Abstract
Resuscitation promoting factor (Rpf) proteins, which hydrolyze the sugar chains in cell-wall peptidoglycan (PG), play key roles in prokaryotic cell elongation, division, and escape from dormancy to vegetative growth. Like other bacteria, Mycobacterium tuberculosis (Mtb) expresses multiple Rpfs, none of which is individually essential. This redundancy has left unclear the distinct functions of the different Rpfs. To explore the distinguishing characteristics of the five Mtb Rpfs, we determined the crystal structure of the RpfE catalytic domain. The protein adopts the characteristic Rpf fold, but the catalytic cleft is narrower compared to Mtb RpfB. Also in contrast to RpfB, in which the substrate-binding surfaces are negatively charged, the corresponding RpfE catalytic pocket and predicted peptide-binding sites are more positively charged at neutral pH. The complete reversal of the electrostatic potential of the substrate-binding site suggests that the different Rpfs function optimally at different pHs or most efficiently hydrolyze different micro-domains of PG. These studies provide insights into the molecular determinants of the evolution of functional specialization in Rpfs.
Keywords: resuscitation promoting factor, lytic transglycosylase, peptidoglycan hydrolase, enzyme specificity, electrostatic complementarity
Introduction
It has been estimated that one-third of world population is infected by Mycobacterium tuberculosis (Mtb). In most infected individuals, Mtb persists in a latent state for years until host conditions favor reactivation.1,2 A family of enzymes, called resuscitation promoting factors (Rpfs), plays a central role in Mtb reactivation.2–4 Rpfs share a lytic-transglycosylase domain5–7 that hydrolyzes the glycan chains in the peptidoglycan (PG) sugar-peptide meshwork that surrounds every bacterial cell and protects it against environmental stresses. Additionally, Rpf proteins are thought to collaborate in releasing a PG fragment that binds the sensor domain of PknB and signals the transition from dormancy to vegetative growth.8,9 During growth, the catalytic activity of Rpfs is crucial for cell elongation and division.10,11
Mtb encodes five Rpf proteins (Rv0867c, Rv1009, Rv1884c, Rv2389c, and Rv2450c named RpfA-E).12 They are generally predicted to be secreted (RpfA,B,E) or anchored to the membrane (RpfD). RpfA, C, and E contain low-complexity segments that are expected to be unfolded. Among the five Rpfs, only RpfB contains additional domains, a predicted G5 cell-wall adhesive domain and a repetitive domain of unknown function (DUF348).7
The Rpfs appear to be functionally redundant, since deletion of each single gene causes no differences in cell growth or cell morphology in rich medium in vitro. Thus, no member of the family is essential for growth in culture.13,14 The combined deletions of at least three rpf genes produce cell-growth defects in vitro, uncovering apparent functional specialization of Rpfs in Mtb.13 Consistent with this idea, the Mtb rpfB deletion mutant fails to resuscitate in mice,3 while rpfE is individually essential for switching mycobacterial cultures from slow to fast growth in a chemostat.15 These results provide functional data suggesting that RpfB and RpfE are the most important Rpf family members.9 To better understand the specialized roles of the Mtb Rpfs, we determined the crystal structure of the RpfE catalytic domain and compared the structure to RpfB.7
Results and Discussion
To obtain the soluble RpfE catalytic domain (residues 98–172), we deleted the predicted signal peptide (1–28) and low complexity region (28–97). The predicted catalytic domain (98–172) was expressed in E. coli, and the protein was exposed to 10:1 mixture of reduced to oxidized glutathione during the purification to form the disulfide bond conserved in this protein family. Purified RpfE crystallized readily in almost 70% of more than 500 conditions screened. Increasing the salt concentration from 0.1M to 0.3M and changing the pH failed to improve the crystal morphology. The best crystals for diffraction were obtained using an additive screen (Silver Bullets HT). The structure was determined by molecular replacement at 2.76 Å resolution (TableI) using the catalytic domain of RpfB (3EO5)7 as the search model. The asymmetric unit contained 6 molecules, with a Cα RMSD of 0.336 Å. The model (R/Rfree = 0.235/0.287) contains the entire sequence of each chain except for Ser98, Arg171, and Gly172 in chains A and C, Ser98, Ala144 in chain B, Ser98, Val99, Arg171 and Gly172 in chains D, E, and F.
Table I.
X-Ray Data Collection, Analysis, and Refinement Statistics for the Crystal Structure of the RpfE
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 34.54 84.00 72.63 |
| α, β, γ (°) | 90, 103.53, 90 |
| Wavelength | 1.11 |
| Resolution (Å) | 42.0–2.756 (2.85–2.76) |
| No. reflections | 10,478 (1021) |
| Rmerge (%) | 12.9% (81.9) |
| I/σI | 109.5/10.2 (13.9/6.9) |
| Completeness (%) | 99.61 (96.32) |
| Redundancy | 3.9 (3.8) |
| Refinement | |
| Resolution (Å) | 36.0–2.76 |
| Rwork/Rfree | 0.235/0.287 |
| No. atoms | 3241 |
| Protein | 3234 |
| Water | 7 |
| Protein residues | 431 |
| B-factors | 49.30 |
| Protein | 49.30 |
| Water | 44.90 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.58 |
| Ramachandran plot | |
| Ramachandran favored (%) | 93 |
| Ramachandran outliers (%) | 1.7 |
| PDB ID | 4CGE |
Comparison between RpfE and RpfB catalytic domains
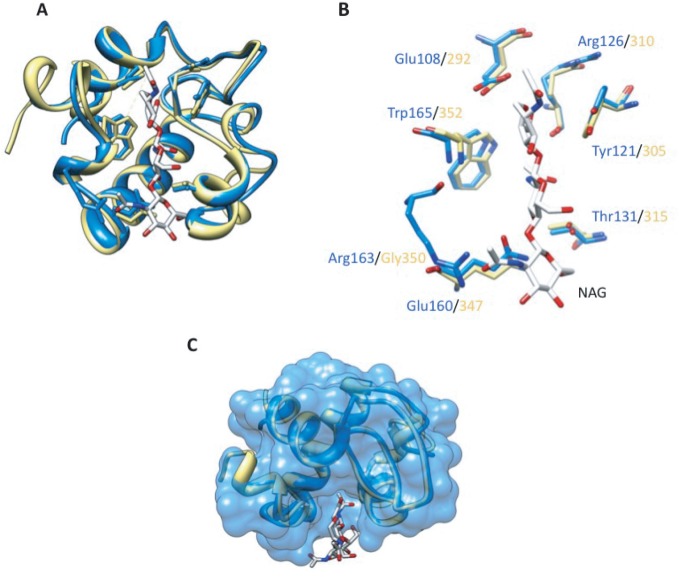
The RpfE catalytic domain adopts the fold of six alpha helices with a single disulfide bond as observed in RpfB7 [Fig. 1(A)], which shares 64% sequence identity. The Cα RMSD between RpfB and the six molecules of the RpfE catalytic domain in the asymmetric unit ranges from 0.52 to 0.78 Å. The RpfE catalytic cleft contains the conserved Glu102 characteristic of lytic transglycosylases. These features suggest that RpfE and RpfB bind the substrates in similar orientation and catalyze hydrolysis by similar mechanisms.
Figure 1.
A: Ribbon representation of superimposed RpfE (blue) and RpfB (yellow) catalytic domain. The conserved, catalytic glutamate is shown in stick representation. B: RpfE and RpfB catalytic-cleft, Cα distance differences. C: Electrostatic potential surfaces (blue, > +5 kT and red, < −5 kT) of RpfE and RpfB calculated using the program Swiss-PdbViewer. RpfE shows positive potential around the predicted peptide-binding surfaces (arrows), while RpfB presents predicted peptide-binding surfaces (arrows) that are calculated to be negatively charged at neutral pH.
Unlike RpfB, however, RpfE does not show the two short 310 helices in the α2–α3 loop. In addition, the active site cleft of RpfE is 0.8–2.3 Å wider than the cleft in RpfB [Fig. 1(B)]. This expansion increases the surface and volume of the catalytic cleft (RpfE 122.5 Å3, 95.8 Å2 and RpfB 82.3 Å3, 94.8 Å2, respectively).
A third large difference between RpfE and RpfB is the calculated electrostatic potential of the surface of the catalytic cleft [Fig. 1(C)]. The RpfE catalytic domain is a basic protein (calculated pI = 9.5), compared to the acidic RpfB catalytic domain (calculated pI = 5.5). This sequence variation produces a dramatic difference in the calculated charge states of surface residues at neutral pH. In particular, charge differences occur on the surfaces in and around the active site. The pocket containing the catalytic glutamate, for example, is calculated to be negatively charged in RpfB and more neutral in RpfE [Fig. 1(C)]. This difference in the predicted pKa of the catalytic glutamate suggests that these enzymes may have different pH optima.
Specialization of substrate binding surfaces
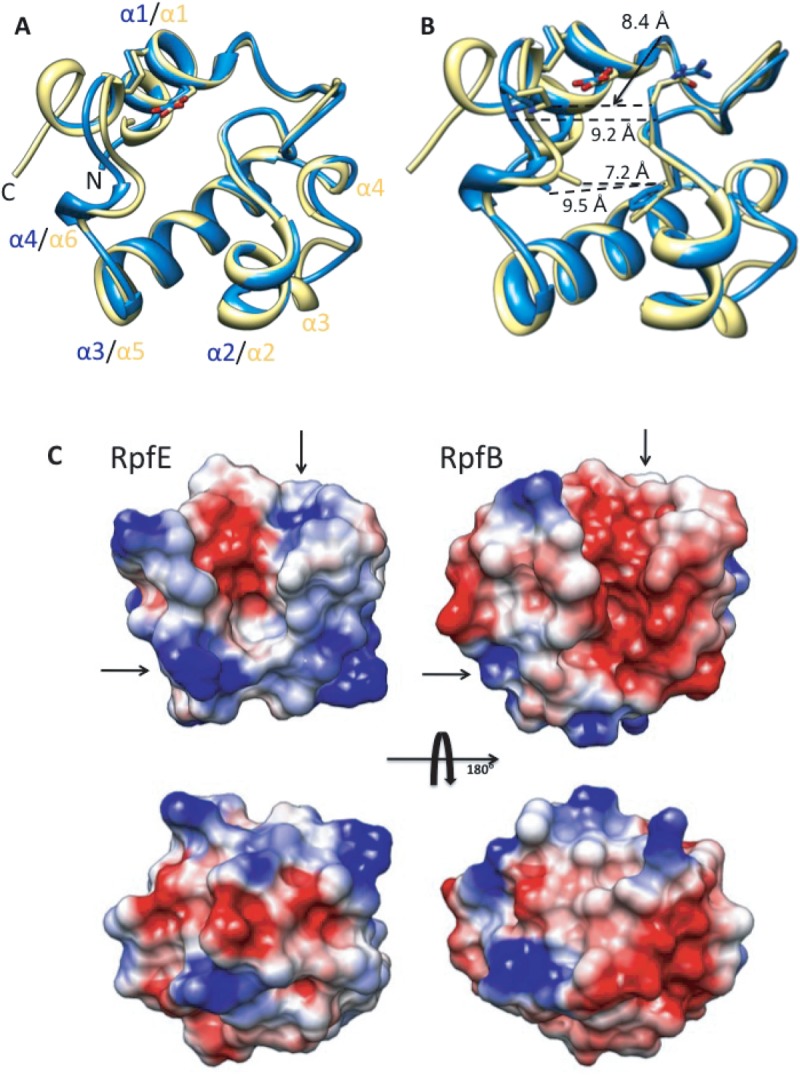
In addition to the catalytic site, the predicted substrate binding surfaces differ in RpfE and RpfB. The structure of RpfB bound to the substrate analog NAG3 (4KPM in PDB)16 supported modeling of the enzyme–substrate complex, which identified likely binding sites not only for the carbohydrate, but also for the peptide crosslinks in PG16 [Fig. 2(A)]. The crosslinks differ in the identity of the second residue (isoglutamate or isoglutamine), the covalently bonded residues (3-4 or 3-3), cleavage of the terminal D-Ala, and other modifications.17 Based on a structural alignment, all but one of the RpfB residues that contact NAG3 are conserved in RpfE [Fig. 2(B)]. RpfB Gly350, however, is replaced in RpfE by Arg163. This residue overlaps with superimposed NAG3, requiring conformational adjustments for substrate binding [Fig. 2(C)].
Figure 2.
A: Superimposed RpfE (blue) and RpfB (yellow) catalytic domain bound to NAG3. B: Superimposed RpfE (blue) and RpfB (yellow) residues in the catalytic cleft involved in binding NAG3. All the residues are conserved between the two enzymes except for Gly350 substituted in RpfE by Arg163. C: RpfE surface with NAG3 modeled in the catalytic cleft, based on a superposition with the structure of the RpfB-NAG3 complex (PDB ID:4KPM). RpfE Arg163 overlaps with substrate, indicating that a conformational adjustment or a different ligand stereochemistry is necessary to accommodate the substrate in the catalytic cleft.
Importantly, the calculated surface potential of RpfE is more basic than that of RpfB in the predicted binding sites for the isoglutamate residue in both interacting peptide crosslinks in bound PG [Fig. 1(C)]. This charge reversal in RpfE compared to RpfB is positioned to complement isoglutamate (RpfE) or isoglutamine (RpfB) in the modeled crosslinks. The sequence variation in the predicted peptide binding groove is observed not only among Rpfs in Mtb, but also in the three homologs in the simplified genome of Mycobacterium leprae18 [Fig. 3(A–C)]. Because the genome of M. leprae has suffered mutations that inactivate all but the most important genes, the conservation of charge reversal in the predicted peptide-crosslink interacting surfaces of the Rpfs supports the potential functional importance of Rpf proteins capable of discriminating between modifications in the peptide moieties in PG.
Figure 3.
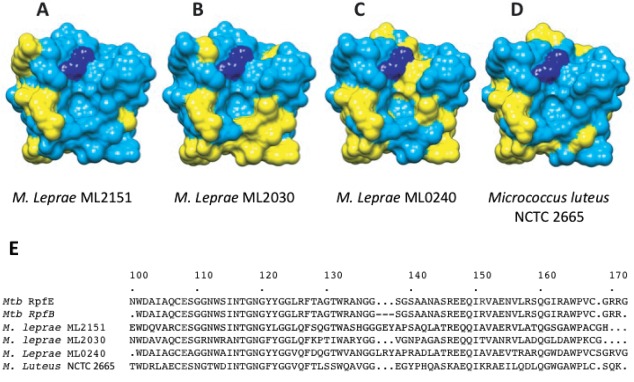
Sequence variation for RpfE and Mycobacterium leprae homologs. Conserved (blue) and variable residues (yellow) in RpfE and ML2151, ML2030, and ML0240, respectively, (A), (B), and (C). Sequence differences are prominent in the predicted peptide-binding groove to the right of the catalytic glutamate (dark blue) and the predicted carbohydrate-binding surface below the active-site pocket. D: Sequence variation between RpfE and the most similar protein in Micrococcus luteus (51% sequence identity) shows homologies in a more distantly related Rpf. E: Sequence aligment of Mtb RpfE and RpfB catalytic domain with M. leprae and M. luteus Rpfs.
The specialization of Rpf proteins
The unique roles of Rpf proteins in bacteria have not yet been elucidated. Among the five Mtb Rpf proteins, the transglycosylase domain is conserved, but the presence of additional domains is predicted to impart different functions. In addition, a prevalent idea is that Rpf genes are regulated independently by distinct mechanisms. Moreover, Rpf proteins may be regulated by association with distinct partners. Rubin and coworkers10,11 reported the interaction between RpfB/E and RipA, a peptidase involved in PG remodeling and cell division. The nature of the interaction is uncertain, however, because the structure of RipA19 revealed that most of the putative Rpf-interacting segment identified using a yeast two-hybrid assay forms a buried β-strand. Nonetheless, a PG fragment released by the action of a hydrolase (Rpf) and the essential peptidase, RipA, is a likely candidate for signaling cell reactivation. The work presented here, describing the structure of an Mtb Rpf protein, reveals structural and chemical differences of the catalytic cleft that may enable recognition of variations in PG or reactions with different pH optima. Overall, specialization in appended domains, gene expression, enzyme regulation, and chemical specificity may contribute to the functional differences among Rpfs.
Materials and Methods
RpfE catalytic domain cloning expression and purification
Nucleotide sequence corresponding to the RpfE catalytic domain (98–172) was amplified by PCR using H37Rv genomic DNA. The PCR product was cloned in a HisMBP expression vector (2MT vector http://www.addgene.org), carrying a tobacco etch virus (TEV) protease cleavage site at the N-terminus of RpfE. The protein was expressed using the Rosetta2(DE3)pLysS expression system (Novagen). Cells were grown at 37°C until optical density (OD)600 0.6 was reached, and cultures were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16°C and shaken overnight for protein expression. The cell pellets were resuspended in Buffer A (300 mM NaCl, 20 mM imidazole, 20 mM Tris pH 8, 10% glycerol, 0.5 mM (tris(2-carboxyethyl)phosphine) (TCEP)), containing protease inhibitor cocktails (E-64, leupeptin, and AEBSF). The cells were sonicated on ice and the lysate centrifuged at 18,000 rpm for 1 h. Supernatant was loaded onto a Ni-affinity column and washed with 10 volumes of Buffer A. The protein was eluted with Buffer B (300 mM NaCl, 20 mM Tris pH 8, 0.5 mM TCEP, 300 mM imidazole, 10% glycerol). TEV protease was added, and the protein was dialyzed against Buffer A (without TCEP) overnight to remove the excess of imidazole. In the same dialysis buffer, a 10:1 ratio of reduced glutathione (10 mM) versus oxidized glutathione (1 mM) was added to promote formation of disulfide bonds.20 The reaction mixture was purified by an additional affinity chromatography step on a Ni column to remove the His_MBP tag and TEV protease. The flow-through fraction was concentrated and further purified on a preparative HiLoad 26/60 Superdex 75 column (GE) in Buffer C (100 mM NaCl, 5% glycerol, and 20 mM Tris pH 8), where RpfE ran as a monomer. Elution fractions were collected and investigated by Coomassie blue-stained SDS PAGE.
RpfE crystallization, data collection and structure determination
The fractions containing RpfE were concentrated to 12 mg/mL, and the protein was screened for crystallization by hanging drop vapor diffusion. RpfE crystallized as small needles in more than 70% of screened conditions.
Several crystals generated in different conditions were screened for diffraction. Crystals that grew in 25% polyethylene glycol (PEG) 3350, 0.1M (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES) sodium pH 6.8, in the presence of Silver Bullets HT F1 condition (0.25% wt/vol methylenediphosphonic acid, 0.25% wt/vol Phytic acid sodium salt hydrate, 0.25% wt/vol sodium pyrophosphate tetrabasic decahydrate, 0.25% wt/vol sodium triphosphate pentabasic, 0.02M HEPES sodium pH 6.8) provided the best diffraction data. Crystals were frozen using 25% PEG 3350, 0.1M HEPES sodium, and 10% PEG 200 as cryoprotectant. Diffraction data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1.21 Data were scaled using HKL2000. The PHENIX software suite22 was used for model building by molecular replacement using the RpfB homolog (3EO5 in PDB) as the search model. Cycles of refinement were performed with PHENIX. Images and structural alignments were generated using Chimera software.23 Protein surface potential was calculated using Swiss-PdbViewer.
Coordinates
The coordinates and structure factors were deposited in the PDB under the accession number 4CGE.
Acknowledgments
Authors thank J. Holton, G. Meigs, and J. Tanamachi at Beamline 8.3.1 at Lawrence Berkeley National Laboratory for help with X-ray data collection. They also thank Abbey Hartland and Scott Gradia from the QB3 Macrolab at University of California, Berkeley. Authors appreciate the support of the TB Structural Genomics Consortium (NIH grant P01 AI068135-06). The authors declare no conflict of interest.
Glossary
- Mtb
Mycobacterium tuberculosis
- NAG3
N,N′,N″-triacetyl-chitotriose
- PG
peptidoglycan
- PDB
Protein Data Bank
- Rpf
resuscitation promoting factor
- RipA
Rpf-interacting protein A
- RMSD
root-mean-square deviation
References
- 1.Loddenkemper R, Hauer B. Drug-resistant tuberculosis: a worldwide epidemic poses a new challenge. Dtsch Arztebl Int. 2010;107:10–19. doi: 10.3238/arztebl.2010.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell-Goldman E, Xu J, Wang X, Chan J, Tufariello JM. A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect Immun. 2008;76:4269–4281. doi: 10.1128/IAI.01735-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tufariello JM, Mi K, Xu J, Manabe YC, Kesavan AK, Drumm J, Tanaka K, Jacobs WR, Jr, Chan J. Deletion of the Mycobacterium tuberculosis resuscitation-promoting factor Rv1009 gene results in delayed reactivation from chronic tuberculosis. Infect Immun. 2006;74:2985–2995. doi: 10.1128/IAI.74.5.2985-2995.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RK, Srivastava R. Resuscitation promoting factors: a family of microbial proteins in survival and resuscitation of dormant mycobacteria. Indian J Microbiol. 2012;52:114–121. doi: 10.1007/s12088-011-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtje JV. Lytic transglycosylases. EXS. 1996;75:425–429. doi: 10.1007/978-3-0348-9225-4_21. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Gonsaud M, Barthe P, Bagneris C, Henderson B, Ward J, Roumestand C, Keep NH. The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat Struct Mol Biol. 2005;12:270–273. doi: 10.1038/nsmb905. [DOI] [PubMed] [Google Scholar]
- 7.Ruggiero A, Tizzano B, Pedone E, Pedone C, Wilmanns M, Berisio R. Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis. J Mol Biol. 2009;385:153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 8.Alber T. Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr Opin Struct Biol. 2009;19:650–657. doi: 10.1016/j.sbi.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol Med Microbiol. 2010;58:39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- 10.Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 2008;4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol. 2007;66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 12.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, Mizrahi V. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tufariello JM, Jacobs WR, Jr, Chan J. Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo. Infect Immun. 2004;72:515–526. doi: 10.1128/IAI.72.1.515-526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beste DJ, Espasa M, Bonde B, Kierzek AM, Stewart GR, McFadden J. The genetic requirements for fast and slow growth in mycobacteria. PLoS One. 2009;4:e5349. doi: 10.1371/journal.pone.0005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Squeglia F, Romano M, Ruggiero A, Vitagliano L, Simone De A, Berisio R. Carbohydrate recognition by RpfB from Mycobacterium tuberculosis unveiled by crystallographic and molecular dynamics analyses. Biophys J. 2013;104:2530–2539. doi: 10.1016/j.bpj.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 19.Ruggiero A, Marasco D, Squeglia F, Soldini S, Pedone E, Pedone C, Berisio R. Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure. 2010;18:1184–1190. doi: 10.1016/j.str.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Annis I, Hargittai B, Barany G. Disulfide bond formation in peptides. Methods Enzymol. 1997;289:198–221. doi: 10.1016/s0076-6879(97)89049-0. [DOI] [PubMed] [Google Scholar]
- 21.MacDowell AA, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, Domning EE, Duarte RM, Kelez N, Plate DW, Cork CW, Earnest TN, Dickert J, Meigs G, Ralston C, Holton JM, Alber T, Berger JM, Agard DA, Padmore HA. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]