Abstract
The SENP proteases regulate the SUMO conjugates in the cell by cleaving SUMO from target proteins. SENP6 and SENP7 are the most divergent members of the SENP/ULP protease family in humans by the presence of insertions in their catalytic domains. Loop1 insertion is determinant for the SUMO2/3 activity and specificity on SENP6 and SENP7. To gain structural insights into the role of Loop1, we have designed a chimeric SENP2 with the insertion of Loop1 into its sequence. The structure of SENP2-Loop1 in complex with SUMO2 was solved at 2.15 Å resolution, and reveals the details of an interface exclusive to SENP6/7 and the formation of unique contacts between both proteins. Interestingly, functional data with SUMO substrates showed an increase of the proteolytic activity in the SENP2-Loop1 chimera for diSUMO2 and polySUMO2 substrates.
Keywords: SUMO, polySUMO chain, ubiquitin-like protein, SENP6, SENP7, SENP2
Introduction
After discovery of ubiquitin, an entire family of small ubiquitin-like proteins has emerged, with new members still being added.1 Of these proteins, SUMO (small ubiquitin-like modifier), which shares 18% sequence identity with ubiquitin, has been one of the most scrutinized.2–4 Since its discovery in 1996, hundreds of SUMO substrates have been identified and it has been implicated in a wide range of cellular processes including but not limited to: nuclear transport, DNA repair, DNA transcription, and the cell cycle.5,6
In humans there are four SUMO isoforms, SUMO1-4. SUMO2 and-3 share 95% sequence identity and are commonly referred to as SUMO2/3, while SUMO1 shares only 50% sequence identity with either of the two. SUMO4 shares 87% sequence identity with SUMO2, but it still remains unknown whether SUMO4 is processed or conjugated to cellular proteins.7 Although SUMO1 and SUMO2/3 share the same overall structure and essentially the same conjugation machinery, the different SUMO isoforms seem functionally distinct; some substrates can be exclusively modified by SUMO1 or SUMO2/3, whereas others can be modified by both SUMO isoforms.8–10 While the vast majority of SUMO1 exists in conjugated species, there is usually a free pool of SUMO2/3 species in cells8 and SUMO2/3 is strongly induced in response to in vivo heat shock and oxidative stress.11
The SUMO conjugation/deconjugation pathway is analogous to that of ubiquitin, represented by an enzyme cascade resulting in the linkage of SUMO to the epsilon amino of a lysine residue from a target protein.5,10 In this pathway the proteolytic enzymes associated with the maturation of the SUMO precursor and with substrate deconjugation belong to the SENP/ULP family, which is comprised by six members in humans (SENP1-3 and SENP5-7) and two members in budding yeast (ULP1 and ULP2).12–14 However, recently two new SUMO proteases have been identified in humans not belonging to the SENP/ULP family, desumoylating isopeptidase 1 & 2 (DESI 1 & 2) and ubiquitin-specific protease-like 1 (USPL1).15,16
Sequence comparison of the conserved cysteine protease catalytic domain can divide the SENP/ULP family into two subgroups: one containing ULP1 (SENP1, SENP2, SENP3, and SENP5); and one containing ULP2 (SENP6 and SENP7).13 The second group consists of proteins containing long amino acid insertions within their catalytic domains, such as in the case of human SENP6 and SENP7, whose catalytic domains contain 150 and 50 amino acid residue inserts, respectively. The C-terminal catalytic domain persists through all of the SENP/ULPs.17 The N-termini are non-conserved and vary between each protease. The N-terminal domains of the SENP/ULP family have been shown to direct sub-cellular localization and substrate specificity but their full function within the cell has yet to be determined.18–21
In vitro studies have shown that the SENP/ULP conserved C-terminal catalytic domain can discriminate between SUMO isoforms in deconjugation (cleavage of SUMO-conjugated substrates) and in processing reactions (maturation of SUMO precursors).22,23 For example, the catalytic domains of SENP1 and SENP2 are able to efficiently both cleave SUMO1 and SUMO2/3 from conjugated substrates in vitro, but display different abilities in processing SUMO precursors, whereby the reaction is mostly dependent on the residues C-terminal to the di-glicine motif of the SUMO precursors.24,25
SENP6 and SENP7 show little in vitro activity in processing reactions of SUMO precursors.26,27 Both enzymes, however, have demonstrated activity in deconjugating reactions with SUMO substrates, in particular against polySUMOylated chains, and show isoform specificity for SUMO2/3 over SUMO1.26–28 As mentioned before, SENP6 and SENP7 are characterized by the presence of long insertions in their catalytic domains. In vitro deconjugation assays with constructs lacking these inserts, however, show similar functional activities, leaving the relevance of these large insertions to be elucidated.26,27 Nevertheless, Loop1 insertion, a short 8-residue insertion unique to SENP6 and SENP7, is essential for the in vitro proteolytic activity and is determinant for the SUMO2/3 isoform specificity of SENP6 and SENP7.
To get the structural details of the interface between Loop1 and SUMO2 and after unfruitful crystallization trials with SENP6 and SENP7, we have constructed a chimeric mutant of SENP2 containing the Loop1 insertion from SENP6 (SENP2-Loop1). The crystal structure of SENP2-Loop1 in complex with SUMO2 was determined at 2.15 Å, and structural examination reveals key shifts of both SUMO2 and SENP2-Loop1 at the interface between both proteins. The disclosed structural details for the Loop1 interaction with SUMO2/3 support previous mutagenesis analysis. New biochemical data indicate an increase of activity for the chimeric SENP2-Loop1 for some SUMO2 substrates, in particular diSUMO2/3 and polySUMO2/3 substrates, suggesting a role of the SENP6/7 Loop1 insertion in chain dismantling.
Results
Production of the SENP2-Loop1 chimera
The proteolytic activity of the four sequence insertions located in the catalytic domain of SENP7 and SENP6 (named Loop1 to Loop4) has been partially characterized in vitro.26,27 Of special interest was the relevance of the Loop1 insertion for activity, unique to SENP6 and SENP7 proteases, which presents only a single amino acid substitution between these two proteases (alanine and threonine for SENP6 and SENP7, respectively) (Fig. 1). Loop1 is comprised of eight residues, consisting of four prolines, forming a poly-proline helix motif, a lysine residue at the center just C-terminal to the poly-proline helix, and two glycine residues (Fig. 1). The crystal structure of the apo SENP7 revealed that Loop1 is structured and its deletion produced marked defects in the proteolytic activities of both SENP6 and SENP7.26,27
Figure 1.
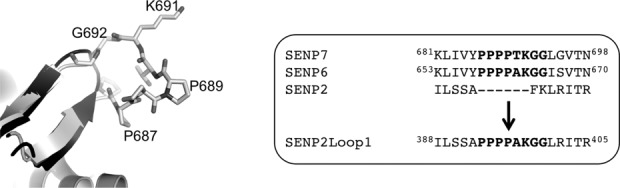
Design of the SENP2-Loop1 chimera. A: Structural comparison between the Loop1 structure of SENP7 (PDB code 3EAY) and SENP2 (PDB code 2IO0). Residues are labeled and shown in stick configuration. Right, sequence alignment of the residues forming the Loop1 insertion in SENP6, SENP7, and SENP2 and the formation of the chimera SENP2-Loop1. Inserted residues are shown in bold character.
To get direct structural details of the Loop1 interface with SUMO2/3, we introduced Loop1 into the catalytic domain of SENP2. The SENP6 Loop1 sequence was inserted at position 392 of SENP2 (PPPPAKGG), resulting in an insertion of six amino acids (PPPPAK) and the mutation of two amino acids (F393G and K394G) in the SENP2 sequence (Fig. 1). Loop1 insertion is located between β1 and β2 strands in SENP6/7, and only represents a two-residue hairpin connector between these two strands in SENP2 (Fig. 1). Insertion of Loop1 into SENP2 at this site would presumably not interfere much structurally at this region, but rather elongate the already present beta sheet loop connector. Activity assays showed that insertion of Loop1 into SENP2 does not compromise and in some cases even enhances the proteolytic activity of SENP2 (see later).
Crystal structure of SENP2-Loop1 insertion in complex with SUMO2
After unfruitful crystallization trials between SUMO2/3 substrates with SENP6 and SENP7 proteases, we were able to obtain the crystal structure of SUMO2 in complex with the chimeric SENP2-Loop1 protease at 2.1 Å resolution. An active site mutant of SENP2 (Cys548 for serine) was used to form a stable complex with the SUMO2 precursor that was subsequently purified by gel filtration. Both components of the complex, SUMO2 and SENP2-Loop1 protease could be completely traced in the electron density maps, including the Loop1 insertion [Fig. 3(C)].
Figure 3.
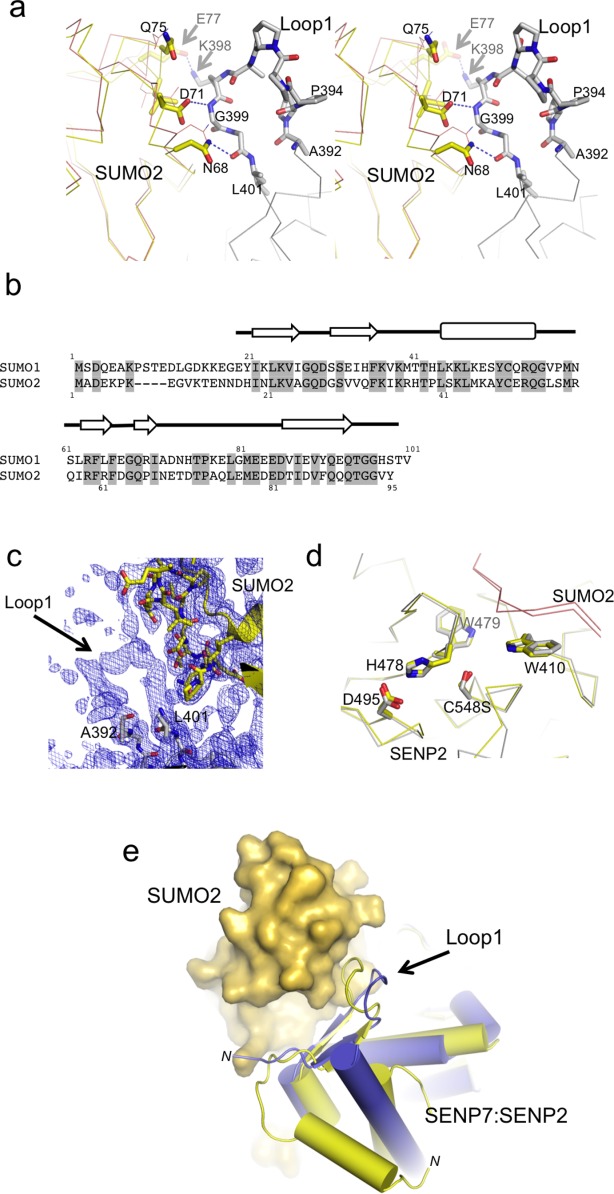
Structural details of the Loop1 interaction with SUMO2. A: Close-up stereo view of the residues involved in the interface between SUMO2 and Loop1 insertion from SENP2. SUMO2 from the present complex is shown superimposed with SUMO2 from the complex with SENP2 (PDB code 2IO0), which is shown in a thin orange line. Major residues involved in the interface are labeled and shown in stick representation. B: Structural alignment of human SUMO1 and SUMO2 proteins. Numbering and secondary structure elements are shown above sequence. C: Composite 2fofc omit map covering the region corresponding to Loop1 contoured at 0.9 σ. D: Comparative stick representation of the active site of SENP2 and SENP2-Loop1 in complex with SUMO2 precursors. E: Ribbon representation of the superposition between SENP2-Loop1/SUMO2 and SENP7 (PDB code 3EAY) depicting the movement of Loop1 upon interaction with SUMO2. SENP7 is colored blue, SENP2 yellow, and SUMO2 is represented as a yellow surface. N-termini of SENP2 and SENP7 are labeled.
Comparison of the structural alignment of SUMO2 in complex with SENP2 and with SENP2-Loop1 reveals a shift of SUMO2 moiety, which we attribute to the presence of interactions between SUMO2 and Loop1. Structural analysis with the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int) showed a larger interface area for SUMO2 in complex with SENP2-Loop1, displaying interfaces of 1153 Å2 and 1046 Å2 for SENP2-Loop1 and SENP2 complexes, respectively. Sixty-two residues are involved in the interface of SENP2-Loop1 with SUMO2, while 55 residues are involved in the SENP2 complex with SUMO2.
Structural alignment of SENP2 indicates that the novel interface produces an approximately 2–3° angle rotation in a quite extended region of SUMO229 (Fig. 2). The major changes are observed within the long turn between Pro66 and Thr83 in contact with Loop1 and in the region between residues Lys33 and Ile58, which includes the short alpha helix of the SUMO2 structure (Fig. 2). The SUMO2 residues in these two regions are displaced around 2–3 Å between both complexes. The alignment reveals a better superposition of SUMO2 residues opposite to the Loop1 interface with SENP2, in contrast to the marked shift of SUMO2 residues next to Loop1. These observations are supported by the B-factor analysis of the complex, displaying higher values in the SUMO elements next to Loop1 [Fig. 2(C)].
Figure 2.
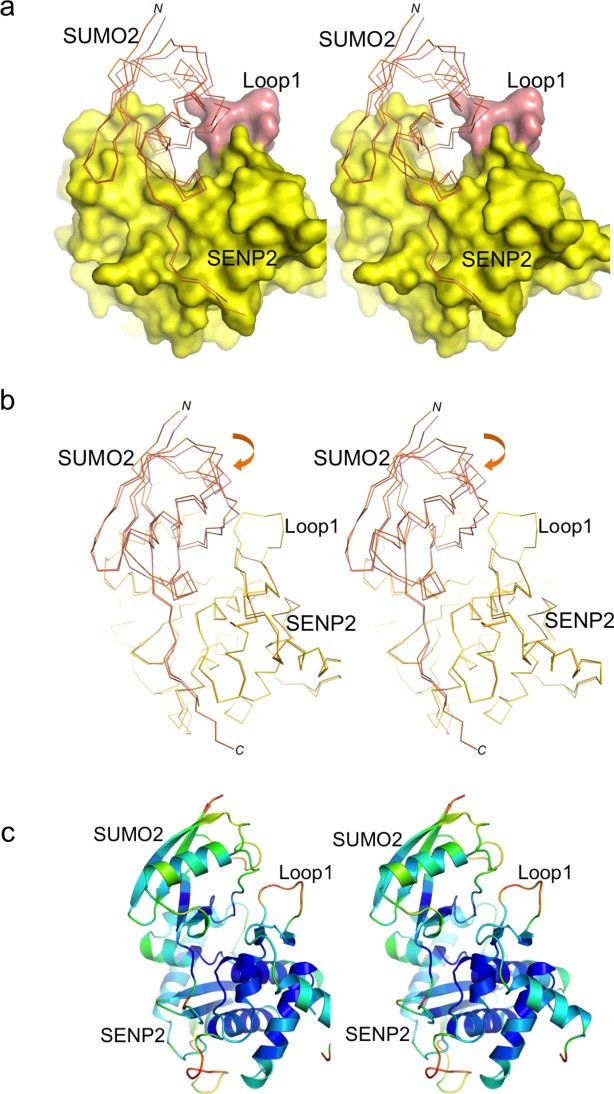
Structural comparison of SUMO2 in complex to SENP2-Loop1 and SENP2. A: Stereo representation of the complex between SENP2-Loop1, shown in as surface representation, and SUMO2, shown as a thin line. SUMO2 is shown superimposed with SUMO2 from the complex with SENP2 (PDB code 2IO0). Loop1 insertion is shown in orange. B: Stereo backbone representation of the superposition between SENP2-Loop1 (yellow) and SENP2 (orange, PDB code 2IO0) in complex with SUMO2. C: B-factor color-based stereo cartoon representation of the SENP2-Loop1:SUMO2 complex.
Structural characterization of the interface between Loop1 insertion and SUMO2
The extended interface includes several polar and van der Waals interactions between SUMO2 and Loop1 insertion, including six SUMO2 residues between Asn68 and Glu77 not observed in the previous SUMO2-SENP2 complex29 [Fig. 3(A)]. Mutagenesis and biochemical analysis revealed the role of Asn68 and Asp71 in the specificity of SENP6/7 for SUMO2/3 over SUMO1.27 The present structure directly reveals hydrogen bond interactions between these two residues with Loop1, in particular the side chain of Asn68 is at 2.9 Å distance to the carbonyl oxygen of Gly400, and the side-chain of Asp71 is at 3.5 Å to the backbone nitrogen of Gly399 (using the SENP2-Loop1 numbering). The chemical nature of these interactions favors the interaction of Loop1 with SUMO2/3, in comparison to alanine and histidine, the corresponding residues in the SUMO1 isoform, thus explaining the SUMO2/3 isoform preference displayed in SENP6/7 proteases [see SUMO alignment in Fig. 3(B)].
Lys398, which is located at the center of the Loop1 insertion and as inferred from its high side-chain B-factors might be flexible, could also interact with a negative pocket created by the side chain of Glu77 (at 2.3 Å) and the carbonyl oxygen of Gln75 (at 3.8 Å). Lys398 (Lys691 in SENP7 numbering) plays an important role in the activity of the SENP6/7, with a dramatic reduction of the activity by a point mutation to glutamic acid and a partial reduction by a point mutation to alanine.27 The present structure suggests that the positive charge of Lys398 might participate in the interaction between Loop1 and SUMO2, perhaps by establishing contacts with the aforementioned negative pocket. It is worth noting that SUMO2 Glu77 is substituted by a glycine residue in SUMO1, perhaps contributing to the reduced proteolytic activities showed by SENP6 and SENP7 against the SUMO1 isoform.
Finally, in addition to the movement of SUMO2 towards the Loop1 region, structural alignments between SENP7 and SENP2-Loop1 reveal a displacement of the Loop1 insertion towards SUMO2 [Fig. 3(E)]. An important structural element contained in the Loop1 insertion is the poly-proline helical motif, in which the substitution of the four consecutive proline residues for glycine produced a dramatic reduction of the proteolytic activity of SENP7 against SUMO2 substrates.27 In the present complex structure, despite the displacement towards the SUMO2 interface, the integrity of the Loop1 insertion seems to be maintained. Thus the formation of the extended interface between Loop1 and SUMO2 still preserves the structural integrity of the poly-proline helical motif described in the apo SENP7 structure.
Comparative proteolytic analysis of the SENP2-Loop1 chimera
Deconjugation assays with SENP2 and SENP2-Loop1 chimera were run using diSUMO2 and polySUMO2 substrates [Fig. 4(A)]. End-point activity assays using 0.05, 0.5, and 5 nM enzyme concentration against diSUMO2 showed complete cleavage by SENP2-Loop1 at only 0.05 nM enzyme concentration after the allotted time period, while only ∼20% of the substrate was cleaved by SENP2 in the same condition. Comparable deconjugation of SUMO2 from diSUMO2 was achieved by SENP2 only at a concentration 100x (5 nM) that of SENP2-Loop1. We next run time-course assays using polySUMO2 as a substrate at 0.5 nM fixed concentration of SENP2 and SENP2-Loop1. As seen in the assays results [Fig. 4(B)], SENP2-Loop1 showed elevated rates of deconjugation compared to that of the SENP2, with complete dismantling of polySUMO2 into constituent SUMO2 after 30 min, while SENP2 was only able to achieve complete deconjugation after 120 min. Interestingly SENP2-Loop1 showed around 80% dismantling of polySUMO2 chains after only 5 min.
Figure 4.
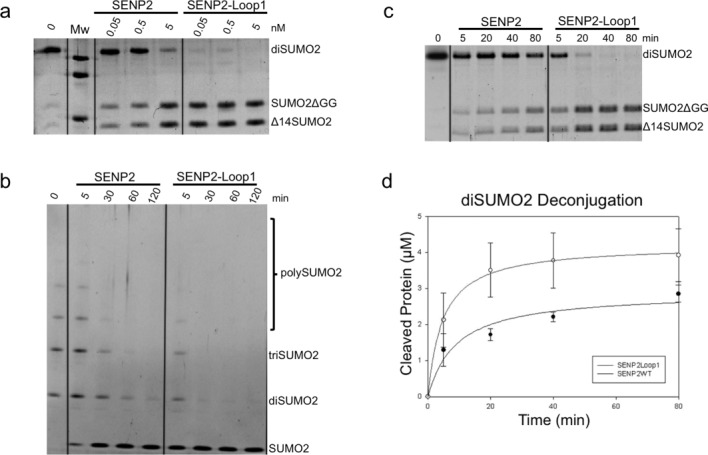
Comparison of the deconjugation of SENP2-Loop1 and SENP2 for diSUMO2 and polySUMO2 chains. A: Deconjugation activity assays of SENP2 and SENP2Loop1 against diSUMO2. Assays were run at 0.05, 0.5, and 5 nM enzyme concentrations and 0.5 μM substrate concentration at 37°C and stopped after 25 min with loading buffer and analyzed by SDS-PAGE. B: Time course deconjugation assays using SENP2 and SENP2-Loop1 against polySUMO2. Reactions were run at 0.5 nM enzyme concentration and time intervals are indicated above each lane in minutes. Reactions were stopped at each respective interval with loading buffer and analyzed by SDS-PAGE. Proteins in both assays were detected by staining with SYPRO (Bio-Rad). C: Time course deconjugation assay using SENP2 and SENP2-Loop1 against diSUMO2. Reactions were run at 0.05 nM enzyme concentration and time intervals are indicated above each lane in minutes. D: Representation of the initial velocity of diSUMO2 deconjugation by SENP2 and SENP2-Loop1 taken at 0, 5, 20, 40 and 80 min. Axes are labeled and error bars were obtained by conducting the assays in triplicate.
Time-course assays using SENP2 and SENP2-Loop1 against diSUMO2 substrate were also run at a fixed enzyme concentration of 0.05 nM [Fig. 4(C,D)]. As previously observed, SENP2-Loop1 showed an elevated activity compared to that of SENP2 and showed ∼20% diSUMO2 cleavage after 5 min and complete cleavage after 20 min. SENP2, on the other hand, achieved only ∼50% cleavage of diSUMO2 after 80 min. These experiments show that the SENP2-Loop1 chimera is able to cleave multi-SUMO2/3ylated species at a rate almost 8x times that of the wild type SENP2. This activity can, presumably, be attributed to the additional interface with SUMO2 provided by the eight residue insert in SENP2-Loop1 and suggests an active role for the Loop1 insertion of SENP6/7 in the dismantling of polySUMO2/3 chains, even in the context of SENP2.
We previously showed that Loop1 is not only indispensable for the activity of SENP6 and SENP7, but that key elements of Loop1 are responsible for the isoform specificity of both enzymes for SUMO2/3 over SUMO1.27 In order to check the effect of Loop1 in SENP2 against SUMO1 and SUMO2/3 isoforms, additional time-course assays using our “canonical” substrates RanGAP1-SUMO1 and RanGAP1-SUMO2 were run (Fig. 5). Although less dramatic than in the diSUMO2 substrate, the Loop1 insertion adversely affected the ability of SENP2 to deconjugate RanGAP1-SUMO1 compared to that of SENP2, but had a positive effect on the ability of the enzyme to deconjugate SUMO2 from RanGAP1-SUMO2. This is presumably due to the ability of Loop1 in SENP2-Loop1 to interact favorably with the residues of SUMO2/3 at the Loop1-SUMO2 interface in a manner comparable to that of SENP6/7.
Figure 5.
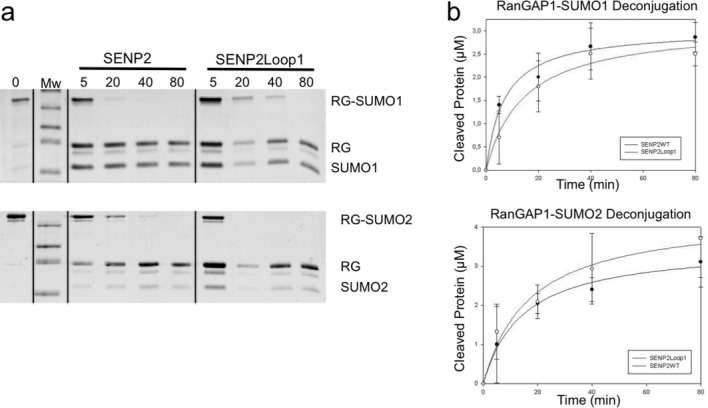
Comparison of the deconjugation of SENP2-Loop1 and SENP2 for RanGAP1-SUMO1 and RanGAP1-SUMO2. A: Time course assay using SENP2 and SENP2-Loop1 against RanGAP1-SUMO1 and RanGAP1-SUMO2. Reactions were run at 0.5 nM enzyme concentration and time intervals are indicated above each lane in minutes. Reactions were stopped at each respective interval with loading buffer and analyzed by SDS-PAGE. B: Representation of the initial velocities of RanGAP1-SUMO1 and RanGAP1-SUMO2 deconjugation by SENP2 and SENP2-Loop1 taken at 0, 5, 20, 40, and 80 min. Axes are labeled and error bars were obtained by conducting the assays in triplicate.
Discussion
SENP6 and SENP7 are the most divergent members of the SENP/ULP protease family, with insertions of varying lengths dispersed throughout their catalytic domains. Of special relevance is the Loop1 insertion, whose deletion or point mutation seriously compromises the proteolytic activity of SENP6 and SENP7. In particular, the structural conformation of Loop1, with the presence of a poly-proline helical motif, and the charge properties of Lys691 (SENP7 numbering) were shown to be key for the correct proteolytic activity of SENP7.27 Additionally, a second role for the Loop1 insertion alluded to its part in determining the isoform preference for SUMO2/3 over SUMO1, a characteristic restricted to SENP6 and SENP7 family members. No other SENP/ULP family members contain the Loop1 insertion in their catalytic domain, but in those cases this lack of sequence does not impede their proteolytic activity, or their ability to distinguish among SUMO isoforms.22,23 Our structural model predicted the formation of specific contacts between the SUMO2/3 surface and the Loop1 insertion of both SENP6 and SENP7.27 Unfortunately, after numerous crystallization trials, we were not able to get the structure of any SUMO2/3 complex with either SENP6 or SENP7. In order to gain structural and functional details of this tentative interface involving the Loop1 insertion, we designed a chimeric SENP2 construct containing the Loop1 insertion of SENP6 inside the catalytic domain of SENP2, mimicking the potential interactions that would take place in the SENP6 and SENP7 proteases. SENP2 is a well-characterized SUMO protease with a broader isoform activity than SENP6/7 and lacking the Loop1 insertion in its sequence.29
Although the crystal structure of the chimeric SENP2-Loop1 in complex with a SUMO2 precursor presented in this work does not occur in the cellular context, we believe that it helps to reveal the structural details of this extended interface between SUMO2 and the Loop1 insertion. Structural comparison with the previous SENP2-SUMO2 structure discloses significant changes produced in the complex interface by the presence of the Loop1 insertion, which might be relevant in the context of SENP6/7. These include minor differences, such as a slight rotation of SUMO2 moiety, and more pronounced differences, such as direct contacts made by SUMO2 residues with the Loop1 insertion, which moves towards the SUMO2 surface. The complex structure shows direct interactions of the side chains of Asn68, Asp71, and Glu77 with the Loop1 insertion, thus unveiling the role of this negative patch in the SUMO2/3 isoform specificity displayed by SENP6/7 (these three residues are substituted by alanine, histidine, and glycine in SUMO1).
The observed geometry of the catalytic active site triad in the SENP2-Loop1:SUMO2 complex is similar to previous SENP2 structures (PDB ID: 2IO0) [Fig. 3(D)]. Despite the nonproductive orientation displayed in both crystal structures by the active site histidine in the catalytic triad, our functional data indicate that the SENP2 proteolytic activity is not compromised by this orientation. Our assays with a variety of SUMO substrates indicate that the presence of Loop1 insertion in SENP2 does not impede and even enhances, in some cases, the proteolytic activity against SUMO2/3 substrates. Particularly interesting is the increase in deconjugation activity for diSUMO2/3 substrates showed by the SENP2-Loop1 chimera, comparable with SENP6/7 against these types of substrates. Several groups have reported a preference of SENP6/7 proteases for the deconjugation of polySUMO2/3 chain substrates,26,30,31 although SENP2 has also been reported to possess a significant activity against polySUMO2/3 chains in vitro.32 In this work we show a significant increase (around 8-fold for diSUMO2/3) of SENP2 deconjugation activity against diSUMO2/3 and polySUMO2 chain substrates upon insertion of Loop1 into the SENP2 sequence. The mechanism by which the SENP2-Loop1 chimera is able to deconjugate better SUMO2/3ylated species remains to be eluded, but additional, specific contacts made with the SUMO2 surface could perhaps improve SENP6/7's recognition that could be important to polySUMO2/3 chain deconjugation. These direct interactions, as can be seen in our SENP2-Loop1:SUMO2 structure, might facilitate the cleavage of the isopeptidic bond between two SUMO moieties by providing a more extensive interaction surface with SENP6/7, perhaps by enabling one SUMO moiety to “dock” while cleavage is taking place.
However, in the case of other SUMO substrates, such as RanGAP1-SUMOs or SUMO precursors (data not shown), alterations in the proteolytic activity for SENP2-Loop1 are not so marked. In the context of SENP2, where the lack of the Loop1 insertion does not impede its proteolytic activity as is the case for SENP6/7, the insertion of Loop1 does not strongly affect the SUMO isoform preference. These results further validate our claim that the Loop1 insertion is a characteristic of the SENP6 and SENP7 members of the SENP/ULP family necessary for the activity of these two enzymes and whose presence likely facilitates the recognition of poly-SUMOylated species, perhaps by providing an additional interface for SUMO chains. Additional structural work will be required to describe the full role for the SENP6/7 Loop1 in the context of polySUMO2/3 chains, perhaps by the structural characterization with a diSUMO2 molecule.
Materials and Methods
Protein mutagenesis and purification
The catalytic domain of human SENP2-(364-589) was produced in E.coli and purified as described.24 All SUMO2 constructs were produced in E. coli as described previously.24
SENP6 Loop1 residues 658-666 (PPPPAKGG) were inserted into SENP2 by inserting PPPP C-terminal to residue 392 of SENP2 and AKGG N-terminal to residue 395 of SENP2 to give the following sequence: EILSSAPPPPAKGGLRIT (SENP2-Loop1), where inserted amino acids are underlined. The mutant construct was amplified by PCR. All constructs were confirmed by DNA sequencing. SENP2 mutants were purified by metal affinity chromatography and gel filtration and concentrated to 1 mg/mL in a buffer containing 25 mM Tris-HCl (pH 8.0), 350 mM NaCl, and 1 mM β-mercaptoethanol.
To prepare diSUMO2 dimer, Δ14-SUMO2 was produced as SUMO donors, and SUMO2ΔGG (deletion of the C-terminal di-Gly motif) was produced as SUMO acceptor. Both proteins were produced and purified in E.coli as described before for the wild-type.24 DiSUMO2 was formed overnight at 37°C in a reaction mixture containing 20 mM HEPES pH 7.5, 5 mM MgCl2, 0.1% Tween, 50 mM NaCl, 1 mM dithiothreitol, 2 mM ATP, 150 nM SAE1/SAE2 (E1), 100 nM Ubc9 (E2), 100 nM IR1 (E3), 32 mM Δ14SUMO2, and 16 mM SUMO2ΔGG in MilliQ water; and purified by gel filtration (Superdex75, GE Healthcare).
Purification of the complex SENP2-Loop1:Δ14-SUMO2 was performed by gel filtration (Superdex200, GE Healthcare) by mixing equimolar amounts of the two components in a buffer containing 25 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM β-mercaptoethanol. Complex peak was pooled and concentrated at 15 mg/mL.
Biochemical and kinetic assays
Titration of diSUMO2 deconjugation activity was measured by incubating diSUMO2 with purified SENP2 and SENP2-Loop1 at three different enzyme concentrations (0.05, 0.5, and 5 nM) at 37°C in a buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% Tween 20, and 2 mM dithiothreitol. Titration with canonical substrates RanGAP1-SUMO1 and RanGAP1-SUMO2 (production described in Ref.24) were measured with the same conditions. Reactions were stopped after 25 min with SDS loading buffer and analyzed by gel electrophoresis (PAGE). Proteins were detected by staining with SYPRO (Bio-Rad). Products were quantified by detecting fluorescence under UV illumination using a Gel-Doc apparatus with associated integration software (Quantity-One; Bio-Rad).
Deconjugation time course reactions were performed with similar buffer conditions as for the end-point reactions. SENP2 and SENP2-Loop1 were incubated with diSUMO2 and polySUMO2 (Boston Biochem) at 0.05 nM and 0.5 nM enzyme concentration, respectively and substrates at 3 μM. Reactions were run at 37 C and stopped at 5, 20, 40 and 80 minutes for diSUMO2 and 5, 30, 60, and 120 for polySUMO2 with SDS loading buffer and analyzed by PAGE. Products were quantified by detecting fluorescence using a Gel-Doc apparatus with associated integration software (Quantity-One; Bio-Rad). All data points were fitted to a hyperbolic curve. All assays were conducted in triplicate. Error bars indicate ±1 standard deviation.
Crystallization and data collection
Crystals of the SENP2-Loop1 catalytic domain in complex with Δ14-SUMO2 were obtained at 18°C by sitting drop vapor diffusion methods. The reservoir solution contained 0.24M sodium malonate, pH 7 and 20% PEG3350. Single crystals appeared after 8 days from equal volumes of protein solution (10 mg/mL in 5 mM Tris-HCl (pH 8.0), 25 mM NaCl) and reservoir solution. Crystals were cryo-protected in reservoir buffer containing 10% glycerol and flash-frozen in liquid nitrogen prior to diffraction analysis. Diffraction data were recorded from cryo-cooled crystals (100K) at the ALBA synchrotron in Barcelona (BL13-XALOC beamline). Data were integrated and merged using XDS33 and scaled, reduced, and further analyzed using CCP4 34 (TableI).
Table I.
Data Collection and Refinement Statistics
| Data collection | SENP2-Loop1:SUMO2 |
|---|---|
| Space group | H32 |
| Cell dimensions | |
| a, b, c (Å) | 145.52, 145.52, 104.67 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å)a | 50–2.15 (2.26–2.15) |
| Rmergeb | 0.036 (0.770) |
| I/σI | 21.0 (2.2) |
| Completeness (%) | 99.8 (99.2) |
| Redundancy | 5.1 (5.0) |
| Refinement | |
| Resolution (Å) | 48–2.15 |
| No. reflections | 22034 |
| Rwork/Rfreec | 18.81/23.62 |
| No. Atoms | 2596 |
| No. aa protein | 255 |
| Water | 49 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.02 |
| Bond angles (°) | 2.34 |
| PDB code | 3ZO5 |
Statistic for highest resolution shell is shown in parentheses.
Rmerge = ∑||Ii − < I >| / ∑ Ii, where Ii is the ith measurement of the intensity of an individual reflection or its symmetry-equivalent reflections and <I> is the average intensity of that reflection and its symmetry-equivalent reflections.
Rwork = ∑||Fo| − |Fc||/∑ |Fo| for all reflections and Rfree = ∑||Fo| − |Fc||/ ∑ |Fo|, calculated based on the 5% of data excluded from refinement.
Structure determination and refinement
The structure of SENP2-Loop1 in complex with Δ14-SUMO2 precursor was determined from the x-ray data by molecular replacement using a previous SENP2-SUMO2 structure (PDB code 2IO0) as a model using the program MolRep.34 The initial electron density maps showed the Loop1 insertion trace which was manually built using the program COOT.35 Model refinement was performed with Refmac34 and Phenix.36 Ramachandran analysis shows 95.44% of residues (293) are in preferred regions, 4.23% of residues (13) are in allowed regions and 0.33% of residues (1) are in outlier regions.35 Refinement and data statistics are provided in tableI. Structural representations were prepared with PyMOL.37
Accession codes
Protein Data Bank
Coordinates and structure factors were deposited in the PDBe databank with accession code 3ZO5.
Acknowledgments
The authors acknowledge Christopher D. Lima for reagents. They acknowledge Jordi Benach and Jordi Juanhuix from the XALOC beamline (BL13) at the ALBA synchrotron for their help in the data collection.
References
- 1.Veen Van der AG, Ploegh HL. Ubiquitin-like proteins. Ann Rev Biochem. 2012;81:323–347. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 2.Hay RT. SUMO: history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Ann Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Geiss-Friedlander R, Melchoir F. Concepts in SUMOylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 5.Johnson ES. Protein modification by SUMO. Ann Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 6.Praefcke GJ, Hofmann K, Dohmen SUMO playing tag with ubiquitin. Trends Biochem Sci. 2012;37:23–31. doi: 10.1016/j.tibs.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Owerbach DA, McKay EM, Yeh ET, Gabbay KM, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 9.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and deconjugation. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossis G, Melchoir F. SUMO: regulating the regulator. Mol Cell. 2006;21:349–357. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;6724:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;6:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y. DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 2012;14:339–346. doi: 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz S, Chachami G, Kozaczkiewicz L, Winter U, Stankovic-Valentin N, Haas P, Hofmann K, Urlaub H, Ovaa H, Wittbrodt J, Meulmeester E, Melchior F. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–938. doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol Chem. 2001;5:727–733. doi: 10.1515/BC.2001.088. [DOI] [PubMed] [Google Scholar]
- 18.Bailey D, O'Hare P. Characterization of the localization and protealytic activity of SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- 19.Bacco Di A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroetz MB, Su D, Hochstrasser M. Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Mol Biol Cell. 2009;20:2196–2206. doi: 10.1091/mbc.E08-10-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goeres J, Chan PK, Mukhopadhyay D, Zhang H, Raught B, Matunis MJ. The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol Biol CelI. 2010;22:4868–4882. doi: 10.1091/mbc.E10-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282:26217–26217. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 23.Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J. 2010;2:335–344. doi: 10.1042/BJ20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;8:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 25.Shen L, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J. 2006;397:297–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem. 2008;28:32045–32055. doi: 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alegre KO, Reverter D. Swapping the small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7. J Biol Chem. 2011;286:36142–36151. doi: 10.1074/jbc.M111.268847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hattersly N, Shen L, Jaffray EG, Hay RT. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol Biol Cell. 2011;1:78–90. doi: 10.1091/mbc.E10-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol. 2006;13:1060–1068. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J. 2009;2:223–230. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- 32.Bekes M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem. 2011;12:10238–10247. doi: 10.1074/jbc.M110.205153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Cryst. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrödinger L. 2010. The PyMOL Molecular Graphics System, Version 1.3.


