 435 |
| 11 | Tubulin beta 1B | Assembly of cytoskeleton structures |
TBA1B_MOUSE | 2 | 0-1-1 | 0 | 302 | 413 |
| 12 | Neural cell adhesion molecule 1 |
Neuronal cell adhesion and axonal growth |
NCAM1_MOUSE | 3 | 2-2-1 | 558, 565, 660 | 558, 660 | 558, 565 |
| 13 | Cell adhesion molecule 3 |
Neuronal cell adhesion and axonal growth |
CAM3_MOUSE | 3 | 1-1-1 | 234 | 234 | 234 |
| 14 | Microtubule associated protein 1B |
Cytoskeleton associated protein |
MAP1B_MOUSE | 3 | 1-1-1 | 1372 | 1372 | 1372 |
| 15 | Microtubule associated protein 6 |
Cytoskeleton associated protein |
MAP16_MOUSE | 2 | 1-1-0 | 244 | 244 | 0 |
| 16 | Creatine kinase B | Energy and metabolism |
KCRB_MOUSE | 3 | 2-2-2 | 352 | 352, 377 | 352 |
| 17 | ATP synthase subunit beta, mitochondrial |
ATP production | ATPB_MOUSE | 2 | 2-0-1 | 442, 478 | 0 | 478 |
| 18 | Alpha internexin | Intermedia filaments (cytoskeleton) |
AINX_MOUSE | 3 | 1-1-1 | 504 | 504 | 504 |
| 19 | 14-3-3 protein gamma | Regulates pathways leading to apoptosis |
1433G_MOUSE | 2 | 0-1-1 | 0 | 165 | 165 |
| 20 | Dihydropyrimidinase- related protein 1 |
Catabolism | DPYL1_MOUSE | 3 | 1-1-3 | 498 | 498 | 215, 437, 498 |
| 21 | Dihydropyrimidinase- related protein 5 |
Catabolism | DPYL5_MOUSE | 3 | 1-1-1 | 507 | 507 | 507 |
| 22 | Adenylyl cyclase-associated protein 1 |
cAMP production | CAP1_MOUSE | 2 | 0-1-1 | 0 | 27 | 27 |
| 23 | Calmodulin | Signaling pathways regulator |
CALM_MOUSE | 2 | 0-1-1 | 0 | 77 | 77 |
| 24 | Microtubule- associated protein tau |
Cytoskeleton | TAU_MOUSE | 2 | 0-1-1 | 0 | 542 | 542 |
| 25 | Stress-induced- phosphoprotein 1 |
Stress death/survival processes |
STIP1_MOUSE | 2 | 0-1-1 | 0 | 1 | 1 |
| 26 | Tubulin beta 2A | Assembly of cytoskeleton structures |
TBB3_MOUSE | 3 | 6-8-7 | 73, 247, 330 |
73, 247, 292, 299, 300, 330 |
73, 292, 299, 300, 330 |
| 27 | Actin cytoplasmic 2 | Assembly of cytoskeleton structures |
ACTG_MOUSE | 3 | 1-3-2 | 325 | 44, 47, 227, 325 | 44, 47, 325 |
| 28 | Dihydropyrimidinase- related protein 2 |
Catabolism | DPYL2_MOUSE | 3 | 3-2-1 | 64, 437 | 64, 437 | 437 |
| 29 | 14-3-3 protein epsilon | Regulates pathways leading to apoptosis |
1433E_MOUSE | 3 | 2-2-1 | 1 | 1, 33 | 1 |
Proteins #1–25 had no oxidized methionines in synaptosomes from WT mice. Proteins #26–29 had oxidized methionines in WT synaptosomes at different residues than in the Hdh140Q/140Q synaptosomes. Underlined residues are also oxidized when WT synaptosomes are exposed to H2O2.
Next we determined if the novel methionine oxidations present in the Hdh140Q/140Q proteins could also occur by ex vivo exposure to H2O2. In WT samples exposed to H2O2 prior to LC-MS/MS, 139 of 375 or 37% identified proteins had oxidized methionines compared to 153 of 622 or 25% of proteins in the untreated WT samples. Only a minority (10 of 29 proteins or 31%) of the proteins modified by methionine oxidation in the Hdh140Q/140Q samples were identified in the WT samples treated with H2O2. The methionines that were oxidized in the WT + H2O2 proteins are underlined in Table 1. The results suggest that oxidation at novel methionine sites in the majority of Hdh140Q/140Q proteins (59%) is occurring in vivo and cannot be due to increased sensitivity to oxidation during preparation and handling.
We showed previously that synaptosomes from the cortex of Hdh140Q/140Q mice have increased activity of the ROS generator NOX2 and that Hdh140Q/140Q primary cortical neurons have elevated ROS [21]. Therefore, we also looked for evidence of protein oxidation in cortical synaptosomes of 12 month old Hdh140Q/140Q mice. A total of 620 and 621 proteins were identified in the WT and Hdh140Q/140Q cortical synaptosomes respectively by mass spectrometry and 608 were the same proteins in WT and Hdh140Q/140Q samples. Novel methionine oxidations in Hdh140Q/140Q proteins were identified using the same criteria described for the analysis of striatal synaptosomes. Table 2 lists 31 proteins in the cortical synaptosomes of Hdh140Q/140Q mice that had oxidized methionines not seen in the same proteins of WT mice. Table 2 also shows the location of the methionine residues with novel oxidation in the proteins of Hdh140Q/140Q synaptosomes. Proteins 1–27 had a total of 36 oxidized methionines in the Hdh140Q/140Q mice and none in the WT mice. Proteins 28–31 had 8 oxidized methionines in the Hdh140Q/140Q mice that were not seen in the WT proteins that did have other sites of methionine oxidation. In most Hdh140Q/140Q proteins, only one novel site was identified. In 6 of the 31 Hdh140Q/140Q proteins 2–4 residues with atypical methionine oxidation were detected (neurofilament light polypeptide, creatine kinase B-type, calcium/calmodulin-dependent proteinase K type II subunit alpha, dihydropyrimidinase-related protein 1, tubulin alpha 1B chain, and EAAC2).
Table 2.
Distribution of novel oxidized proteins in cortical synaptosomes of 12 month Hdh140Q/140Q mice
| # | Protein* | Function | Gene code accession# |
# mice affected |
# of peptides with oxidized methionines |
Sites of oxidized methionines HD1 |
Sites of oxidized methionines HD2 |
Sites of oxidized methionines HD3 |
|---|---|---|---|---|---|---|---|---|
| 1 | Synapsin 2 | Synaptic vesicle protein | SYN2_MOUSE | 3 | 1-1-1 | 466 | 466 | 466 |
| 2 | Synaptophysin | Synaptic vesicle protein | SYPH_MOUSE | 3 | 1-1-1 | 170 | 170 | 170 |
| 3 | Vesicle-fusing ATPase | Vesicle transport and fusion |
NSF_MOUSE | 2 | 0-1-1 | 0 | 504 | 504 |
| 4 | Excitatory amino acid transporter 2 |
Uptake of excitatory amino acids |
EAA2_MOUSE | 2 | 0-1-1 | 0 | 18 | 10 |
| 5 | V-type proton ATPase subunit G |
Regulates ion balance in vesicles (synaptic) |
VATG2_MOUSE | 3 | 1-1-1 | 73 | 73 | 73 |
| 6 | V-type proton ATPase subunit B |
Regulates ion balance in vesicles (synaptic) |
VATB2_MOUSE | 2 | 1-0-1 | 410 | 0 | 410 |
| 7 | Calcium/calmodulin- dependent protein kinase type II subunit alpha |
Signaling pathways regulator |
KCC2A_MOUSE | 3 | 1-2-3 | 281 | 281, 307 | 281, 307, 373 |
| 8 | Neurofilament light polypeptide |
Assembly of cytoskeleton structures |
NFL_MOUSE | 3 | 3-3-2 | 64, 326, 435 | 326, 435 | 326, 435 |
| 9 | Tubulin alpha 1B chain | Assembly of cytoskeleton structures |
TBA1B_MOUSE | 2 | 0-1-1 | 0 | 302 | 413 |
| 10 | Cell adhesion molecule 3 | Neuronal cell adhesion and axonal growth |
CADM3_MOUSE | 3 | 1-1-1 | 234 | 234 | 234 |
| 11 | Microtubule-associated protein 6 OS |
Cytoskeleton associated protein |
MAP6_MOUSE | 2 | 1-1-0 | 244 | 244 | 0 |
| 12 | Creatine kinase B-type | Energy and metabolism | KCRB_MOUSE | 3 | 1-3-1 | 352 | 352, 377 | 352 |
| 13 | Alpha-internexin | Intermediate filaments (cytoskeleton) |
AINX_MOUSE | 3 | 1-1-1 | 504 | 504 | 504 |
| 14 | 14-3-3 protein gamma | Regulates pathway leading to apoptosis |
1433G_MOUSE | 2 | 0-1-1 | 0 | 165 | 165 |
| 15 | Dihydropirimidinase related protein 1 |
Catabolism | DPYL1_MOUSE | 3 | 1-1-3 | 498 | 498 | 215, 437, 498 |
| 16 | Dihydropirimidinase related protein 5 |
Catabolism | DPYL5_MOUSE | 3 | 1-1-1 | 507 | 507 | 507 |
| 17 | Adenylyl cyclase-associated protein 1 |
cAMP production | CAP1_MOUSE | 2 | 1-1-0 | 27 | 27 | 0 |
| 18 | Calmodulin | Signaling pathways regulator |
CAL_MOUSE | 2 | 0-1-1 | 0 | 77 | 77 |
| 19 | Microtubule-associated protein tau |
Cytoskeleton | TAU_MOUSE | 2 | 0-1-1 | 0 | 542 | 542 |
| 20 | Stress-induces phospho-protein 1 |
Stress death/survival processes |
STIP1_MOUSE | 2 | 0-1-1 | 0 | 1 | 1 |
| 21 | Microtubule-associated protein 1A |
Cytoskeleton associated protein |
MAP1A_MOUSE | 3 | 1-1-1 | 1372 | 1372 | 1372 |
| 22 | Hemoglobin subunit alpha |
Oxygen transport | HBA_MOUSE | 3 | 1-1-1 | 33 | 33 | 33 |
| 23 | Hemoglobin subunit beta1 |
Oxygen transport | HBB1_MOUSE | 2 | 1-2-0 | 56 | 56 | 0 |
| 24 | Fructose-biphosphate- aldolase A |
Catabolism | ALDOA_MOUSE | 2 | 0-1-1 | 0 | 165 | 165 |
| 25 | Heat shock cognate 71 kD protein |
Repressor of transcriptional activation |
HSPC_MOUSE | 3 | 1-1-1 | 61 | 61 | 61 |
| 26 | Band 4.1-like protein 3 | Promotes apoptosis and tumor suppression |
E41L3_MOUSE | 2 | 0-1-1 | 0 | 703 | 703 |
| 27 | Complexin 1 | Regulates synaptic vesicular exocytosis |
CPLX1_MOUSE | 2 | 0-1-1 | 0 | 86 | 86 |
| 28 | Spectrin alpha chain non-erytrocitic 1 |
Membrane associated protein (axons and dendrite) |
SPTN1_MOUSE | 3 | 2-1-1 | 541 | 541 | 541 |
| 29 | Actin-cytoplasmic 2 | Assembly of cytoskeleton structures |
ACTG_MOUSE | 3 | 1-3-2 | 325 | 44, 47, 227, 325 | 44, 47, 325 |
| 30 | 14-3-3 protein epsilon | Regulates pathways leading to apoptosis |
1433E_MOUSE | 3 | 2-2-1 | 1 | 1, 33 | 1 |
| 31 | Ras-related protein Rab3A |
Regulates late steps in synaptic vesicular function |
RAB3A_MOUSE | 2 | 0-1-1 | 0 | 187 | 187 |
Proteins #1–27 had no oxidized methionines in synaptosomes from WT mice. Proteins #28–31 had oxidized methionines in WT synaptosomes at different residues than in the Hdh140Q/140Q synaptosomes. Underlined residues are also oxidized when WT synaptosomes are exposed to H2O2.
Of interest, 18 of 29 (62%) affected proteins identified in the striatum of 6 month old Hdh140Q/140Q mice were also affected in the cortex of the 12 month old Hdh140Q/140Q mice (18 of 31 or 58%). Among the commonly affected proteins were those involved in vesicle function and neurotransmission (calmodulin, synapsin 2, synaptophysin, vesicle-fusing ATPase, v type proton ATPase subunit G and EAAC2), proteins related to cytoskeleton assembly and function (MAP1A, MAP6, neurofilament light polypeptide), adhesion (CAM, NCAM) and signaling (14-3-3 protein epsilon and gamma, adenylyl cyclase associated protein 1). These results suggest that a similar group of proteins undergo oxidative modification at cortical and striatal synapses of Hdh140Q/140Q mice.
Finally, 13 of the 31 proteins found oxidized at methionines in the Hdh140Q/140Q samples (42%) were also oxidized at one or more of the same methionines in the WT samples treated with H2O2. The methionines that were also oxidized in the WT + H2O2 proteins are underlined in Table 2. From this data, we conclude that the majority of Hdh140Q/140Q cortical proteins with novel methionine oxidization (58%) are due to in vivo changes and not to increased sensitivity to ex vivo preparation or handling.
DISCUSSION
Disruption of striatal circuitry is an early and fundamental problem in HD. In this study we show that synaptosomal fractions, which are enriched for neuronal synaptic proteins, provide a useful and efficient preparation for conducting parallel biochemical, enzymatic and mass spectrometry assays. The Hdh140Q/140Q synaptosomes we studied expressed endogenous full length mhtt. Full length mhtt has been previously detected in human brain synaptosomal fractions and immunoreactive htt localization in presynaptic endings distal from the synaptic cleft has been observed at the electron microscopic level [1]. It is noteworthy that at 6 months we found higher levels of full length mhtt compared to WT htt in synaptosomes. The Ab1 antibody used for the detection may be sensitive to an altered conformation of mhtt that accumulates in synaptic compartments. However CAG repeat expansion may also increase htt translation into protein [29]. We found that striatal synaptosomes from 6 month old mice showed marked changes in the levels of synaptic proteins, altered glutamate release, and novel methionine oxidation. Our findings suggest that young HD homozygous mice expressing full-length mhtt at endogenous levels have robust alterations at the synaptic level.
In accord with previous immunohistochemical analysis in Hdh140Q/140Q mice [13], we observed by Western blot a reduction in the levels of DARPP32, a dopamine- and cAMP-regulated phosphoprotein and highly specific marker of medium spiny neurons. DARPP32 levels declined progressively from 6 months (50% of WT) to 12 months of age (20% of WT). The loss of signal for DARPP32 in synaptosomes may reflect the reduction in neuronal processes belonging to medium spiny neurons. In contrast to DARPP32, other altered proteins in Hdh140Q/140Q synaptosomes did not show progressive changes with age. Levels of syntaxin-1 and calmodulin, which were the other proteins markedly reduced at 6 months, were unchanged compared to WT at 12 months. These findings suggest that a dynamic process of neurode-generative and compensatory/regenerative events is occurring at the synaptic level in HD brain. Not surprising, striatal synaptosomes from 12 month old Hdh140Q/140Q mice had the most proteins with reduced levels including the SNARE proteins SNAP-25 and VAMP1. The levels of these proteins were also reduced in the cortex at the same age. Syntaxin 1, SNAP-25, and VAMP1 (synaptobrevin), are components of the N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex that functions in vesicle fusion and synaptic transmission at the presynaptic zone. Consistent with the loss of SNAP-25 in cortex of 12 month old Hdh140Q/140Q mice, early decline in SNAP-25 was described in the postmortem cortex of HD patients [30]. The reduction in SNAP-25 protein may arise from lower mRNA levels as reported in another HD mouse model, or from oxidative damage and degradation of SNAP-25 (observed by Smith et al, in the cortex [30]). No change in mRNA levels of the SNARE genes (SNAP-25, syntaxin 1A and synaptobrevin-2) were found by in situ hybridization in the brains of R6/2 mice [31] suggesting that the decline we see in SNARE proteins in Hdh140Q/140Q could be post-translational. The atrophy of neuronal processes and loss of synapses may account for some of the decline in levels of SNAP-25 and VAMP1.
Synaptophysin is a highly abundant synaptic vesicle protein that is frequently used as a marker of synaptic density. Striatal synaptosomes from 12 month old Hdh140Q/140Q mice had reduced levels of synaptophysin. A decline in immunoreactive synaptophysin has been seen by immunohistochemisty in the striatum of HD patient postmortem brain (and was considered to be evidence for synaptic loss) [32], additionally in 11–15 week old R6/2 HD mice in concert with severely abnormal spontaneous excitatory post-synaptic potential activity [33]. Although one study reported no difference in levels of synaptophysin in cortex and hippocampus of HD compared to WT mice, synaptophysin levels did rise with environmental enrichment of both genotypes [34]. The increase in VAMP2 that coincided with the decline in VAMP1 in Hdh140Q/140Q striatum at 12 months was not surprising. Increases in VAMP2 were noted in the R6/1 HD mouse model in parallel with an increased probability of synaptic vesicle release from stimulated neuromuscular junctions [35]. The decline in VAMP1 levels and rise in VAMP2 levels in Hdh140Q/140Q synaptosomes may reflect a response to axonal pathology. After experimental axotomy of lower motor neurons, the loss of VAMP1 mRNA is accompanied by increases in VAMP2 mRNA [36].
Calmodulin is a small calcium binding protein that is critical for synaptic vesicle mobilization, neuro-transmission and synaptic plasticity. Though reduced in synaptosomes from 6 month old Hdh140Q/140Q mice, calmodulin levels were comparable to WT at 12 months. A slower migrating band that appeared on Western blot with anti-calmodulin antibody suggests that the protein is modified post-translationally. The loss and modification of calmodulin in HD may be a neuronal response to remove calmodulin-mhtt associations from the synapse. Calmodulin interacts with mhtt [37-40] and a reduction in calmodulin -mhtt fragment interaction in vitro reduces cellular toxicity [39].
Surprisingly, a marked decline in levels of the htt binding partner HAP40 in synaptosomes of 12 month old Hdh140Q/140Q mice was observed (about 80% reduction). Previous studies have reported a rise in the levels of HAP40 in HD patient fibroblasts that is thought to contribute to an abnormal cytoskeletal dependent function of Rab5 positive early endosomes [41, 42]. The decline in HAP40 may help neurons reduce the burden of early endosome dysfunction in HD.
Synaptosomes of 12 month Hdh140Q/140Q mice had reduced levels of proteins that affect presynaptic and postsynaptic events important for glutamatergic transmission and synaptic plasticity. The transporters VGlut1 and VGlut2 take up glutamate for synaptic release and are known to be selectively enriched in cortical and thalamic axons that innervate the striatum, respectively [43, 44]. The loss of these proteins in Hdh140Q/140Q mice at 12 months by 30–40% may reflect the decline in afferent inputs and/or be an intrinsic neuronal response that reduces the loading of glutamate into synaptic vesicles. Reduced levels of Na+/K+-ATPase by more that 30% were also observed in 12 month old Hdh140Q/140Q mice. We previously found levels of Na+/K+-ATPase reduced in Hdh140Q/140Q neurons compared to WT neurons in signaling domains known as lipid rafts [9]. If the decline in protein levels of Na+/K+-ATPase in HD reflects diminished enzyme activity there may be increased glutamate transmission. Inhibiting Na+/K+-ATPase activity in striatal neurons was shown to increase glutamate dependent depolarization [45].
Proteins that play a role in anchoring NMDA receptors at the postsynaptic membrane and actin cytoskeleton –PSD95 and alpha-actinin –were also reduced in the synaptosomes of 12 month old Hdh140Q/140Q mice. Reduced protein levels of PSD95 and alpha-actinin 2 were reported in the cortex and hippocampus of R6/2 mice [46]. The decline in levels of PSD95 in hippocampus of R6/1 mice was slowed along with improvement in spatial learning when mice were exposed to an enriched environment [34]. Changes in alpha-actinin levels affect spine morphology [47], which is known to be altered in medium spiny neurons in patients with Huntington’s disease [48]. TrkA is a growth factor receptor for NGF, with less affinity for BDNF. The marked decline in levels of TrkA (60% reduction) in the striatum and cortex was unexpected since no changes in TrkA protein have been previously reported in HD. NGF-TrkA dependent signaling is important for neurite outgrowth. In PC12 cells and in primary neurons, mhtt disrupts this pathway by interfering with the stability of TrkA through an abnormal interaction with its binding partner huntingtin interacting protein 1 (HAP1) [49].
Based on electrophysiological studies conducted in HD transgenic mouse models that express full length mtt, glutamatergic transmission on striatal medium spiny neurons is increased in the early stages of HD [50, 51]. Of interest in 1.5 month old YAC128 HD mice there is an increase in glutamate neurotrans-mission in dopamine D1 receptor-containing neurons that form the direct output pathway whereas at 12 months in both YAC128 HD and BAC HD mice there is a decrease in glutamate neurotransmission. Changes in neurotransmission may arise from multiple factors including alterations that affect the molecular machinery for vesicle fusion and release. We found glutamate release from striatal synaptosomes to be elevated in Hdh140Q/140Q synaptosomes of 6 month old mice. In accord with our findings, increased neurotransmitter release at the neuromuscular junction was seen in a Drosophila model of HD that expressed full-length mhtt with 128 CAG repeats and was caused by an increased probability of vesicle release. The deficit was ameliorated by reducing the levels of SNAP-25 [11]. In our study of Hdh140Q/140Q synaptosomes, SNAP-25 levels were lower than normal at 12 months of age. The decline may be a compensatory response to normalize glutamate release, which was abnormal at 6 months. Down regulation in expression of VGlut1 and VGlut2 seen at 12 months in the synaptosomes of Hdh140Q/140Q mice might also be part of a homeostatic neuronal response to reduce the loading of glutamate into synaptic vesicles.
Methionine oxidation in proteins can affect their biological activity by altering protein structure [52-56]. Our proteomics analysis suggested that about 4% of proteins identified by mass spectrometry in Hdh140Q/140Q synapses are targets of abnormal methionine oxidation. Vesicle membrane proteins including synaptophysin, synapsin 2, syntaxin 1 and calmodulin, as well as cytoskeletal proteins cytoplasmic actin 2, neurofilament, and tubulin had oxidation at methionine residues that were not oxidized in WT mice. Abnormal methionine oxidation induces protein dysfunction. Two proteins in the Hdh140Q/140Q mice were oxidized at methionine residues that are known to affect their function–calmodulin at methionine 77 and cytoplasmic actin 2 at methionine 44 [52, 53]. Studies are lacking at this point to know if other proteins identified with novel methionine oxidation in HD synaptosomes are functionally impaired. It is noteworthy that the striatum and cortex showed a considerable overlap (about 66%) in the proteins with oxidized methionines. Proteins with oxidation at residues other than methionine were not studied and may also be affected in HD. Nevertheless, our findings suggest that in the two regions most affected in HD (striatum and cortex), a similar group of proteins important for synaptic function are vulnerable to oxidative damage.
In summary, our findings show that multiple changes including altered protein levels, increased glutamate transmission and methionine oxidation are detected in striatal and cortical synaptosomes of Hdh140Q/140Q mice up to one year of age. The success of preclinical studies in HD mouse models depends in part on the use of simple reliable quantitative measures to monitor disease progression and efficacy of treatment. Since changes in HD synaptosomes are robust, study of synaptosomes in mice should provide an efficient way to monitor the effects of therapeutic interventions assuming a proper power analysis for treatment-induced changes is determined. Depending on the therapy, some changes found in HD synaptosomes may be better than others for detecting improvements.
ACKNOWLEDGMENTS
This work was supported by the CHDI Foundation to MD and NA, NIH NS074381 to MD, and NIH NS38194 to NA. NA is an active member of the UMMS DERC.
Footnotes
The authors have no conflicts of interest to report.
REFERENCES
- 1.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14(5):1075–81. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 2.Smith R, Brundin P, Li JY. Synaptic dysfunction in Huntington’s disease: A new perspective. Cell Mol Life Sci. 2005;62(17):1901–12. doi: 10.1007/s00018-005-5084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozas JL, Gomez-Sanchez L, Tomas-Zapico C, Lucas JJ, Fernandez-Chacon R. Presynaptic dysfunction in Hunting-’s disease. Biochem Soc Trans. 2010;38(2):488–92. doi: 10.1042/BST0380488. [DOI] [PubMed] [Google Scholar]
- 4.Nithianantharajah J, Hannan AJ. Dysregulation of synaptic proteins, dendritic spine abnormalities and pathological plasticity of synapses as experience-dependent mediators of cognitive and psychiatric symptoms in Huntington’s disease. Neuroscience. 2013;251:66–74. doi: 10.1016/j.neuroscience.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Block-Galarza J, Chase KO, Sapp E, Vaughn KT, Vallee RB, DiFiglia M, Aronin N. Fast transport and retrograde movement of huntingtin and HAP 1 in axons. Neuroreport. 1997;8(9-10):2247–51. doi: 10.1097/00001756-199707070-00031. [DOI] [PubMed] [Google Scholar]
- 6.Xu Q, Huang S, Song M, Wang CE, Yan S, Liu X, Gaertig MA, Yu SP, Li H, Li S, Li XJ. Synaptic mutant huntingtin inhibits synapsin-1 phosphorylation and causes neurological symptoms. J Cell Biol. 2013;202(7):1123–38. doi: 10.1083/jcb.201303146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kegel-Gleason KB. Huntingtin interactions with membrane phospholipids: Strategic targets for therapeutic intervention. J Huntington’s Disease. 2013;2:239–50. doi: 10.3233/JHD-130068. [DOI] [PubMed] [Google Scholar]
- 8.Kegel KB, Sapp E, Alexander J, Valencia A, Reeves P, Li X, Masso N, Sobin L, Aronin N, DiFiglia M. Polyglutamine expansion in huntingtin alters its interaction with phospholipids. J Neurochem. 2009;110(5):1585–97. doi: 10.1111/j.1471-4159.2009.06255.x. [DOI] [PubMed] [Google Scholar]
- 9.Valencia A, Reeves PB, Sapp E, Li X, Alexander J, Kegel KB, Chase K, Aronin N, DiFiglia M. Mutant huntingtin and glycogen synthase kinase 3-beta accumulate in neuronal lipid rafts of a presymptomatic knock-in mouse model of Huntington’s disease. J Neurosci Res. 2010;88(1):179–90. doi: 10.1002/jnr.22184. [DOI] [PubMed] [Google Scholar]
- 10.Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, Cha GH, Ukani L, Chepanoske CL, Zhen Y, Sahasrabudhe S, Olson J, Kurschner C, Ellerby LM, Peltier JM, Botas J, Hughes RE. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3(5):e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero E, Cha GH, Verstreken P, Ly CV, Hughes RE, Bellen HJ, Botas J. Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length huntingtin accumulating in the cytoplasm. Neuron. 2008;57(1):27–40. doi: 10.1016/j.neuron.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 13.Hickey MA, Kosmalska A, Enayati J, Cohen R, Zeitlin S, Levine MS, Chesselet MF. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington’s disease mice. Neuroscience. 2008;157(1):280–95. doi: 10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer B, Jenny M, Fresser F, Uberall F, Baier G. AKT1/PKBalpha is recruited to lipid rafts and activated downstream of PKC isotypes in CD3-induced T cell signaling. FEBS Lett. 2003;541(1-3):155–62. doi: 10.1016/s0014-5793(03)00287-4. [DOI] [PubMed] [Google Scholar]
- 15.Huo H, Guo X, Hong S, Jiang M, Liu X, Liao K. Lipid rafts/caveolae are essential for insulin-like growth factor-1 receptor signaling during 3T3-L1 preadipocyte differentiation induction. J Biol Chem. 2003;278(13):11561–9. doi: 10.1074/jbc.M211785200. [DOI] [PubMed] [Google Scholar]
- 16.Pike LJ, Han X, Gross RW. Epidermal growth factor receptors are localized to lipid rafts that contain a balance of inner and outer leaflet lipids: A shotgun lipidomics study. J Biol Chem. 2005;280(29):26796–804. doi: 10.1074/jbc.M503805200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill MM, Feng J, Hemmings BA. Identification of a plasma membrane Raft-associated PKB Ser473 kinase activity that is distinct from ILK and PDK1. Curr Biol. 2002;12(14):1251–5. doi: 10.1016/s0960-9822(02)00973-9. [DOI] [PubMed] [Google Scholar]
- 18.Arcaro A, Aubert M, Espinosa del Hierro ME, Khanzada UK, Angelidou S, Tetley TD, Bittermann AG, Frame MC, Seckl MJ. Critical role for lipid raft-associated Src kinases in activation of PI3K-Akt signalling. Cell Signal. 2007;19(5):1081–92. doi: 10.1016/j.cellsig.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Valencia A, Sapp E, Masso N, Alexander J, Reeves P, Kegel KB, Aronin N, Difiglia M. Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J Neurosci. 2010;30(13):4552–61. doi: 10.1523/JNEUROSCI.5865-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajos F. An improved method for the preparation of synaptosomal fractions in high purity. Brain Res. 1975;93(3):485–9. doi: 10.1016/0006-8993(75)90186-9. [DOI] [PubMed] [Google Scholar]
- 21.Valencia A, Sapp E, Kimm JS, McClory H, Reeves PB, Alexander J, Ansong KA, Masso N, Frosch MP, Kegel KB, Li X, Difiglia M. Elevated NADPH oxidase activity contributes to oxidative stress and cell death in Huntington’s disease. Hum Mol Genet. 2013;22(6):1112–31. doi: 10.1093/hmg/dds516. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.McClure-Begley TD, King NM, Collins AC, Stitzel JA, Wehner JM, Butt CM. Acetylcholine-stimulated [3H]GABA release from mouse brain synaptosomes is modulated by alpha4beta2 and alpha4alpha5beta2 nicotinic receptor sub-types. Mol Pharmacol. 2009;75(4):918–26. doi: 10.1124/mol.108.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill MP, Brotchie JM. Control of glutamate release by calcium channels and kappa-opioid receptors in rodent and primate striatum. Br J Pharmacol. 1999;127(1):275–83. doi: 10.1038/sj.bjp.0702523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valencia A, Sapp E, Reeves PB, Alexander J, Masso N, Li X, Kegel KB, DiFiglia M. Reagents that block neuronal death from Huntington’s disease also curb oxidative stress. Neuroreport. 2012;23(1):10–5. doi: 10.1097/WNR.0b013e32834d92e6. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8(12):2759–69. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal Chem. 2009;81(18):7757–65. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Searle BC. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10(6):1265–9. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 28.Radulovic D, Jelveh S, Ryu S, Hamilton TG, Foss E, Mao Y, Emili A. Informatics platform for global proteomic profiling and biomarker discovery using liquid chromatographytandem mass spectrometry. Mol Cell Proteomics. 2004;3(10):984–97. doi: 10.1074/mcp.M400061-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Krauss S, Griesche N, Jastrzebska E, Chen C, Rutschow D, Achmuller C, Dorn S, Boesch SM, Lalowski M, Wanker E, Schneider R, Schweiger S. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1-PP2A protein complex. Nat Commun. 2013;4:1511. doi: 10.1038/ncomms2514. [DOI] [PubMed] [Google Scholar]
- 30.Smith R, Klein P, Koc-Schmitz Y, Waldvogel HJ, Faull RL, Brundin P, Plomann M, Li JY. Loss of SNAP-25 and rabphilin 3a in sensory-motor cortex in Huntington’s disease. J Neurochem. 2007;103(1):115–23. doi: 10.1111/j.1471-4159.2007.04703.x. [DOI] [PubMed] [Google Scholar]
- 31.Freeman W, Morton AJ. Regional and progressive changes in brain expression of complexin II in a mouse transgenic for the Huntington’s disease mutation. Brain Res Bull. 2004;63(1):45–55. doi: 10.1016/j.brainresbull.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Goto S, Hirano A. Synaptophysin expression in the striatum in Huntington’s disease. Acta Neuropathol. 1990;80(1):88–91. doi: 10.1007/BF00294227. [DOI] [PubMed] [Google Scholar]
- 33.Cepeda C, Hurst RS, Calvert CR, Hernandez-Echeagaray E, Nguyen OK, Jocoy E, Christian LJ, Ariano MA, Levine MS. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J Neurosci. 2003;23(3):961–9. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nithianantharajah J, Barkus C, Murphy M, Hannan AJ. Gene-environment interactions modulating cognitive function and molecular correlates of synaptic plasticity in Huntington’s disease transgenic mice. Neurobiol Dis. 2008;29(3):490–504. doi: 10.1016/j.nbd.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Rozas JL, Gomez-Sanchez L, Tomas-Zapico C, Lucas JJ, Fernandez-Chacon R. Increased neurotransmitter release at the neuromuscular junction in a mouse model of polyglutamine disease. J Neurosci. 2011;31(3):1106–13. doi: 10.1523/JNEUROSCI.2011-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobsson G, Piehl F, Meister B. VAMP-1 and VAMP-2 gene expression in rat spinal motoneurones: Differential regulation after neuronal injury. Eur J Neurosci. 1998;10(1):301–16. doi: 10.1046/j.1460-9568.1998.00050.x. [DOI] [PubMed] [Google Scholar]
- 37.Bao J, Sharp AH, Wagster MV, Becher M, Schilling G, Ross CA, Dawson VL, Dawson TM. Expansion of polyglutamine repeat in huntingtin leads to abnormal protein interactions involving calmodulin. Proc Natl Acad Sci U S A. 1996;93(10):5037–42. doi: 10.1073/pnas.93.10.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai Y, Dudek NL, Li Q, Fowler SC, Muma NA. Striatal expression of a calmodulin fragment improved motor function, weight loss, and neuropathology in the R6/2 mouse model of Huntington’s disease. J Neurosci. 2009;29(37):11550–9. doi: 10.1523/JNEUROSCI.3307-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudek NL, Dai Y, Muma NA. Neuroprotective effects of calmodulin peptide 76-121aa: Disruption of calmodulin binding to mutant huntingtin. Brain Pathol. 2010;20(1):176–89. doi: 10.1111/j.1750-3639.2008.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zainelli GM, Ross CA, Troncoso JC, Fitzgerald JK, Muma NA. Calmodulin regulates transglutaminase 2 cross-linking of huntingtin. J Neurosci. 2004;24(8):1954–61. doi: 10.1523/JNEUROSCI.4424-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal A, Severin F, Hopfner S, Zerial M. Regulation of endosome dynamics by Rab5 and Huntingtin-HAP40 effector complex in physiological versus pathological conditions. Methods Enzymol. 2008;438:239–57. doi: 10.1016/S0076-6879(07)38017-8. [DOI] [PubMed] [Google Scholar]
- 42.Pal A, Severin F, Lommer B, Shevchenko A, Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington’s disease. J Cell Biol. 2006;172(4):605–18. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doig NM, Moss J, Bolam JP. Cortical and thalamic innervation of direct and indirect pathway medium-sized spiny neurons in mouse striatum. J Neurosci. 2010;30(44):14610–8. doi: 10.1523/JNEUROSCI.1623-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wojcik SM, Rhee JS, Herzog E, Sigler A, Jahn R, Takamori S, Brose N, Rosenmund C. An essential role for vesicular glutamate transporter 1 (VGLUT1) in postnatal development and control of quantal size. Proc Natl Acad Sci U S A. 2004;101(18):7158–63. doi: 10.1073/pnas.0401764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calabresi P, De Murtas M, Pisani A, Stefani A, Sancesario G, Mercuri NB, Bernardi G. Vulnerability of medium spiny striatal neurons to glutamate: Role of Na+/K + ATPase. Eur J Neurosci. 1995;7(8):1674–83. doi: 10.1111/j.1460-9568.1995.tb00689.x. [DOI] [PubMed] [Google Scholar]
- 46.Luthi-Carter R, Apostol BL, Dunah AW, DeJohn MM, Farrell LA, Bates GP, Young AB, Standaert DG, Thompson LM, Cha JH. Complex alteration of NMDA receptors in transgenic Huntington’s disease mouse brain: Analysis of mRNA and protein expression, plasma membrane association, interacting proteins, and phosphorylation. Neurobiol Dis. 2003;14(3):624–36. doi: 10.1016/j.nbd.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa T, Engler JA, Sheng M. The dynamic turnover and functional roles of alpha-actinin in dendritic spines. Neuropharmacology. 2004;47(5):734–45. doi: 10.1016/j.neuropharm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 48.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227(4688):770–3. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 49.Rong J, McGuire JR, Fang ZH, Sheng G, Shin JY, Li SH, Li XJ. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J Neurosci. 2006;26(22):6019–30. doi: 10.1523/JNEUROSCI.1251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andre VM, Cepeda C, Fisher YE, Huynh M, Bardakjian N, Singh S, Yang XW, Levine MS. Differential electrophysiological changes in striatal output neurons in Huntington’s disease. J Neurosci. 2011;31(4):1170–82. doi: 10.1523/JNEUROSCI.3539-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andre VM, Fisher YE, Levine MS. Altered balance of activity in the striatal direct and indirect pathways in mouse models of Huntington’s disease. Front Syst Neurosci. 2011;5:46. doi: 10.3389/fnsys.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalle-Donne I, Rossi R, Giustarini D, Gagliano N, Di Simplicio P, Colombo R, Milzani A. Methionine oxidation as a major cause of the functional impairment of oxidized actin. Free Radic Biol Med. 2002;32(9):927–37. doi: 10.1016/s0891-5849(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 53.Lim JC, Kim G, Levine RL. Stereospecific oxidation of calmodulin by methionine sulfoxide reductase A. Free Radic Biol Med. 2013;61C:257–64. doi: 10.1016/j.freeradbiomed.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoshi T, Heinemann S. Regulation of cell function by methio-nine oxidation and reduction. J Physiol. 2001;531(Pt 1):1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu YR, Narhi LO, Spahr C, Langley KE, Lu HS. In vitro methionine oxidation of Escherichia coli-derived human stem cell factor: Effects on the molecular structure, biological activity, and dimerization. Protein Sci. 1996;5(6):1165–73. doi: 10.1002/pro.5560050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein JC, Moen RJ, Smith EA, Titus MA, Thomas DD. Structural and functional impact of site-directed methionine oxidation in myosin. Biochemistry. 2011;50(47):10318–27. doi: 10.1021/bi201279u. [DOI] [PMC free article] [PubMed] [Google Scholar]