Abstract
Mutations in mitochondrial DNA (mtDNA) are reported to be responsible for the pathogenesis of maternally inherited hearing loss. Complete mtDNA sequencing may detect pathogenic mutations, but whether they are indeed pathogenic can be difficult to interpret because of normal ethnic-associated haplogroup variation and other rare variations existing among control populations. In this study, we performed systemic mutational analysis of mtDNA in 394 Japanese patients with hearing loss. Two different cohorts were analyzed in this study: Cohort 1, 254 maternally inherited patients; and Cohort 2, 140 patients with various inheritance modes. After screening of the entire mtDNA genome with direct sequencing, we evaluated the frequency of previously reported mutations and the frequency and pathogenicity of the novel variants. As a result, the ‘Confirmed' mitochondrial mutations were found predominantly in Cohort 1 rather than in Cohort 2 (14.6 vs 0.7%). 1555A>G (n=23) is the most common mutation, followed by the 3243A>G (n=11) mutations. On the basis of prediction analysis, we detected 10 novel homoplasmic mitochondrial variants. After further classification, the 3595A>G and 6204A>G variants were found to be new candidate mutations possibly associated with hearing loss.
Keywords: mitochondrial mutation, non-syndromic hearing loss
Introduction
Hearing impairment is one of the most common sensory handicaps, with a frequency of at least 1/1000 at birth, and 50% of these cases can be attributed to genetic causes. Furthermore, causative mitochondrial DNA (mtDNA) mutations have been found in 5–10% of patients with postlingual non-syndromic hearing loss.1
Among mitochondrial mutations, 1555A>G mutations in the mitochondrial 12S rRNA are found frequently (0.6–5.3%, depending on the ethnic group) in aminoglycoside-induced and late-onset non-syndromic hearing loss.2, 3, 4 A 1494C>T mutation in 12S rRNA is also associated with aminoglycoside-induced and non-syndromic hearing loss.5 A 3243A>G mutation in the tRNALeu(UUR) is associated with maternally inherited diabetes combined with deafness,6 and mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS), which frequently present with hearing loss. 7445A>C/G/T,7, 8 7472insC and 7510T>C9 mutations in the tRNASer(UCN) are also associated with aminoglycoside-induced or non-syndromic hearing loss.
Moreover, additional mutations in 12S rRNA (827A>G,10 961T>C, 961delT+Cn, 1005T>C and 1095T>C11) have been reported as mitochondrial hearing loss mutations. Although there were growing numbers of reports of various novel mtDNA mutations associated with hearing loss, most focused on a few limited nucleotide positions or only the 12SrRNA region.12 Therefore, we conducted a whole mitochondrial genome mutational analysis by direct sequencing using samples from 254 maternally inherited and 140 non-syndromic Japanese hearing loss probands with various inheritance modes, and summarized the frequencies of the mutations, as well as the spectrum and phenotypes found in the hearing loss patients with mtDNA mutations.
Materials and methods
Subjects
Two cohorts were used in this study: Cohort 1, 254 Japanese maternally (or possibly autosomal dominant with affected mother and one or more affected children) inherited sensorineural hearing loss (SNHL) subjects; and Cohort 2, 140 Japanese SNHL subjects with various inheritance modes (14 autosomal dominant or mitochondrial inherited, 126 autosomal recessive inherited or sporadic cases), both collected from 33 ENT departments nationwide in Japan. All subjects gave prior written informed consent for participation in the project, which was approved by the ethical committee of each hospital. The control group consisted of 192 unrelated Japanese healthy individuals with normal hearing evaluated by auditory testing.
Mutation analysis
Whole mtDNA from each patient was amplified into two long fragments, A and B, by LA Taq DNA polymerase (TaKaRa BIO, Shiga, Japan) as described elsewhere.13 In brief, each genomic DNA sample was amplified by long PCR for 1 min at 94 °C, followed by 30 three-step cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 6 min, with a final extension at 72 °C for 5 min, ending with a holding period at 4 °C.
After the PCR amplification, resulting products were purified and direct sequenced with ABI Big Dye terminators and ABI 3130 autosequencer (Applied Biosystems, Carlsbad, CA, USA). Sequencing reaction was performed with 50 primers for the whole mitochondrial genome, designed with mitoSEQr Resequencing System (Applied Biosystems).
Sequencing data were analyzed by SeqScape ver2.6 and SeqAnalysis (Applied Biosystems). The sequencing result from each patient was compared with the rCRS (Reversed Cambridge Reference Sequence) to identify mtDNA mutations. Mitochondrial DNA mutations included in the mtSNP (http://mtsnp.tmig.or.jp/mtsnp/index.shtml), MITOMAP (http://www.mitomap.org/MITOMAP) or Uppsala mtDB (http://www.genpat.uu.se/mtDB/) databases were excluded as non-pathogenic variants when we search to novel variants.
We evaluated mutations according to evaluation criteria derived from a previous report by Zaragoza et al.14
Prediction of pathogenicity of mtDNA mutations
Initially, we measured the frequencies of each mutation found in healthy controls in our study (n=192) and in the mtSNP database (n=2153, including: centenarians in Gifu and Tokyo, type 2 diabetes mellitus patients (with or without vascular disorders), overweight and non-overweight young adult males, Parkinson's disease patients and Alzheimer's disease patients in Japan). The nucleotide conservation in each gene from humans and 60 mammalian species (Artibeus jamaicensis, NC_002009; Balaenoptera musculus, NC_001601; Balaenoptera physalus, NC_001321; Bos taurus, NC_006853; Canis familiaris, NC_002008; Cavia porcellus, NC_000884; Cebus albifrons, NC_002763; Ceratotherium simum, NC_001808; Chalinolobus tuberculatus, NC_002626; Dasypus novemcinctus, NC_001821; Didelphis virginiana, NC_001610; Dugong dugon, NC_003314; Echinops telfairi, NC_002631; Echinosorex gymnura, NC_002808; Equus asinus, NC_001788; Equus caballus, NC_001640; Erinaceus europaeus, NC_002080; Felis catus, NC_001700; Gorilla gorilla, NC_001645; Halichoerus grypus, NC_001602; Hippopotamus amphibious, NC_000889; Hylobates lar, NC_002082; Isoodon macrourus, NC_002746; Lama pacos, NC_002504; Loxodonta africana, NC_000934; Macaca sylvanus, NC_002764; Macropus robustus, NC_001794; Mus musculus, NC_005089; Myoxus glis, NC_001892; Nycticebus coucang, NC_002765; Ochotona collaris, NC_003033; Ornithorhynchus anatinus, NC_000891; Orycteropus afer, NC_002078; Oryctolagus cuniculus, NC_001913; Ovis aries, NC_001941; Pan paniscus, NC_001644; Pan troglodytes, NC_001643; Papio hamadryas, NC_001992; Phoca vitulina, NC_001325; Physeter catodon, NC_002503; Pongo pygmaeus, NC_002083; Pongo pygmaeus abelii, NC_002083; Pteropus dasymallus, NC_002612; Pteropus scapulatus, NC_002619; Rattus norvegicus, NC_001665; Rhinoceros unicornis, NC_001779; Sciurus vulgaris, NC_002369; Soriculus fumidus, NC_003040; Sus scrofa, NC_000845; Tachyglossus aculeatus, NC_003321; Talpa europaea, NC_002391; Tarsius bancanus, NC_002811; Thryonomys swinderianus, NC_002658; Trichosurus vulpecula, NC_003039; Tupaia belangeri, NC_002521; Ursus americanus, NC_003426; Ursus arctos, NC_003427; Ursus maritimus, NC_003428; Volemys kikuchii, NC_003041; Vombatus ursinus, NC_003322) was evaluated by the ClustalW method or the mtSNP database (mtSAP Evaluation; http://mtsnp.tmig.or.jp/mtsnp/search_mtSAP_evaluation.html). The mutations were considered to be possibly pathogenic if the original amino acid or base was conserved in >50% of the species (31 or more of 61 species).15
Results
Direct sequence screening of the 254 probands of Japanese maternally inherited SNHL families and 140 non-syndromic hearing loss probands with various severities of hearing loss revealed 634 single-nucleotide polymorphisms in whole mitochondrial genome. Among those single-nucleotide polymorphisms, 19 were previously reported as associated with hearing loss: 792C>T (n=1), 827A>G (n=10), 856A>G (n=3), 961T>C (n=3), 1005T>C (n=2), 1095T>C (n=1), 1310C>T (n=3), 1494C>T (n=1), 1555A>G (n=23), 3243A>G (n=11), 3398T>C (n=1), 3421G>A (n=2), 5628T>C (n=1), 7511T>C (n=3), 8108A>G (n=1), 8348A>G (n=1), 11696G>A (n=4), 14693A>G (n=1) and 15927G>A (n=4) (Tables 1 and 2). In this study, based on the MITOMAP database, status was considered to be ‘Confirmed' if at least two or more independent laboratories had published reports on the pathogenicity of a specific mutation (Table 1). More ambiguous substitutions were categorized as ‘Unclear,' ‘Reported' or ‘Point mutation/polymorphism' (Table 2). ‘Reported' status indicates that one or more reports have considered the mutation as possibly pathologic. ‘Point mutation/polymorphism' status indicates that some reports have determined the mutation to be a non-pathogenic polymorphism. In all, 14.6% (37/254) of the patients in Cohort 1 (maternally inherited patients) were associated with the ‘Confirmed' mutations. Only 0.7% (1/140) of the patients had the ‘Confirmed' mutations in Cohort 2 (patients with various inherited modes) (Table 1). Ambiguous-status substitutions were associated in 7.5% (19/254) of Cohort 1, in contrast to 13.6% (19/140) of Cohort 2 (Table 2).
Table 1. ‘Comfirmed' mitochondrial mutations associated with sensorineural hearing loss found in this study.
|
Case |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Locus | Statusa | Disease | Total (/394) | Cohort 1 (/254) | Cohort 2 (/140) | Control (/192) | Hearing characteristics | Progression of hearing loss | Tinnitus | Vertigo | Associated symptom | Reference |
| C1494T | 12S rRNA | Confirmed | SNHL | 1 | 0 | 1 | 0 | High frequency | 1/1 | 1/1 | 0/1 | 0 | 5 |
| A1555G | 12S rRNA | Confirmed | SNHL | 23 | 23 | 0 | 0 | High frequency | 15/21 | 13/16 | 6/16 | 0 | 2 |
| A3243G | tRNA Leu (UUR) | Confirmed | SNHL/DM/FSGS/Cardiac dysfunction | 11 | 11 | 0 | 0 | Flat | 10/10 | 6/10 | 6/10 | Diabetes mellitus (8/10) | 6 |
| T7511C | tRNA Ser (UCN) | Confirmed | SNHL | 3 | 3 | 0 | 0 | High frequency | 1/2 | 3/4 | 0/4 | 0 | 23 |
| Total | 37/254 (14.6%) | 1/140 (0.7%) | 27/34 | 23/31 | 12/31 | ||||||||
Abbreviations: DM, diabetes mellitus; FSGS, focal segmental glomerulosclerosis; SNHL, sensorineural hearing loss.
Based on the MITOMAP database; ‘Confirmed' status indicates that at least two or more independent laboratories have published reports on the pathogenicity of a specific mutation.
Table 2. Ambiguous-status mitochondrial substitutions associated with sensorineural hearing loss found in this study.
|
Case |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Locus | Statusa | Disease | Total (/394) | Cohort 1 (/254) | Cohort 2 (/140) | Control (/96) | Hearing characteristics | Progression of hearing loss | Tinnitus | Vertigo | Associated symptom | Reference |
| C792T | 12S rRNA | Reported | SNHL | 1 | 1 | 0 | 0 | Flat | 1/1 | 1/1 | 1/1 | 0 | 24 |
| A827G | 12S rRNA | Conflicting reports | SNHL | 10 | 5 | 5 | 1 | High frequency | 4/11 | 6/11 | 2/11 | 0 | 10 |
| A856G | 12S rRNA | Reported | SNHL/LHON/AD | 3 | 3 | 0 | 0 | Flat | 1/1 | 1/1 | 1/1 | 0 | 25 |
| T961C | 12S rRNA | Unclear | SNHL/LVNC | 3 | 3 | 0 | 2 | Profound | 1/1 | 1/1 | 1/1 | 0 | 26 |
| T1005C | 12S rRNA | Unclear | SNHL | 2 | 1 | 1 | 1 | Low frequency | 2/2 | 1/1 | 1/1 | 0 | 26 |
| T1095C | 12S rRNA | Unclear | SNHL | 1 | 1 | 0 | 0 | Flat | 1/1 | 1/1 | 1/1 | 0 | 11 |
| C1310T | 12S rRNA | Reported | SNHL | 3 | 0 | 3 | 0 | unknown | 1/3 | 0/3 | 0/3 | 0 | 24 |
| T3398C | ND1 | Reported | SNHL/DM/HCM/GDM/LVNC/Cardiomyopathy | 1 | 1 | 0 | 0 | Profound | 1/1 | 1/1 | 0/1 | 0 | 27 |
| G3421A | ND2 | Reported | SNHL | 2 | 1 | 1 | 0 | Profound | 1/1 | 1/1 | 0/1 | 0 | 28 |
| T5628C | tRNAAla | Reported | SNHL/CPEO | 1 | 1 | 0 | 1 | Profound | 1/1 | 0/1 | 1/1 | 0 | 29 |
| A8108G | CO2 | Reported | SNHL | 1 | 1 | 0 | 0 | Low frequency | 1/1 | 1/1 | 1/1 | 0 | 30 |
| A8348G | tRNALys | Reported | SNHL/Cardiomyopathy/HT | 1 | 0 | 1 | 0 | Low frequency | 1/1 | 0/1 | 1/1 | 0 | 31 |
| G11696A | ND4 | Reported | SNHL/LHON/LDYT/HT | 4 | 0 | 4 | 2 | Profound | 1/4 | 1/4 | 0/4 | 0 | 32 |
| A14693G | tRNAGlu | Reported | SNHL/MELAS/LHON/HT | 1 | 0 | 1 | 1 | Profound | 0/1 | 0/1 | 0/1 | 0 | 33 |
| G15927A | tRNAThr | Point mutation/Polymorphism | SNHL/MS | 4 | 1 | 3 | 4 | High frequency | 3/4 | 0/4 | 0/4 | 0 | 34 |
| Total | 19/254 (7.5%) | 19/140 (13.6%) | 20/34 | 15/33 | 10/33 | ||||||||
Abbreviations: AD, Alzheimer's disease; DM, diabetes mellitus; FSGS, focal segmental glomerulosclerosis; HT, hypertension; LDYT, Leber's hereditary optic neuropathy and dystonia; LHON, Leber hereditary optic neuropathy; LVNC, left ventricular non-compaction; MELAS, mitochondrial encephalomyopathy lactic acidosis, and stroke-like episodes; MIDD, maternally inherited diabetes and deafness; MS, multiple sclerosis; SNHL, sensorineural hearing loss.
‘Point mutation/Polymorphism' status indicates that some published reports have determined the mutation to be a non-pathogenic polymorphism.
Based on the MITOMAP database; ‘Reported' status indicates that one or more reports have considered the mutation as possibly pathologic.
With regard to the audiogram configuration, various types were found. In all, 69% (79% in Cohort 1 and 59% in Cohort 2) of the patients had progressive hearing loss and 59% (74% in Cohort 1 and 45% in Cohort 2) had tinnitus, while 34% (39% in Cohort 1 and 30% in Cohort 2) of the patients were associated with vertigo (Tables 1 and 2). Concerning clinical symptoms other than hearing loss, 80% (8/10) of the patients with the 3243A>G mutation had diabetes mellitus, but no other clinical symptoms were noticed (Table 1).
Ten novel variants that were not included in the public mtDNA databases were found in this study and they were located in the 16S rRNA, ND1, COI, ATPase6, ND4L, ND5, and Cytb regions (Table 3). All new variants were found in only one different family each.
Table 3. Ten novel mitochondrial SNPs.
| Location | Mutation | Conservation in 61 species (base) (/61) | Conservation rate (base) (%) | Amino-acid change | Conservation in 61 species (amino acid) (/61) | Conservation rate (amino acid) (%) | Amino-acid number/all amino acid of locus | Control (/192) | Mode of inheritance | Type of hearing loss |
|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA | 2069T>C | 16 | 31.4 | — | — | — | — | 0 | AD or Mita | High frequency |
| 16S rRNA | 2285T>C | 22 | 43.1 | — | — | — | — | 0 | AD or Mita | High frequency |
| 16S rRNA | 2285T>G | 22 | 43.1 | — | — | — | — | 0 | Sporadic | Dish shaped |
| 16S rRNA | 2634T>C | 34 | 66.7 | — | — | — | — | 0 | Sporadic | Profound |
| ND1 | 3595A>G | 54 | 88.5 | Asn>Asp | 54 | 88.5 | 97/318 | 0 | AD or Mita | High frequency |
| COI | 6204A>G | 61 | 100 | Ser>Gly | 61 | 100 | 101/513 | 0 | AD or Mita | High frequency |
| ATPase6 | 9124A>G | 60 | 98.4 | Thr>Ala | 59 | 96.7 | 200/226 | 0 | Sporadic | Unilateral |
| ND4L | 10680G>A | 59 | 96.7 | Ala >Thr | 59 | 96.7 | 71/98 | 0 | Sporadic | Unknown |
| ND5 | 13153A>G | 44 | 72.1 | Ile >Val | 35 | 57.4 | 273/603 | 0 | Sporadic | High frequency |
| Cytb | 15003G>C | 61 | 100 | Gly >Ala | 61 | 100 | 86/380 | 0 | Sporadic | Profound |
Abbreviation: SNPs, single-nucleotide polymorphisms
AD or Mit; autosomal dominant inheritance or maternal inheritance.
Four of the novel variants were found in the 16S rRNA gene: 2069T>C, 2285T>G, 2285T>C and 2634T>C. Although the 2634T>C variant had a high conservation rate (66.7%), the 2069T>C, 2285T>G and 2285T>C variants had low conservation rates: 31.4, 43.1 and 43.1%, respectively.
The remaining six novel variants were located in the protein coding regions: 3595A>G in NADH dehydrogenase 1 gene (MTND1 (MIM 516000), 6204A>G in cytochrome oxydase I gene (MTCOI (MIM 516030)), 9124A>G in ATPase 6 gene (MTATP6 (MIM 516060)), 10680G>A in NADH dehydrogenase 4L gene (MTND4L (MIM 516004)), 13153A>G in NADH dehydrogenase 5 gene (MTND5 (MIM 516005)) and 15003G>C in cytochrome b gene (MTCYB (MIM 516020)).
These variants are found in very well-conserved gene positions (57.4–100%).
The conservation rates in all ‘Confirmed' mtDNA mutations were high (Table 4).
Table 4. Conservation rate of ‘Confirmed' mitochondrial mutations.
| Location | Mutation | Conservation in 61 species (base) (/61) | Conservation rate (base) (%) |
|---|---|---|---|
| 12S rRNA | 1494A>G | 61 | 100.0 |
| 12S rRNA | 1555A>G | 56 | 91.8 |
| tRNALeu (UUR) | 3243A>G | 60 | 98.4 |
| tRNALeu (UUR) | 3291T>C | 58 | 95.0 |
| tRNASer (UCN) | 7445A>G | 42 | 68.9 |
| tRNASer (UCN) | 7511T>C | 60 | 98.4 |
| tRNALys | 8363G>A | 49 | 80.3 |
| tRNAHis | 12147G>A | 61 | 100.0 |
| tRNAGlu | 14709T>C | 58 | 95.0 |
However, as in Table 3, the 9124A>G, 10680G>A, 13153A>G and 15003G>C variants were found in sporadic cases which are not generally compatible with mitochondrial deafness. On the basis of the above evaluations, we categorized 3595A>G, and 6204A>G as possibly pathogenic mutants, and the remaining eight others as uncertain pathogenic mutants.
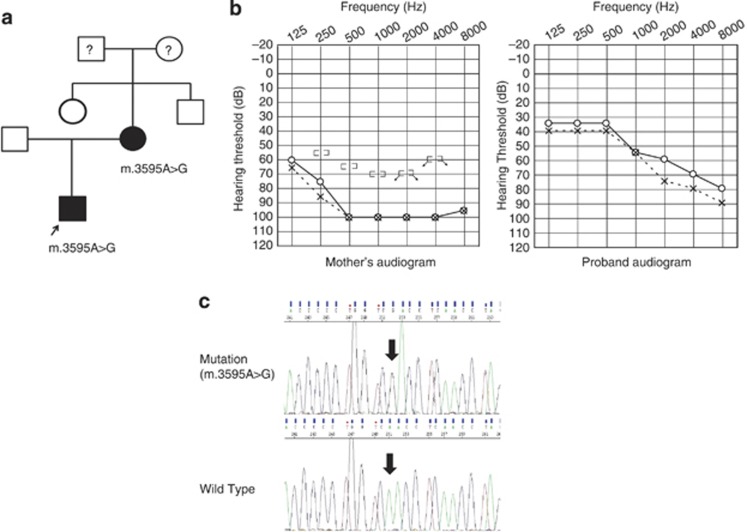
The homoplasmic mutation 3595A>G in the ND1 was found in a 4-year-old male patient with prelingual, severe hearing loss of high frequencies (Figure 1). He was suspected to have hearing impairment when he was about 1 year old, but ABR testing and Computed Tomography resulted in a diagnosis of normal hearing. However, when he was 3 years old, his mother again suspected that he had hearing impairment and testing confirmed it. The mother, who had the same mutation, also had hearing impairment as well as progressive bilateral tinnitus and occasional vertigo from childhood.
Figure 1.
Clinical features of the proband carrying the homoplasmic 3595A>G variant. (a) Family pedigree. Individuals with hearing loss are indicated by filled symbols. The arrow indicates the proband. (b) Audiograms of the proband and mother. (c) Electropherogram depicting the 3595A>G sequence and its flanks. Arrow indicates the position of the 3595A>G variant.
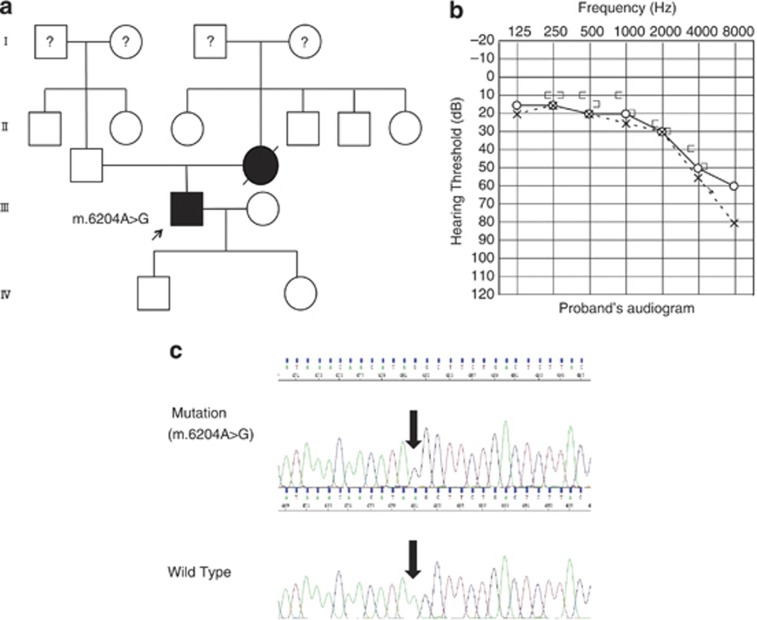
The homoplasmic mutation 6204A>G in the COI gene was found in a 62-year-old male with mild hearing loss of high frequencies (Figure 2). He noticed his hearing loss at the age of 50 and suffered from tinnitus, and mild diabetes mellitus. His mother also had hearing impairment that gradually progressed with age. DNA samples were not obtained from other family members.
Figure 2.
Clinical features of the proband carrying the homoplasmic 6204A>G variant. (a) Family pedigree. Individuals with hearing loss are indicated by filled symbols. The arrow indicates the proband. (b) Audiogram of the proband. (c) Electropherogram depicting the 6204A>G sequence and its flanks. Arrow indicates the position of the 6204A>G variant.
Discussion
Nineteen known mitochondrial mutations were found predominantly in the maternally inherited group (Tables 1 and 2). Clarification of pathogenecity of mitochondrial substitutions was hampered by low penetrance (probably due to heteroplasmy). Therefore, based on the MITOMAP database, they were classified as ‘Confirmed' or ‘Ambiguous-status' substitutions (Tables 1 and 2). The ‘Confirmed' mitochondrial mutations were found predominantly in Cohort 1 rather than in Cohort 2 (14.6 vs 0.7%), supporting the pathogenicity of these mutations. Frequencies of 1555A>G and 3243A>G mutations were significantly high, indicating that these two mutations are important causes of maternally inherited hearing loss. In general, patients with these mitochondrial mutations showed more or less similar clinical characteristics, that is, progressive hearing loss with tinnitus (Table 1).
Among the 10 novel variants (Table 3), two, the ND1 mutation 3595A>G and COI mutation 6204A>G, are thought to be possibly pathogenic, because (1) they are found in autosomal dominant or maternal inheritance (some of the others are found as sporadic cases); (2) the conservation rate of the variation at the position among mammals is at least over 50%, as is the conservation rate in all confirmed mtDNA mutations associated with phenotypes (Table 4) and (3) they are associated with high frequency hearing loss; the characteristic hearing type of mitochondrial hearing loss. These mutations affected a conserved nucleotide in the mitochondrial gene in primates and other species and had a conservation index of >50% (88.5 and 100%, respectively). None of these mutations were found in the controls or in the databases, further indicating that they are associated with hearing loss, however, no conclusion can be drawn without enzymatic analysis. Unfortunately, this study was a retrospective study using collected DNA samples from 1995 to 2012, so it was impossible to contact the patients and to get muscle or living samples from them. Therefore, enzymatic analysis of these mtDNA samples was not feasible.
In this study, we found one novel possibly pathogenic mutation in the ND1 hydrophobic arm region, in a patient with a homoplasmic 3595A>G mutation and hearing loss of the high frequencies from age 3 without complications. The family members of this patient did not have diabetes mellitus.
On the other hand, the novel possibly pathogenic mutation 6204A>G was located in the COI gene. The amino-acid conservation rate of this position was 100% (61/61 mammals). In previous reports, more than 20 pathogenic mutations in the MT-ND1 gene were reported in patients with LHON (Leber's hereditary optic neuropathy) and MELAS. Also, ND1 mutation-related hearing impairment has been reported: 3308T>C causing MELAS with deafness,16 3395A>G causing hypertrophic cardiomyopathy with profound SNHL,17 and 3396T>C and 3421G>A causing maternally inherited diabetes and deafness.18, 19 Three COI mutations related to hearing loss have also been reported (7443A>G,20 7444G>A21 and 7445A>G7, 8). Our results taken with these previous reports support the possibility that mutations in the ND1 and COI regions are associated with hearing impairment.
Most of the mtDNA mutations associated with hearing loss indicate low penetrance explained as a mild biochemical defect indicating that the mutation itself is not sufficient to produce the clinical phenotype. Thus, other modifying factors including nuclear backgrounds, environmental factors and mitochondrial haplotypes are necessary for the phenotypic manifestation of the mutation. The degree of hearing loss from mtDNA mutation can be similar within individual families but varied among different family groups, probably due to the modifier effect by nuclear genes.22
Conclusion
Nineteen known mitochondrial mutations were found predominantly in the maternally inherited group. Among them, frequencies of 1555A>G and 3243A>G mutations were significantly high, indicating that these two mutations are important causes of maternally inherited hearing loss. In addition to the previously reported mitochondrial mutations, we detected 10 novel homoplasmic mutations in the mitochondrial genes related to hearing loss by direct sequencing of whole mitochondrial genomes in Japanese patients. Two of them, 3595A>G and 6204A>G, are possibly associated with hearing loss.
Acknowledgments
We thank the participants of the Deafness Gene Study Consortium: Drs Norihito Takeichi and Satoshi Fukuda (Hokkaido University), Drs Atsushi Namba and Hideichi Shinkawa (Hirosaki University), Drs Yumiko Kobayashi and Hiroaki Sato (Iwate Medical University), Drs Tetsuaki Kawase and Toshimitsu Kobayashi (Tohoku University), Drs Tomoo Watanabe, Tsukasa Ito and Masaru Aoyagi (Yamagata University), Drs Hiroshi Ogawa and Koichi Omori (Fukushima Medical University), Drs Kotaro Ishikawa and Keiichi Ichimura (Jichi Medical University), Drs Kyoko Nagai and Nobuhiko Furuya (Gunma University), Drs Shuntaro Shigihara, Yasuyuki Nomura and Minoru Ikeda (Nihon University School), Drs Tetsuo Ikezono and Toshiaki Yagi (Nippon Medical School), Dr Shunichi Tomiyama (Nippon Medical School Tama Nagayama Hospital), Drs Hiromi Kojima, Yuika Sakurai and Hiroshi Moriyama (Jikei University), Dr Kozo Kumakawa (Toranomon Hospital), Drs Hajime Sano and Makito Okamoto (Kitasato University), Dr Satoshi Iwasaki (Hamamatsu Medical University), Dr Kazuhiko Takeuchi (Mie University), Dr Masako Nakai (Shiga Medical Center for Children), Drs Masahiko Higashikawa and Hiroshi Takenaka (Osaka Medical College), Drs Yuko Saito and Masafumi Sakagami (Hyogo College of Medicine), Dr Yasushi Naito (Kobe City Medical Center General Hospital), Drs Keiji Fujihara, Akihiro Sakai and Noboru Yamanaka (Wakayama Medical University), Drs Kunihiro Fukushima and Kazunori Nishizaki (Okayama University), Drs Kazuma Sugahara and Hiroshi Yamashita (Yamaguchi University), Drs Naoto Hato and Kiyofumi Gyo (Ehime University), Drs Yasuhiro Kakazu and Shizuo Komune (Kyushu University), Drs Mayumi Sugamura and Takashi Nakagawa (Fukuoka University), Dr Haruo Takahashi (Nagasaki University), Dr Yukihiko Kanda (Kanda ENT Clinic), Drs Hirokazu Kawano and Tetsuya Tono (Miyazaki Medical College), Drs Ikuyo Miyanohara andYuichi Kurono (Kagoshima University), Drs Akira Ganaha and Mikio Suzuki (Ryukyus University) for providing samples of their patients. We thank all the families who participated in the present study. We would also like to thank Ms. S Matsuda for technical assistance and Ms AC Apple-Mathews for help in preparing the manuscript.
References
- Jacobs H. T., Hutchin T. P., Kappi T., Gillies G., Minkkinen K., Walker J., et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur. J. Hum. Genet. 2005;13:26–33. doi: 10.1038/sj.ejhg.5201250. [DOI] [PubMed] [Google Scholar]
- Prezant T. R., Agapian J. V., Bohlman M. C., Bu X., Oztas S., Qiu W. Q., et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Fischel-Ghodsian N., Prezant T. R., Bu X., Oztas S. Mitochondrial ribosomal RNA gene mutation in a patient with sporadic aminoglycoside ototoxicity. Am. J. Otolaryngol. 1993;14:399–403. doi: 10.1016/0196-0709(93)90113-l. [DOI] [PubMed] [Google Scholar]
- Usami S., Abe S., Akita J., Namba A., Shinkawa H., Ishii M., et al. Prevalence of mitochondrial gene mutations among hearing impaired patients. J. Med. Genet. 2000;37:38–40. doi: 10.1136/jmg.37.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Li R., Wang Q., Yan Q., Deng J. H., Han D., et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ouweland J. M., Lemkes H. H. P., Ruitenbeek W., Sandkjujl L. A., deVijlder M. F., Struyvenberg P. A., et al. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- Reid F. M., Vernham G. A., Jacobs H. T. A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum. Mutat. 1994;3:243–247. doi: 10.1002/humu.1380030311. [DOI] [PubMed] [Google Scholar]
- Fischel-Ghodsian N., Prezant T. R., Fournier P., Stewart I. A., Maw M. Mitochondrial mutation associated with nonsyndromic deafness. Am. J. Otolaryngol. 1995;16:403–408. doi: 10.1016/0196-0709(95)90078-0. [DOI] [PubMed] [Google Scholar]
- Hutchin T. P., Parker M. J., Young I. D., Davis A. C., Pulleyn L. J., Deeble J. A novel mutation in the mitochondrial tRNA(Ser(UCN)) gene in a family with non-syndromic sensorineural hearing impairment. J. Med. Genet. 2000;37:692–694. doi: 10.1136/jmg.37.9.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Greinwald J. H., Jr., Yang L., Choo D. I., Wenstrup R. J., Guan M. X., et al. Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J. Med. Genet. 2004;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan D., Bressman S., Bruno C., Przedborski S., Shanske S. A novel mitochondrial 12SrRNA point mutation in parkinsonism, deafness, and neuropathy. Ann. Neurol. 2000;48:730–736. [PubMed] [Google Scholar]
- Mutai H., Kouike H., Teruya E., Takahashi-Kodomari I., Kakishima H., Taiji H., et al. Systematic analysis of mitochondrial genes associated with hearing loss in the Japanese population: dHPLC reveals a new candidate mutation. BMC. Med. Genet. 2011;12:135. doi: 10.1186/1471-2350-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneux S., Fransen E., Van Eyken E., Van Laer L., Huyghe J., Van de Heyning P., et al. Inherited mitochondrial variants are not a major cause of age-related hearing impairment in the European population. Mitochondrion. 2011;11:729–734. doi: 10.1016/j.mito.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Zaragoza M. V., Brandon M. C., Diegoli M., Arbustini E., Wallace D. C. Mitochondrial cardiomyopathies: how to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny. Eur. J. Hum. Genet. 2011;19:200–207. doi: 10.1038/ejhg.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveque M., Marlin S., Jonard L., Procaccio V., Reynier P., Amati-Bonneau P., et al. Whole mitochondrial genome screening in maternally inherited non-syndromic hearing impairment using a microarray resequencing mitochondrial DNA chip. Eur. J. Hum. Genet. 2007;15:1145–1155. doi: 10.1038/sj.ejhg.5201891. [DOI] [PubMed] [Google Scholar]
- Li X., Fischel-Ghodsian N., Schwartz F., Yan Q., Friedman R. A., Guan M. X. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids. Res. 2004;32:867–877. doi: 10.1093/nar/gkh226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamkha I., Mkaouar-Rebai E., Aloulou H., Chabchoub I., Kifagi C., Fendri-Kriaa N., et al. A novel m.3395A>G missense mutation in the mitochondrial ND1 gene associated with the new tRNA(Ile) m.4316A>G mutation in a patient with hypertrophic cardiomyopathy and profound hearing loss. Biochem. Biophys. Res. Commun. 2011;404:504–510. doi: 10.1016/j.bbrc.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Mkaouar-Rebai E., Tlili A., Masmoudi S., Belguith N., Charfeddine I., Mnif M., et al. Mutational analysis of the mitochondrial tRNALeu(UUR) gene in Tunisian patients with mitochondrial diseases. Biochem. Biophys. Res. Commun. 2007;355:1031–1037. doi: 10.1016/j.bbrc.2007.02.083. [DOI] [PubMed] [Google Scholar]
- Chen F. L., Liu Y., Song X. Y., Hu H. Y., Xu H. B., Zhang X. M., et al. A novel mitochondrial DNA missense mutation at G3421A in a family with maternally inherited diabetes and deafness. Mutat. Res. 2006;602:26–33. doi: 10.1016/j.mrfmmm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Pandya A., Xia X. J., Erdenetungalag R., Amendola M., Landa B., Radnaabazar J., et al. Heterogenous point mutations in the mitochondrial tRNASer(UCN) precursor coexisting with the A1555G mutation in deaf students from Mongolia. Am. J. Hum. Genet. 1999;65:1803–1806. doi: 10.1086/302658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. D., Voljavec A. S., Lott M. T., Torroni A., Yang C. C., Wallace D. C. Mitochondrial DNA complex I and III mutations associated with Leber's hereditary optic neuropathy. Genetics. 1992;130:163–173. doi: 10.1093/genetics/130.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Campos Y., Rengo C., Sellitto D., Achilli A., Magri C., et al. Mitochondrial DNA haplogroups do not play a role in the variable phenotypic presentation of the A3243G mutation. Am. J. Hum. Genet. 2003;72:1005–1012. doi: 10.1086/373936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue C. M., Tanji K., Hadjigeorgiou G., Andreu A. L., Nishino I., Krishna S., et al. Maternally inherited hearing loss in a large kindred with a novel T7511C mutation in the mitochondrial DNA tRNA(Ser(UCN)) gene. Neurology. 1999;52:1905–1908. doi: 10.1212/wnl.52.9.1905. [DOI] [PubMed] [Google Scholar]
- Lu J., Li Z., Zhu Y., Yang A., Li R., Zheng J., et al. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion. 2010;10:380–390. doi: 10.1016/j.mito.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano T., Tanaka M., Ohno K., Yoneda M., Ota Y., Terasaki H., et al. Mitochondrial DNA mutations associated with the 11778 mutation in Leber's disease. Biochem. Mol. Bio. Int. 1996;38:693–700. [PubMed] [Google Scholar]
- Li Z., Li R., Chen J., Liao Z., Zhu Y., Qian Y., et al. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum. Genet. 2005;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaksch M., Hofmann S., Kaufhold P., Obermaier-Kusser B., Zierz S., Gerbitz K. D. A novel combination of mitochondrial tRNA and ND1 gene mutations in a syndrome with MELAS, cardiomyopathy, and diabetes mellitus. Hum. Mutat. 1996;7:358–360. doi: 10.1002/(SICI)1098-1004(1996)7:4<358::AID-HUMU11>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Chen F. L., Liu Y., Song X. Y., Hu H. Y., Xu H. B., Zhang X. M., et al. A novel mitochondrial DNA missense mutation at G3421A in a family with maternally inherited diabetes and deafness. Mutat. Res. 2006;602:26–33. doi: 10.1016/j.mrfmmm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Spagnolo M., Tomelleri G., Vattemi G., Filosto M., Rizzuto N., Tonin P. A new mutation in the mitochondrial tRNA(Ala) gene in a patient with ophthalmoplegia and dysphagia. Neuromuscul. Disord. 2001;11:481–484. doi: 10.1016/s0960-8966(01)00195-x. [DOI] [PubMed] [Google Scholar]
- Wang Q., Li R., Zhao H., Peters J. L., Liu Q., Yang L., et al. Clinical and molecular characterization of a Chinese patient with auditory neuropathy associated with mitochondrial 12S rRNA T1095C mutation. Am. J. Med. Genet. A. 2005;133A:27–30. doi: 10.1002/ajmg.a.30424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki F., Tanaka M., Kawamura K., Kanzaki Y., Okabe M., Hayashi T., et al. A case of cardiomyopathy showing progression from the hypertrophic to the dilated form: association of Mt8348A—>G mutation in the mitochondrial tRNA(Lys) gene with severe ultrastructural alterations of mitochondria in cardiomyocytes. Jpn. Circ. J. 2001;65:691–694. doi: 10.1253/jcj.65.691. [DOI] [PubMed] [Google Scholar]
- De Vries D. D., Went L. N., Bruyn G. W., Scholte H. R., Hofstra R. M., Bolhuis P. A., et al. Genetic and biochemical impairment of mitochondrial complex I activity in a family with Leber hereditary optic neuropathy and hereditary spastic dystonia. Am. J. Hum. Genet. 1996;58:703–711. [PMC free article] [PubMed] [Google Scholar]
- Tzen C. Y., Thajeb P., Wu T. Y., Chen S. C. Melas with point mutations involving tRNALeu (A3243G) and tRNAGlu(A14693g) Muscle Nerve. 2003;28:575–581. doi: 10.1002/mus.10473. [DOI] [PubMed] [Google Scholar]
- Chen B., Sun D., Yang L., Zhang C., Yang A., Zhu Y., et al. Mitochondrial ND5 T12338C, tRNA (Cys) T5802C, and tRNA (Thr) G15927A variants may have a modifying role in the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese pedigrees. Am. J. Med. Genet. A. 2008;146A:1248–1258. doi: 10.1002/ajmg.a.32285. [DOI] [PubMed] [Google Scholar]