Abstract
Recent structures of DNA polymerase complexes with dGMPCPP/dT and dCTP/dA mispairs at the insertion site have shown that they adopt Watson-Crick geometry in the presence of Mn2+ indicating that the tautomeric or ionization state of the base has changed. To see whether the tautomeric or ionization state of base-pair could be affected by its microenvironment, we determined 10 structures of an RB69 DNA polymerase quadruple mutant with dG/dT or dT/dG mispairs at position n-1 to n-5 of the Primer/Template duplex. Different shapes of the mispairs, including Watson-Crick geometry, have been observed, strongly suggesting that the local environment of base-pairs plays an important role in their tautomeric or ionization states.
Keywords: RB69 DNA polymerase, tautomerization, thymine-guanine mispairs, Watson-Crick base-pairs
Introduction
Replicative DNA polymerases (pols) are essential for transmission and maintenance of genetic information. They exhibit a high degree of base selectivity, making less than one mistake per 106 insertion events.1 Their fidelity is increased by an additional 102 to 103 fold for pols having exonuclease activity.2 Several factors that contribute to the high fidelity of replicative pols have been identified, including base-pair geometry, interbase hydrogen bonding (HB) and prechemistry conformational transitions.3,4 Nevertheless, mistakes occur at a very low frequency when pols incorporate incorrect nucleotides into the growing primer strand.3 If they remain uncorrected, the resulting mutations often have profound negative effects, associated with many pathological conditions, some of them fatal.5,6 Consequently, the mechanism of misincorporation has been an active subject of research and debate.7,8
In the 1950's, Watson and Crick proposed that replication errors were caused by bases adopting one of its rare tautomeric forms.9 Such base-pairs resemble the shape of canonical Watson-Crick base-pairs, however, no structural data has been available to support this rare tautomer hypothesis until recently. In 2011, Kunkel's group reported a crystal structure of a DNA pol λ derivative, which has a five amino acids deletion in a loop close to the pol active site, with a nonhydrolysable dNTP analog, dGMPCPP, opposite a templating dT.10 The dGMPCPP/dT mispair adopts Watson-Crick like geometry mimicking the shape of the cognate base-pair, dGTP/dC. Subsequently, Beese's group observed a dCTP/dA mispair in the active site of a Bacillus stearothermophilus DNA polymerase I large fragment (BF) double mutant that mimics the shape of the cognate base-pair dTTP/dA.11 These structures provide the first direct evidence to support Watson-Crick's rare tautomer hypothesis. Interestingly, both tautomeric base-pairs observed in the active site of pol λ and BF were formed in the presence of Mn2+, a metal ion that has been reported to decrease the base selectivity of DNA pols.12 In contrast, the dCTP/dA mispair adopts wobble geometry at the insertion site of BF in the presence of Mg2+. Therefore, it seems that Mn2+ can stabilize the nucleic acid bases in their less stable tautomeric form, such as the enol form of thymine and guanine or the imino form of adenine and cytosine.
Alternatively, the Watson-Crick-like dGMPCPP/dT mispair could bear ionized bases as well.10 Beese's group reported two dT/dG mispairs at position n-3 and n-4 of the primer/template (P/T) duplex in complex with BF.13 The dT/dG mispair adopts anti-wobble geometry suggesting that the tautomeric or ionization state of the bases has changed. However, the exact cause of rare tautomer formation and the change in ionization states remain elusive. It is unknown whether the tautomerization or ionization states of nucleic acid bases are affected solely by the presence of Mn2+ or by the microenvironment surrounding the nucleic acid base. In addition, previous kinetic and structural studies have shown that pols asymmetrically recognize pyrimidines and purines.14 Thus, an intriguing question is whether changing the microenvironment of a nucleic acid bases by replacing purine with pyrimidine at the same position of the P/T duplex or vice versa would affect the tautomeric or ionization state of the bases. If changes are observed, how can the tautomers be stabilized or destabilized by their microenvironment? To answer this question, we designed 10 sets of DNA duplexes with either dG/dT or dT/dG mispairs at positions n-1 to n-5 of the P/T duplex (Supporting Information Fig. S1), and then subsequently determined 10 high-resolution structures with each set of DNA duplexes using an engineered RB69 DNA pol (RB69pol) variant, a high-fidelity model family B pol. This RB69pol variant is a quadruple mutant (qm, L415A/L561A/S565G/Y567A), which was previously designed to capture all 12 mispairs at the insertion site of RB69pol.15 Our structures show that the microenvironment of base-pairs plays an important role in their tautomeric or ionization states.
Results and Discussion
The 10 structures were determined with resolutions ranging from 1.85 Å to 2.20 Å and Rfree values spanning 20.6–24.1% (TableI, Supporting Information S1, S2, S3). The overall structures of all 10 ternary complexes are almost identical to that of the dCTP/dG-containing wt RB69pol ternary complex.16 By superimposing all 10 structures with the wt structure, we found that the sugar ring and the triphosphate tail of the incoming dNTPs overlay perfectly well with one another. The root-mean-square deviations of 930 Cα atoms (except residues 250–260) varied between 0.16 and 0.18 Å. Residue 250 to 260 is a flexible β-hairpin structure located in the exonuclease domain. At position n-1 of the P/T duplex, the dG/dT mispair adopts wobble geometry with dG at the primer end receding into the minor groove [Fig. 1(A)]. A very similar wobble pair was observed for the dGTP/dT mispair at the insertion site of the RB69pol qm ternary complex, indicating that translocation from the insertion site to the post-insertion site does not disturb the geometry of the dG/dT mispair. Interestingly, the dG/dT mispairs at positions n-2 and n-3 adopt Watson-Crick base-pair geometry that is virtually indistinguishable from a canonical base-pair [Fig. 1(C,E)]. Superposition of this qm structure with that of the wt ternary complex shows that the dG/dT mispair can overlay perfectly well with the dA/dT base-pair at position n-2 of the P/T duplex in the wt structure [Fig. 2(C)]. The distances between N1 of dG and N3 of dT of the dG/dT mispair at position n-2 and n-3 are 2.97 and 2.85 Å respectively. This suggests that either the tautomeric state or ionization state of bases has changed. However, it is not possible to distinguish between the two alternatives. At position n-4 of the P/T duplex, the dG/dT mispair resumes its wobble geometry, similar to the dG/dT mispair observed at position n-1 [Fig. 1(G)]. In contrast, an anti-wobble conformation was observed when the dG/dT mispair is located at position n-5 of the P/T duplex [Fig. 1(I)]. The corresponding pyrimidine base at the template strand tilts toward the minor groove. The resulting distance between N3 of dT and N1 of dG is 2.99 Å, indicating a change of tautomeric or ionization state of the bases. Similar anti-wobble geometry of the dG/dT mispair has been observed in BF binary complexes at positions n-3 and n-4 of the P/T duplex in the presence of Mn2+.13 Since all our structures were obtained in the presence of Ca2+, the change in the tautomeric or ionization state of bases does not solely depend on the presence of Mn2+.
Table I.
Refinement and Geometry Statistics for 10 RB69pol qm Ternary Complexes
| Position | P/T | PDB code | Resolution (Å) | R (%) | Rfree (%) | d(C1′ − C1′) (Å) | λ(primer) (deg) | λ(template) (deg) | Base-pair geometry |
|---|---|---|---|---|---|---|---|---|---|
| n-1 | dG/dT | 4M3Y | 1.85 | 18.5 | 22.0 | 10.3 | 67.3 | 45.5 | Wobble |
| dT/dG | 4M3R | 2.07 | 17.7 | 22.0 | 10.3 | 44.7 | 66.3 | Wobble | |
| n-2 | dG/dT | 4M3Z | 1.85 | 17.8 | 20.6 | 10.9 | 51.5 | 53.5 | Watson-Crick |
| dT/dG | 4M3T | 1.90 | 17.5 | 20.8 | 10.9 | 49.7 | 51.1 | Watson-Crick | |
| n-3 | dG/dT | 4M41 | 2.15 | 19.4 | 23.7 | 10.7 | 50.2 | 52.7 | Watson-Crick |
| dT/dG | 4M3U | 2.07 | 17.7 | 22.3 | 10.3 | 47.1 | 69.3 | Wobble | |
| n-4 | dG/dT | 4M42 | 2.04 | 18.4 | 22.3 | 10.6 | 63.4 | 41.4 | Wobble |
| dT/dG | 4M3W | 2.10 | 17.7 | 22.0 | 10.3 | 45.7 | 67.6 | Wobble | |
| n-5 | dG/dT | 4M45 | 1.89 | 18.4 | 22.3 | 11.4 | 38.9 | 62.0 | Anti-wobble |
| dT/dG | 4M3X | 2.20 | 19.1 | 24.1 | 10.5 | 45.3 | 65.4 | Wobble |
Note: The crystallographic statistics for data collection and structure refinement are shown in Supporting Information Tables S1 and S2. λ(primer) and λ(template) are defined as the angle between the glycosidic bond of the primer or template nucleotide and the line drawn between the C1' atoms of the base pair.
Figure 1.
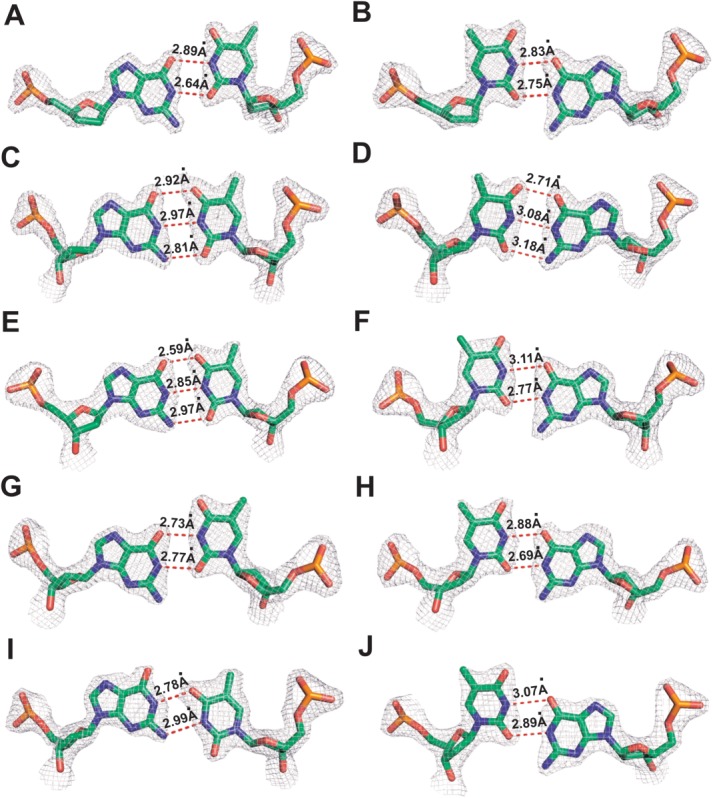
Final 2Fo − Fc electron density map for dG/dT and dT/dG mispairs contoured at 2.0 σ: A, C, E, G, and I are dG/dT mispairs at positions n − 1 to n − 5 of the P/T duplex respectively. B, D, F, H, and J are dT/dG mispairs at positions n − 1 to n − 5 of the P/T duplex, respectively.
Figure 2.
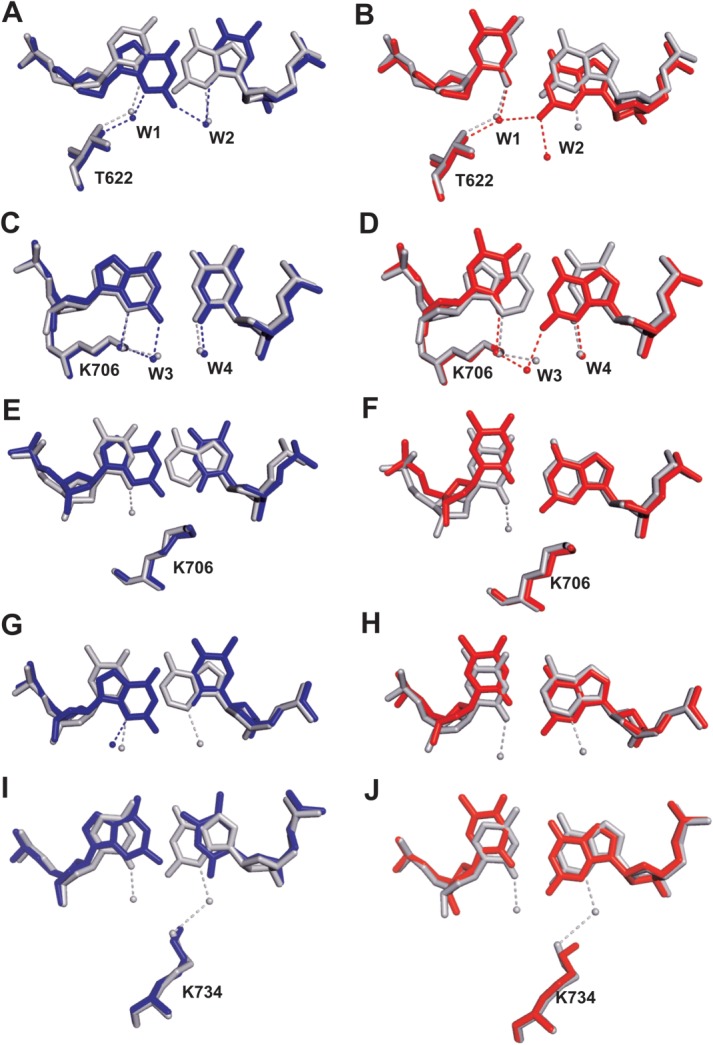
Superpositon of the structure of dG/dT (shown in blue) or dT/dG-containing (shown in red) qm with that of dCTP/dG-containing wt RB69 pol (shown in gray). A and B are overlays of dC/dG with dG/dT or dT/dG at position n − 1 respectively; C and D are overlays of dA/dT with dG/dT or dT/dG at position n − 2, respectively. E and F are overlays of dT/dA with dG/dT or dT/dG at position n − 3, respectively. G and H are overlays of dT/dA with dG/dT or dT/dG at position n − 4, respectively. I and J are overlays of dC/dG with dG/dT or dT/dG at position n − 5, respectively.
When dG is located in the template strand and dT is located in the primer strand, the dT/dG mispair adopts wobble geometry at positions n-1, n-3, n-4, and n-5 of the P/T duplex respectively [Fig. 1(B,F,H,J)]. In contrast, at position n-2 of the P/T duplex, the dT/dG mispair adopts canonical Watson-Crick geometry, mimicking the shape of a dT/dA base-pair [Fig. 2(D)]. The resulting distance between N3 of dT and N1 of dG is 3.08 Å. It is interesting to point out some similarities between the dG/dT mispair and dT/dG mispair namely: (i) both mispairs form Watson-Crick base-pair geometries at position n-2 of the P/T duplex and; (ii) both mispairs form wobble geometry at positions n-1 and n-4 of the P/T duplex, respectively. Therefore, based on our observations with all 10 structures, the shape of guanine-thymine mispair in the P/T duplex depends on: (i) its exact position in the P/T duplex and; (ii) the identity of the base in the template or primer strand; that is, whether it is a purine or a pyrimidine.
Meanwhile, two intriguing questions arise: (i) why do guanine-thymine mispairs tend to have Watson-Crick-like geometry at position n-2 of the P/T duplex but display wobble geometry at positions n-1 or n-4 of the P/T duplex and; (ii) why do dG/dT and dT/dG mispairs have different geometries at positions n-3 or n-5 of the P/T duplex when they are in complex with RB69pol? We believe the microenvironment of the mispair plays an important role that governs its resulting geometry. As shown in Figure 2(A), the N3 hydrogen of dG at position n-1 of the primer strand forms a HB to the hydroxyl group of T622 via an ordered water molecule (w1). The O2 of dT at position n-1 of the template strand forms a HB to another ordered water molecule (w2) which is also within HB distance to the N2 hydrogen of dG on the primer strand [Fig. 2(A)]. When dG is at position n-1 of the template strand, the purine base recedes into the minor groove so that its N2 hydrogen forms a HB to the w1 water molecule, which is also hydrogen bonded to the hydroxyl group of T622 and to O2 of dT at position n-1 of the primer strand [Fig. 2(B)]. Therefore, the water mediated HB network at the minor groove stabilizes dG/dT and dT/dG mispairs with wobble geometry at position n-1 of the P/T duplex. K706 is a conserved amino acid in B family pols.17 The ε-amino group of K706 forms one hydrogen bond (HB) with N3 of dG at position n-2 of the primer strand and another HB with the w3 water molecule [Fig. 2(C)]. This w3 water molecule also forms an HB with the N2 hydrogen of dG at position n-2 of the template strand. Together, the HB interactions among the mispairs, the side chain of K706 and the w3 water molecule prevent the purine base of dG from receding into the minor groove to form wobble base-pairs with dT. When dT is at position n-2 of the primer strand, the ε-amino group of K706 is not only hydrogen bonded to the O2 of dT in the primer strand directly, but also to the N2 hydrogen of dG at position n-2 in the template strand via the w3 water molecule [Fig. 2(D)]. Therefore, the HB network including the K706 side chain and the bridging water molecules at the minor groove, stabilize the guanine-thymine mispair in Watson-Crick base-pair geometry at position n-2 of the P/T duplex. In contrast, no protein side chain is directly located at the minor groove of position n-4 of the P/T duplex to prevent the purine base of dG from shifting downward [Fig. 2(G,H)]. Besides, wobble geometry is commonly observed for guanine-thymine mispairs in hetero-duplex DNA. That could explain why both dG/dT and dT/dG mispairs have wobble geometries at position n-4 of the P/T duplex.
At the minor groove of the P/T duplex, the Cδ of K706 and the ε-amino group K734 are located right below the center of the base-pairs at positions n-3 and n-5, respectively [Fig. 2(E,F,I,J)]. When dG is in the primer strand, the wobble geometry of the dG/dT mispair creates a steric clash between the N2 hydrogen of dG and the Cδ of K706 at position n-3 [Fig. 2(E)] or between the N2 hydrogen of dG and the ε-amino group of K734 at position n-5 [Fig. 2(F)], which is probably the reason why wobble geometry is not observed for dG/dT mispair at either position n-3 or n-5 of the P/T duplex. When dG is in the template strand, the purine base of dG is in a conformation similar to that observed with a Watson-Crick base-pair [Fig. 2(F,J)]. Therefore, dT/dG mispairs are able to adopt wobble geometries at positions n-3 and n-5 of the P/T duplex. Overall, the different geometry of guanine-thymine mispair suggests that RB69pol asymmetrically recognizes pyrimidines and purines at position n-1 to n-5 of a P/T duplex.
In summary, we report 10 structures of qm RB69pol ternary complexes with guanine-thymine mispairs at position n-1 to n-5 of the P/T duplex. At certain locations of the P/T duplex, the guanine-thymine mispairs adopt Watson-Crick base-pair geometry. Our structures strongly suggest that microenvironment of base-pairs plays an important role in the tautomeric or ionization state of the bases. The different geometry of dG/dT or dT/dG mispairs within the P/T duplex is the result of dynamic interactions among base-pairs, protein side chains and ordered water molecules at the minor groove.
Materials and Methods
Crystallization of dATP/dT-containing ternary complexes of RB69pol qm
The RB69pol quadruple mutant (qm), in an exonuclease-deficient background (D222A and D327A), was expressed, purified, and stored as previously described.15 All oligonucleotides were synthesized at the Yale Keck facilities and purified via polyacrylamide gel electrophoresis. The sequence of the primer-templates (P/Ts) used in this study are shown in Supporting Information Figure S1. The crystallization conditions used in this article are very similar to those previously described.15,16 Briefly, the RB69pol qm was mixed in an equimolar ratio with a freshly annealed P/T to give a final protein concentration of 120 µM. dATP was then added to give a final concentration of 2 mM. A solution of 150 mM CaCl2, 10% (w/v) PEG 350 monomethyl ether (MME), and 100 mM sodium cacodylate (pH 6.5) was mixed with an equal volume of the protein complex. The square rod-shaped crystals grew in 3 days at 20°C and had dimensions of ∼150 × 50 × 50 µm. Crystals were transferred to a cryoprotectant solution with 30% w/v PEG 350 MME prior to freezing in liquid nitrogen.
X-ray diffraction data collection, structure determination, and refinement
Data were collected at 110K using the synchrotron radiation sources at beam line 24ID-E of the Northeast Collaborative Access Team (NECAT), Advanced Photon Source (APS), Argonne National Laboratory (ANL, Chicago, IL). Data were processed using the HKL2000 program suite.18 The structures were determined by molecular replacement with a previously determined wild-type RB69pol structure (3NCI), and refined using REFMAC5.19 The P/T duplexes and dNTPs were subsequently built using the program COOT.20 Structure refinement statistics are summarized in TableI, Supporting Information Tables S1 and S2. All figures were prepared in Pymol (Schrodinger, LLC).21 Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4M3R, 4M3T, 4M3U, 4M3W, 4M3X, 4M3Y, 4M3Z, 4M41, 4M42, and 4M45.
Acknowledgments
The authors thank the staff of the NE-CAT beamline 24-ID-E at the Advanced Photon Source of Argonne National Laboratory.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA. Exonucleolytic proofreading. Cell. 1988;53:837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 4.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 6.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 8.Harris VH, Smith CL, Jonathan Cummins W, Hamilton AL, Adams H, Dickman M, Hornby DP, Williams DM. The effect of tautomeric constant on the specificity of nucleotide incorporation during DNA replication: support for the rare tautomer hypothesis of substitution mutagenesis. J Mol Biol. 2003;326:1389–1401. doi: 10.1016/s0022-2836(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 9.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 10.Bebenek K, Pedersen LC, Kunkel TA. Replication infidelity via a mismatch with Watson-Crick geometry. Proc Natl Acad Sci USA. 2011;108:1862–1867. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci USA. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirover MA, Loeb LA. Infidelity of DNA synthesis in vitro: screening for potential metal mutagens or carcinogens. Science. 1976;194:1434–1436. doi: 10.1126/science.1006310. [DOI] [PubMed] [Google Scholar]
- 13.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 14.Lund TJ, Cavanaugh NA, Joubert N, Urban M, Patro JN, Hocek M, Kuchta RD. B family DNA polymerases asymmetrically recognize pyrimidines and purines. Biochemistry. 2011;50:7243–7250. doi: 10.1021/bi2006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia S, Wang J, Konigsberg WH. DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J Am Chem Soc. 2013;135:193–202. doi: 10.1021/ja3079048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Xia S, Blaha G, Steitz TA, Konigsberg WH, Wang J. Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2011;50:581–590. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia S, Christian TD, Wang J, Konigsberg WH. Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA. Biochemistry. 2012;51:4343–4353. doi: 10.1021/bi300416z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrodinger. LLC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


