Abstract
Circadian systems have evolved in plants, eubacteria, neurospora, and the metazoa as a mechanism to optimize energy acquisition and storage in synchrony with the rotation of the Earth on its axis. In plants, circadian clocks drive the expression of genes involved in oxygenic photosynthesis during the light and nitrogen fixation during the dark, repeating this cycle each day. In mammals, the core clock in the suprachiasmatic nucleus (SCN) functions to entrain extra-SCN and peripheral clocks to the light cycle, including regions central to energy homeostasis and sleep, as well as peripheral tissues involved in glucose and lipid metabolism. Tissue-specific gene targeting has shown a primary role of clock genes in endocrine pancreas insulin secretion, indicating that local clocks play a cell-autonomous role in organismal homeostasis. A present focus is to dissect the consequences of clock disruption on modulation of nuclear hormone receptor signaling and on posttranscriptional regulation of intermediary metabolism. Experimental genetic studies have pointed toward extensive interplay between circadian and metabolic systems and offer a means to dissect the impact of local tissue molecular clocks on fuel utilization across the sleep–wake cycle.
Circadian clocks represent a network of internal oscillators that maintain ~24-h periodicity in behavioral and physiological systems. In mice, electrolytic ablation studies localized an area of hypothalamus superior to the optic chiasm (suprachiasmatic nucleus, SCN) as necessary for circadian locomotor activity rhythms (Mohawk and Takahashi 2011). The SCN pacemaker neurons are entrained by light and sustain internal circadian alignment with the environmental cycle through both direct neural projections and secreted factors (Hastings and Herzog 2004; Saper 2006; Mohawk and Takahashi 2011). Forward genetic approaches originally in flies, and subsequently in neurospora and mice, led to the discovery of the first clock genes and established that the core oscillator is encoded by a transcription feedback loop composed of basic helix–loop–helix PAS (Per Arnt Sim) domain transcription factors, in addition to proline- and acid-rich (PAR) basic region leucine zipper (bZIP) factors (reviewed in Zhou et al. 1997; Takahashi et al. 2008; Rosbash 2009; Dibner et al. 2010). Interestingly, these transcription factor superfamilies function as both receptors and DNA binding proteins and are responsive to environmental flux, such as changes in oxygen, xenobiotics, and lipids (McIntosh et al. 2010). A paradigm shift in studies of animal clocks was triggered by the discovery that clock transcription factors are expressed and oscillate not only within pacemaker neurons, but also within peripheral cells. At the local tissue level, the observation that a majority of the 49 known neuroendocrine nuclear hormone receptors show strong circadian oscillation points toward interdependence of pathways involved in energy homeostasis and circadian rhythmicity. Over the past decade, the availability of mice with loss- and gain-of-function mutations in clock genes has enabled systematic analysis of the role of clocks in processes ranging from cell cycle control to feeding, neuroendocrine function, vascular biology, and glucose metabolism. The purpose of this chapter is to highlight how investigation into the molecular organization of circadian systems has begun to enlighten our understanding of mammalian energy homeostasis and the interdependence of circadian and metabolic processes.
RHYTHMIC FEEDBACK OSCILLATORS IN METABOLIC PATHWAYS AND CLOCK TRANSCRIPTION NETWORKS
Molecular Organization of Circadian Clocks: Relationship to Metabolic Oscillators
Metabolic oscillations have been recognized in a wide variety of organisms for many years, with classical studies focused on glycolytic cycles by Britton Chance in the 1960s (Ghosh and Chance 1964; Betz and Chance 1965; Bass and Takahashi 2011; Chandra et al. 2011). Recognition that many metabolic cycles show ~24-h periodicity even in prokaryotes led McKnight and colleagues to suggest that rhythmic cycling may couple energetic, growth, and reproductive pathways (Tu and McKnight 2006). Just as distinct organelles enable spatial separation of incompatible biochemical processes within the cell, so too temporal partitioning prevents incompatible energetic processes from occurring simultaneously. Circadian cycles represent a subset of rhythmic oscillators in light-sensitive organisms that couple internal biological processes to the rising and setting of the sun. For photo-synthetic organisms, including cyanobacteria and plants, the coupling of ~24-h oscillators to energy homeostasis appears to be direct because photons of light are captured during the oxygenic phase of the daily cycle, whereas nitrogen fixation converts acquired energy to usable fuel during the dark. That circadian oscillators provide a selective advantage has been suggested by so-called resonance studies in cyanobacteria (Ouyang et al. 1998) and plants (Dodd et al. 2005), in which survival and reproduction have been shown to directly correlate with phase alignment (or resonance) of the external light–dark cycle with internal period length. The observations that nuclear hormone receptors show robust circadian rhythms and that there is convergence between metabolic transcription factors and core clock genes (e.g., Rev-erbα/β; Pgc1α/Pparα; Grimaldi et al. 2010) suggest the presence of extensive coordination of fuel and nutrient cycles in vertebrates (Yang et al. 2006; So et al. 2009; Schmutz et al. 2010). Further, the expression of nearly 10% of the genome has been found to display rhythmicity, including the transcription of key genes involved in glycolytic and oxidative fuel cycles, in addition to lipogenic pathways (Panda et al. 2002; Storch et al. 2002; Hatanaka et al. 2010; Rey et al. 2011).
Despite intensive focus on the transcriptional interconnections between circadian and metabolic networks, a related line of investigation has posed a “chicken or the egg” puzzle, that is: Is gene transcription necessary to produce ~24-h metabolic cycles? A surprising result in cyanobacteria was the demonstration that in vitro expression of just three proteins (in the absence of nuclei or transcription) was sufficient to reconstitute 24-h patterns of phosphorylation (Nakajima et al. 2005). Recent studies of peroxiredoxins, members of a conserved family of redox-sensitive antioxidants proteins, has shown that the oscillation between oxidized and reduced states of the enzyme persists with a 24-h periodicity even in the absence of gene transcription in both prokaryotes and in human red blood cells. These studies pose the question as to whether genetic programming is dispensable for circadian function (Bass and Takahashi 2011; O’Neill and Reddy 2011; O’Neill et al. 2011). They also raise provocative theoretical points concerning the metabolic sensors responsible for gaging “time”—suggesting that redox state per se may be the most basic signal. Although these experimental observations raise intriguing questions concerning periodic phenomenon, in the intact organism, metabolic cycles are coupled to and dependent on circadian transcription cycles. In summary, metabolic oscillations can be reproduced in vitro and observed in living prokaryotic and eukaryotic cells where they are tied to pathways involved in fuel utilization, reproduction, and growth (Tu and McKnight 2006; Cai et al. 2011). Indeed clock genes have evolved four different times, and as such appear to confer a unique selective advantage; whether this arises as a result of energetic benefits remains a question for further investigation.
Integration of Circadian Systems, Sleep, and Feeding in Animals
In metazoa, the hypothesis that circadian cycles confer energetic advantage is complicated by the complexity of feeding as a means of energy acquisition. Long-term energy constancy is now best understood as the net balance between the production of anabolic and adipostatic hormones generated in the nutrient replete state versus the suppression of anabolic factors and activation of catabolic signals during fasting and starvation. Indeed the daily fasting–feeding cycle is closely coupled with the sleep–wake cycle, and both processes are influenced by a combination of circadian and nutrient signals (Laposky et al. 2008a; Green et al. 2008). Disruption of the clock, feeding time, and/or light cycles alters nutrient balance and increases susceptibility to obesity (Arble et al. 2009; Fonken et al. 2010). Conversely, changes in metabolism or the nutrient status of the organism leads to behavioral and molecular disruption of the clock (Fig. 1). Indeed, leptin, a central anorectic signal produced in proportion to adipose stores, also impacts sleep architecture (Laposky et al. 2006; Laposky et al. 2008b). Clock genes also influence both sleep and feeding, and clock disruption leads to fragmented night-eating, akin to the human night-eating syndrome (Dudley et al. 2003; Stunkard and Lu 2010). In humans, sleep restriction influences appetite (Spiegel et al. 2004; Taheri et al. 2004) and, at a population level, sleep loss is correlated with increases in obesity (Bass and Turek 2005; Spiegel et al. 2009). Orexin/hypocretin, a neuropeptide produced in lateral hypothalamic neurons that stimulates wakefulness (Adamantidis et al. 2007) and which is deficient in narcolepsy (Lin et al. 1999), is also involved in feeding and energy balance (Sakurai et al. 1998; Funato et al. 2009). Indeed, disruption of circadian clock function has also been shown to affect reproduction in mice (Miller et al. 2004), indicating that reproductive fitness, energy balance, sleep, and circadian homeostasis may be interdependent in animals.
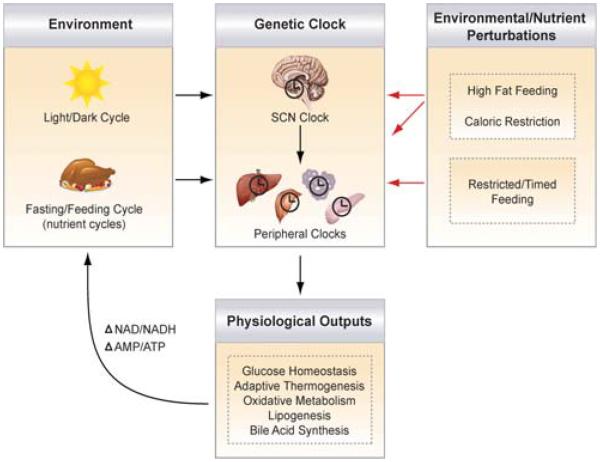
Figure 1.
Interplay between clock, environment, and nutrient availability. The main pacemaker clock within the suprachiasmatic nucleus (SCN) receives photic input from the environment and synchronizes downstream extra-SCN and peripheral metabolic oscillators to optimize energy storage and utilization pathways (listed as physiological outputs). The daily cycles of fasting and feeding entrain the peripheral clocks, perhaps through changes in glucose, fatty acids, NAD/NADH and AMP/ATP ratios, or hormones (peptidergic or steroidal). Changes in food availability, such as in conditions of high-fat feeding, caloric restriction, and restricted feeding, also affect the clock. Although restricted feediing entrains just peripheral clocks, a hypocaloric diet (caloric restriction) has been shown to entrain both peripheral and SCN clocks. Thus, the circadian network optimizes the timing of downstream metabolic processes to coordinate with daily light–dark cycles and the nutrient millieu.
STUDIES IN GENETIC MODELS OF CLOCK DISRUPTION: EXPERIMENTAL CONSIDERATIONS
Whereas clinical studies in human subjects have shown that many aspects of endocrine physiology, including appetitive behavior and glucose metabolism (Spiegel et al. 2009; Kalsbeek et al. 2011), show pronounced circadian oscillation, analyses in genetically modified mice have begun to reveal interconnections between circadian and metabolic systems at the molecular level. Several conceptual considerations in experimental design are important to note in this new field. First, in mice, strong modifiers modulate the penetrance and phenotype of circadian genes with regard to effects on both locomotor activity and physiological endpoints; indeed strain modifiers provide potential new insight into gene network interactions. For instance, the lengthened period of the ClockΔ19 mutant mouse, originally generated by mutagenesis on the C57BL/6J strain, shows shortened period when introgressed into the Balbc/J strain (Shimomura et al. 2001). Similar strain modifier effects have been well documented in studies of the Egf receptor on embryogenesis and placental formation by Threadgill and colleagues (Strunk et al. 2004). Indeed a major effort of experimental genetics has been to delineate the biological basis for inbred strain effects on cell signaling and behavior (Justice et al. 1992). In an analogous way, characterization of monogenic mutations in families with advanced and delayed sleep phase syndromes has provided a molecular and genetic entry point to evaluate clock gene function in humans (Ptacek et al. 2007). Interestingly, studies of monogenic diabetes and obesity syndromes in both mice and humans have similarly proved central to understanding the core principles of nutrient homeostasis and glucose metabolism (Bell and Polonsky 2001).
A second issue in genetic analyses of clock function has been the use of multitissue mutants as a primary model (Rudic et al. 2004; Turek et al. 2005). However, it is difficult to tease apart the opposing effects of circadian disruption in distinct tissues in multitissue mutants, which involve clock defects in both brain and peripheral tissues. For instance, clock function in liver and muscle appears to mask the effects of clock in pancreas—indeed diabetes is weaker in ClockΔ19 and Bmal1−/− global animals compared with islet-specific mutants. In the future, understanding the interrelationship between brain–behavioral properties of the clock and peripheral tissues will necessitate selective ablation within distinct neuronal subpopulations.
Third, phenotypes observed in particular models may reflect “epiphenomena” rather than temporal aspects of physiology. Two points relate to this caveat: First, comparison of the effects on output pathways of loss (or gain) of function of clock activator genes (CLOCK/BMAL/NPAS2) versus clock repressors (CRYs/PERs) provides clues concerning the net effects on downstream outputs. That is, when opposite effects on physiology are observed following loss or gain of function in the activator versus repressor loop, then a reasonable inference would be that there is coupling of timing and physiology. An example of such an opposing outcome is the effect of loss of Clock or Bmal1 in liver (resulting in hypoglycemia) versus loss of the Cry genes (resulting in hyperglycemia). Similarly, at the molecular level, loss of Clock or Bmal1 results in deficiency of NAD+ and NAD+-dependent deacetylase activity, whereas loss of Cry exerts the opposite effects on NAD+. A full understanding of this yin–yang between clock activators and repressors will also require analysis of the different molecular actions of these two arms of the clock (i.e., CLOCK and BMAL1 are direct DNA binding transcription factors whereas PER/CRY function indirectly to repress transcription). Second, to understand whether physiological disorders in circadian mutants arise because of altered “timing” or instead because of altered feeding and behavior, it will be necessary to study these animals under identical nutrient states (i.e., fasting) at different circadian times to neutralize the nutrient modulation/masking of feeding. A final point deserves discussion: In all instances studied to date, a key aspect involves dynamic challenging; that is, in the steady unperturbed state certain phenotypes might not occur (or might be less measurable). Indeed clinical phenotyping of endocrine disorders in humans follows similar principles in that dynamic testing is a mainstay of decision algorithms and therapeutics.
Appreciation of reciprocal interaction between circadian and metabolic systems in vertebrates has been especially advanced through experimental genetic studies in animals harboring either multitissue or tissue-specific alterations in circadian gene function. Although a complete overview of the physiological effects of circadian gene disruption is beyond the scope of this review, several examples underscore the integral link between clock systems and energy homeostasis. In ClockΔ19 mutant mice, a gradual, albeit subtle, increase in body weight is observed when comparing littermate animals of mutant and wild-type genotypes (Turek et al. 2005). This observation prompted dynamic testing using high-fat calorically dense food to compare the relative weight gain and metabolic response to diet in ClockΔ19 and wild-type animals. Here, the difference between genotypes was exaggerated, with ClockΔ19 mutants displaying hyperphagia, visceral adiposity, and a shift in feeding behavior with more food consumed during the normal rest (sleep) period. Interestingly, despite the relative obesity of high-fat-fed ClockΔ19 mutants, these animals never developed hyperinsulinemia—suggesting a primary role of CLOCK at the level of endocrine pancreas glucose-stimulated insulin secretion. Indeed, selective ablation of Bmal1 within endocrine pancreas leads to profound hyperglycemia because of impaired insulin exocytosis (Turek et al. 2005). The relatively mild phenotype of the multitissue ClockΔ19 mutant can be explained by the observation that ablation of circadian gene activators (CLOCK and BMAL1) within liver results in hypoglycemia and increased insulin sensitivity (Lamia et al. 2008); thus, in multitissue mutants, the composite effects of clock function in different tissues (i.e., pancreas vs. liver) give rise to the net “glucose” phenotype. Conversely, multitissue ablation of the clock repressor CRY results in hyperglycemia, owing in part to derepression of the gluconeogenic actions of glucagon (Zhang et al. 2010). Ultimately, systematic ablation of clock function within each distinct metabolic depot (e.g., liver, skeletal muscle, adipose, and brain) will be necessary to more fully understand the genetic basis for circadian control of glucose homeostasis in both postprandial and fasting conditions. Although clock function in glucose homeostasis has been subject to intensive investigation, additional emergent work has also indicated a primary role for clock transcription factors in other aspects of metabolic control, including lipid absorption (Pan and Hussain 2009), adipogenesis (Kawai et al. 2011), adrenal glucocorticoid production (Oster et al. 2006), vascular response to injury (Anea et al. 2009), fibrinolytic homeostasis (Somanath et al. 2011), and renal sodium handling (Gumz et al. 2009). Thus a major outcome of circadian disruption in animals relates to the accelerated development of abnormalities in fuel handling, metabolic homeostasis, and cardiovascular disease.
NUTRIENT SENSORS LINKING METABOLISM TO THE CIRCADIAN CLOCK
Although we have discussed above how the circadian clock regulates downstream metabolic processes including gluconeogenesis and glucose-stimulated insulin secretion, for the remainder of the review we focus on how changes in nutrients and metabolism reciprocally affect the clock. Organisms have evolved to be able to anticipate and respond to daily changes not only in the light–dark cycle, but also in the availability of food. We are only now beginning to get a sense of the number of nutrient sensors, including the redox state, NAD+-dependent deacetylases, protein kinases, and nuclear hormone receptors, that convey information regarding the metabolic state of the cell to the core circadian clock to coordinate downstream clock-controlled events with the nutrient status of the organism.
NAD+-Dependent Sensors
One of the first indications that the clock could be influenced by the metabolic state of the cell came from early in vitro studies that showed that the cellular redox status, represented by the ratio of the nicotinamide adenine dinucleotide cofactors NAD+(H) and NADP+ (H), modulates the transcriptional activity of CLOCK/BMAL1 and NPAS2/BMAL1 (Rutter et al. 2001). The reduced forms (NADH and NADPH) increase binding, while the oxidized forms (NAD+ and NADP+) decrease binding of the clock heterodimers to their E-box targets. More recent studies have further shown that levels of the metabolite NAD+ display daily rhythmicity in liver, probably owing in part to direct circadian transcriptional regulation of the gene encoding the rate-limiting enzyme in NAD+ biosynthesis, NAMPT (nicotinamide ribosyl-transferase; Nakahata et al. 2009; Ramsey et al. 2009). Although levels of Nampt RNA and NAD+ are reduced in livers from Clock and Bmal1 mutant mice, their levels are increased in livers of mice deficient for both Cry1 and Cry2, suggesting that Nampt and NAD+ production is indeed downstream of CLOCK/BMAL1. It is important to keep in mind, however, that other factors may also contribute to daily changes in NAD+ levels, including processes such as fluctuations in glycolysis or activation of the NAD+-consuming DNA repair enzyme PARP-1. Furthermore, it will be important to determine the relative importance of cycling NAD+ within the different subcellular compartments, such as mitochondria versus nucleus, on downstream metabolic outputs.
Although NAD+ is involved in cellular redox reactions within the mitochondria, it also serves as a substrate for the nutrient-responsive sirtuin (SIRT)1 deacetylase, which removes acetyl groups from proteins in an NAD+-dependent manner, producing a deacetylated protein, nicotinamide, and O-acetyl-ADP-ribose as by-products. Thus, the activity of SIRT1 is directly coupled to the redox status of the cell. Intriguingly, SIRT1 has recently been implicated as a major regulator of the core clock machinery, as SIRT1 binds to and inhibits the activity of CLOCK/BMAL1 (Asher et al. 2008; Nakahata et al. 2008) in addition to its role as a regulator of a host of downstream metabolic processes, including gluconeogenesis, fat metabolism, insulin secretion, and apoptosis (reviewed in Haigis and Guarente 2006; Dali-Youcef et al. 2007). Therefore, both clock and nutrient-dependent regulation of SIRT1 probably leads to modulation of such downstream metabolic targets as PPARa, PGC-1α, FOXOs, and CRTC2/TORC2 (Brunet et al. 2004; Rodgers et al. 2005; Liu et al. 2008; Purushotham et al. 2009). PGC-1α has also been shown to be tightly linked to circadian clock function, as it is rhythmically expressed in liver and skeletal muscle, and mice lacking PGC-1α display abnormal rhythms of locomotor activity, metabolic rate, and body temperature because of altered clock gene expression (Liu et al. 2007). Further, PGC-1α acts as a co-activator for the ROR nuclear hormone receptors, which in turn stimulate Bmal1 (Liu et al. 2007). Thus, CLOCK/BMAL1 regulates the activity of NAMPT and SIRT1, which in turn negatively regulate the activity of CLOCK/BMAL1, thereby generating a negative feedback loop intertwining the core clock and metabolic networks.
Of note, a second NAD+-dependent protein that acts as both a nutrient sensor and a modulator of the clock is the poly(ADP-ribose) polymerase PARP-1, an NAD+-dependent ADP ribosyltransferase. PARP-1 activity has been shown to be circadian in liver and regulated by feeding. Furthermore, during the light/rest phase, PARP-1 binds to and poly(ADP-ribosyl)ates CLOCK, regulating its ability to bind to DNA, particularly during the light/rest phase (Asher et al. 2010). However, it should be noted that cyclic NAD+ levels may not directly regulate PARP-1 activity, as PARP-1 has been shown to be able to retain its rhythmic activity even in the absence of a functional clock in hepatocytes. However, both SIRT1 and PARP-1 represent prime examples of how changes in the nutrient milieu can translate to modulation of the core clock to keep the feeding and circadian cycles in sync with one another. It will be interesting to determine the relative role of ribosylation versus acetylation in the pathophysiology of circadian mutant mice.
AMPK
Another cellular energy sensor that has recently been linked to the clock is AMP-activated protein kinase (AMPK), which senses the AMP/ATP ratio and is activated following increases in AMP to maintain cellular energy stores by elevating ATP levels through increasing oxidative metabolism and other catabolic pathways. AMPK has been shown to directly phosphorylate and activate CKIε, resulting in increased PER2 degradation and a phase advance of clock gene expression patterns (Um et al. 2007). AMPK also directly phosphorylates and destabilizes the core clock repressor CRY1 (Lamia et al. 2009). Interestingly, AMPK has also been shown to modulate NAMPT and SIRT1 activity following glucose restriction (Fulco et al. 2008; Canto et al. 2009); thus it is tempting to speculate that AMPK might also modulate circadian systems indirectly via activation of NAMPT and SIRT1.
Nutrient-Sensitive Transcription Factors
Nuclear hormone receptors (NHRs) are another family of nutrient-sensitive proteins that lie at the intersection of metabolic and circadian pathways. As mentioned above, more than half of the NHRs, which are transcription factors that are activated on binding of lipids and fat-soluble hormones, display rhythmic patterns of expression in multiple metabolic tissues (Yang et al. 2006). Several NHRs regulate not only downstream metabolic events but are also key components of the core molecular clock feedback loop. REV-ERBα, a critical regulator of gluconeogenesis, adipocyte differentiation, and lipid metabolism, represses Bmal1 transcription (Preitner et al. 2002), while RORα and PPARα, regulators of lipogenesis and glucose metabolism, positively regulate Bmal1 transcription (Sato et al. 2004; Akashi and Takumi 2005; Canaple et al. 2006). In addition to their regulation of Bmal1 transcription, they have also been recently found to directly interact with PER2 to modulate its activity (Schmutz et al. 2010). Indeed, mice lacking REV-ERBα or RORα display disrupted circadian rhythms of locomotor activity and clock gene expression (Preitner et al. 2002; Sato et al. 2004). Therefore, fatty acids, sterols, and other hormones may be able to communicate information regarding nutrient status to the clock via their activation of the NHRs.
The discovery that nutrient-responsive families of proteins such as the sirtuins and the NHRs lie at the intersection of metabolic and circadian systems sheds light on the necessity of the organism to be able to coordinate regulation of fuel utilization with not only the rising and setting of the sun, but also with the acute daily changes in nutrient availability. Although the clock dictates baseline daily oscillations of sirtuin activity and NHR expression to be in sync with the light–dark cycle, acute changes in response to food consumption throughout the day further fine-tunes their ability to regulate downstream circadian and metabolic processes. For example, in addition to the clock-driven daily rhythms of expression, Nampt is further up-regulated in response to decreased glucose levels in skeletal muscle in an AMPK-dependent manner (Fulco et al. 2008), whereas SIRT1 activity is also augmented following fasting or caloric restriction in numerous metabolic tissues (reviewed in Imai 2009; Canto and Auwerx 2009). Thus, clock-driven regulation of nutrient-sensitive metabolic transcription factors, which in turn feed back to fine-tune the clock, is key to understanding the complex interplay between clocks and metabolism.
RESPONSE OF CIRCADIAN CLOCK TO CHANGES IN NUTRIENT AVAILABILITY
As discussed above, daily changes in nutrient availability translate critical information about the nutritional status of the cell to the circadian clock, allowing the clock to maintain “organismal homeostasis” and keeping physiological processes in sync with food availability. Normally, the cycles of fasting and feeding are in phase with the cycles of sleep and activity. However, a key question emerges as to the consequences on circadian rhythms when the nutrient milieu of an organism is significantly altered, such as in conditions of nutrient deprivation, nutritional excess, or when food is presented at the “wrong” time of the day.
Conditions of Reduced Energy Availability—Restricted Feeding and Caloric Restriction
Restricted feeding (RF) and caloric restriction (CR) exemplify different types of nutrient deprivation that entrain the biological clock. Whereas RF limits the timing and duration of food availability, CR restricts the total number of calories consumed (without malnutrition). Restricting food to a particular time of day during RF results in profound behavioral and physiological changes (reviewed in Escobar et al. 2009). Briefly, mice display food anticipatory activity (FAA), characterized by increased locomotor activity, body temperature, and corticosterone secretion 2–4 h before the presentation of food (Nelson et al. 1975; Duffy et al. 1990; Cambras et al. 1993; Challet et al. 1997). Although the clock probably plays a critical role in FAA, as mice deficient in Per2 do not display the anticipatory increase in activity (Feillet et al. 2006), FAA does persist in mice lacking Bmal1 (Pendergast et al. 2009; Storch and Weitz 2009). On a molecular level, RF entrains molecular clock oscillations in peripheral tissues, such as liver and kidney, without affecting the clock rhythms in the central pace-maker in the SCN (Damiola et al. 2000; Stokkan et al. 2001). Of note, RF shifts many of these physiological and molecular processes even in SCN-lesioned animals (Clarke and Coleman 1986; Marchant and Mistlberger 1997; Yoshihara et al. 1997), suggesting the existence of a food-entrainable oscillator (FEO) independent of the SCN. However, the anatomic location of the FEO is a topic of great debate, as several studies have suggested that the dorsomedial hypothalamus (DMH), a region of the brain involved in food intake and satiety, is critical to induce food-entrainable circadian rhythms (Gooley et al. 2006; Fuller et al. 2008), whereas others have questioned its role as the FEO (Landry et al. 2006). Another important question is what is the nature of the extracellular signals that are able to reset the peripheral clocks—is the cue from changes in redox state, food metabolites (glucose, cholesterol, fatty acids), hormones (ghrelin, leptin, cholecystokinin, insulin), or body temperature?
CR, similar to RF, likewise induces FAA and entrains peripheral clocks. However, unlike RF, CR also entrains the SCN clock (Cambras et al. 1993; Challet et al. 1998; Challet et al. 2003; Mendoza et al. 2005; Resuehr and Olcese 2005; Mendoza et al. 2007), suggesting that hypocaloric feeding itself is responsible for modulating the master pacemaker. When interpreting the effects of CR versus RF on the circadian clock, it is important to note that some of the effects of CR may actually be due to the altered timing of feeding rather than the reduced total caloric intake, as many CR protocols provide a set amount of food at a given time during the light period (i.e., the normal rest period for mice), similar to the timing of food provided during RF. To identify the effects of reduced caloric intake on the clock and metabolism independent of its effects on the shift in feeding time, it will be important to evaluate and perform CR studies whereby food is administered exclusively during the dark compared with the light period (Duffy et al. 1990; Challet et al. 1998).
CR is one of the most consistently proven methods to increase life span and delay age-associated pathologies, such as diabetes and cancer. Although multiple theories prevail as to the mechanism by which CR increases life span (reviewed in Masoro 2005; Sinclair 2005), the NAD+-dependent SIRT1 deacetylase has been shown to be required for the life span–extending effect of CR in multiple species (Lin et al. 2000; Lin et al. 2002; Rogina and Helfand 2004; Wood et al. 2004), in addition to its ability increase life span independent of CR (Kaeberlein et al. 1999; Tissenbaum and Guarente 2001; Rogina and Helfand 2004). Given the known effect of SIRT1 on the molecular clock (Asher et al. 2008; Nakahata et al. 2008), it is intriguing to speculate that SIRT1 is required for the effect of CR on the clock, and that the increased robustness of circadian rhythms following CR (Duffy et al. 1990) contributes to increased longevity of the organism. Interestingly, the sirtuin pathway has already been shown to be required for the increased activity observed in response to CR (Chen et al. 2005). Critical questions emerge: Can CR entrain the molecular clock in mice lacking SIRT1, and can CR still increase life span in mice lacking the circadian clock? Tissue-specific and whole body knockout models will be necessary to determine the relative role of brain versus peripheral tissues in the response of the clock to CR. A link between CR, circadian rhythms, and life span has also been shown in transgenic aMUPA mice, which overexpress a urokinase-type plasminogen activator (uPA), a serine protease involved in tissue remodeling and brain development (Froy et al. 2006). These mice are spontaneously calorically restricted, consuming only 70%–80% of the number of calories that controls eat, and share several characteristics with CR-fed mice, namely increased life span, reduced body weight, reduced IGF-1, and reduced tumor incidence. Interestingly, they also show increased robustness in the amplitude of several clock genes, including Per1, Per2, Cry1, and Clock, in liver, along with higher amplitude rhythms of food intake and body temperature, both clock-controlled events. Thus, taken together, it is likely that feeding regimens, circadian rhythms, and life span are intimately linked processes.
Conditions of Nutritional Excess and Genetic Models of Obesity
Perhaps an even more relevant question in today’s society of increasing waistbands is to understand the consequences of an overabundance of nutrients on circadian rhythms. Although it has been known for more than 20 years that fatty acids influence the period length of spore formation rhythms in Neurospora (Lakin-Thomas and Brody 1985), the molecular mechanisms underlying the link between nutrients and the clock have remained unclear. More recently, a growing body of evidence has revealed that alterations in metabolism—either environmentally or genetically induced—disrupt circadian rhythms and the sleep–wake cycle. For instance, feeding mice a high-fat diet has been shown to lead to rapid changes in the period of the locomotor activity, diurnal feeding patterns, and rhythmic expression of clock and nuclear hormone receptor genes within both brain and peripheral metabolic tissues, suggesting that a hypercaloric diet influences the function of the mammalian circadian clock (Kohsaka et al. 2007). High-fat feeding also leads to increased sleep time, but decreased sleep consolidation (Jenkins et al. 2006), and altered clock synchronization to light (Mendoza et al. 2008). Genetic models of obesity, such as the leptin-deficient ob/ob mice, similarly have decreased locomotor activity rhythms, increased NREM sleep time, and decreased sleep consolidation (Laposky et al. 2006).
Together, these data show the existence of a vicious cycle, whereby disruption of the circadian clock (either genetic or induced by high-fat diet) leads to increased risk of metabolic disorders such as diabetes and altered sleep; conversely, changes in metabolism or diet lead to changes in circadian control of metabolic processes and sleep.
CONCLUSIONS
The last decade has shed much light on the interconnectedness of circadian and metabolic networks, especially with the discovery that several nutrient-responsive NHRs, including REV-ERBα and RORα, are both regulated by and directly regulate the clock itself. The availability of knockout and transgenic mouse models further sheds light on the close relationship between the clock and metabolism, as mice with circadian defects (i.e., clock gene knockouts or altered feeding time) have revealed that a plethora of metabolic disorders arise as a consequence of clock disruption. Reciprocally, several mouse models of metabolic disruption (i.e., Pgc-1α knockouts or high-fat diet) have revealed disrupted circadian clock function. Given the vast number of oscillating RNAs in peripheral tissues, it will be critical for investigators to carefully consider the time of day when analyzing knockout mice, as phenotypes may differ depending on time of day. In view of the ever expanding number of nutrient sensors found to be linked to the clock, it will likewise be informative to examine effects on circadian rhythms themselves, although dynamic testing, such as challenges with a high-fat diet or caloric restriction, may be required to elicit such phenotypes. In summary, in addition to regulation of downstream metabolic events, the circadian clock itself remains in sync with the nutrient milieu because of the combinatorial effect of numerous metabolic/nutrient sensors. Thus, the circadian system participates in energy homeostasis through both entrainment to the environmental light cycle and by responding to the organismal feeding, sleep, and activity cycles.
REFERENCES
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Anea CB, Zhang M, Stepp DW, Simkins GB, Reed G, Fulton DJ, Rudic RD. Vascular disease in mice with a dysfunctional circadian clock. Circulation. 2009;119:1510–1517. doi: 10.1161/CIRCULATIONAHA.108.827477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian rhythms: Redox redux. Nature. 2011;469:476–478. doi: 10.1038/469476a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Turek FW. Sleepless in America: A pathway to obesity and the metabolic syndrome? Arch Intern Med. 2005;165:15–16. doi: 10.1001/archinte.165.1.15. [DOI] [PubMed] [Google Scholar]
- Bell GI, Polonsky KS. Diabetes mellitus and genetically programmed defects in b-cell function. Nature. 2001;414:788–791. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- Betz A, Chance B. Phase relationship of glycolytic intermediates in yeast cells with oscillatory metabolic control. Arch Biochem Biophys. 1965;109:585–594. doi: 10.1016/0003-9861(65)90404-2. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambras T, Vilaplana J, Diez-Noguera A. Effects of long-term restricted feeding on motor activity rhythm in the rat. Am J Physiol. 1993;265:R467–R473. doi: 10.1152/ajpregu.1993.265.2.R467. [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor a defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challet E, Pevet P, Vivien-Roels B, Malan A. Phase-advanced daily rhythms of melatonin, body temperature, and locomotor activity in food-restricted rats fed during daytime. J Biol Rhythms. 1997;12:65–79. doi: 10.1177/074873049701200108. [DOI] [PubMed] [Google Scholar]
- Challet E, Solberg LC, Turek FW. Entrainment in calorie-restricted mice: Conflicting zeitgebers and free-running conditions. Am J Physiol. 1998;274:R1751–R1761. doi: 10.1152/ajpregu.1998.274.6.R1751. [DOI] [PubMed] [Google Scholar]
- Challet E, Caldelas I, Graff C, Pevet P. Synchronization of the molecular clockwork by light- and food-related cues in mammals. Biol Chem. 2003;384:711–719. doi: 10.1515/BC.2003.079. [DOI] [PubMed] [Google Scholar]
- Chandra FA, Buzi G, Doyle JC. Glycolytic oscillations and limits on robust efficiency. Science. 2011;333:187–192. doi: 10.1126/science.1200705. [DOI] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005:310–1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Coleman GJ. Persistent meal-associated rhythms in SCN-lesioned rats. Physiol Behav. 1986;36:105–113. doi: 10.1016/0031-9384(86)90082-x. [DOI] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: The ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- Duffy PH, Feuers RJ, Hart RW. Effect of chronic caloric restriction on the circadian regulation of physiological and behavioral variables in old male B6C3F1 mice. Chronobiol Int. 1990;7:291–303. doi: 10.1080/07420529009064635. [DOI] [PubMed] [Google Scholar]
- Escobar C, Cailotto C, Angeles-Castellanos M, Delgado RS, Buijs RM. Peripheral oscillators: The driving force for food-anticipatory activity. Eur J Neurosci. 2009;30:1665–1675. doi: 10.1111/j.1460-9568.2009.06972.x. [DOI] [PubMed] [Google Scholar]
- Feillet CA, Ripperger JA, Magnone MC, Dulloo A, Albrecht U, Challet E. Lack of food anticipation in Per2 mutant mice. Curr Biol. 2006;16:2016–2022. doi: 10.1016/j.cub.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Fonken LK, Workman JL, Walton JC, Weil ZM, Morris JS, Haim A, Nelson RJ. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci. 2010;107:18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O, Chapnik N, Miskin R. Long-lived aMUPA transgenic mice exhibit pronounced circadian rhythms. Am J Physiol Endocrinol Metab. 2006;291:E1017–E1024. doi: 10.1152/ajpendo.00140.2006. [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato H, Tsai AL, Willie JT, Kisanuki Y, Williams SC, Sakurai T, Yanagisawa M. Enhanced orexin receptor-2 signaling prevents diet-induced obesity and improves leptin sensitivity. Cell Metab. 2009;9:64–76. doi: 10.1016/j.cmet.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Chance B. Oscillations of glycolytic intermediates in yeast cells. Biochem Biophys Res Commun. 1964;16:174–181. doi: 10.1016/0006-291x(64)90357-2. [DOI] [PubMed] [Google Scholar]
- Gooley JJ, Schomer A, Saper CB. The dorsomedial hypothalamic nucleus is critical for the expression of food-entrainable circadian rhythms. Nat Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARg. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumz ML, Stow LR, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Weaver DR, Wingo CS. The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice. J Clin Invest. 2009;119:2423–2434. doi: 10.1172/JCI36908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Herzog ED. Clock genes, oscillators, and cellular networks in the suprachiasmatic nuclei. J Biol Rhythms. 2004;19:400–413. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S. SIRT1 and caloric restriction: An insight into possible trade-offs between robustness and frailty. Curr Opin Clin Nutr Metab Care. 2009;12:350–356. doi: 10.1097/MCO.0b013e32832c932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JB, Omori T, Guan Z, Vgontzas AN, Bixler EO, Fang J. Sleep is increased in mice with obesity induced by high-fat food. Physiol Behav. 2006;87:255–262. doi: 10.1016/j.physbeh.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Jenkins NA, Copeland NG. Recombinant inbred mouse strains: Models for disease study. Trends Biotechnol. 1992;10:120–126. doi: 10.1016/0167-7799(92)90193-y. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A, Scheer FA, Perreau-Lenz S, La Fleur SE, Yi CX, Fliers E, Buijs RM. Circadian disruption and SCN control of energy metabolism. FEBS Lett. 2011;585:1412–1426. doi: 10.1016/j.febslet.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-g nuclear translocation. Proc Natl Acad Sci. 2011;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas PL, Brody S. Circadian rhythms in Neurospora crassa: Interactions between clock mutations. Genetics. 1985;109:49–66. doi: 10.1093/genetics/109.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry GJ, Simon MM, Webb IC, Mistlberger RE. Persistence of a behavioral food-anticipatory circadian rhythm following dorsomedial hypothalamic ablation in rats. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1527–R1534. doi: 10.1152/ajpregu.00874.2005. [DOI] [PubMed] [Google Scholar]
- Laposky AD, Shelton J, Bass J, Dugovic C, Perrino N, Turek FW. Altered sleep regulation in leptin deficient mice. Am J Physiol Regul Integr Comp Physiol. 2006;290:R894–R903. doi: 10.1152/ajpregu.00304.2005. [DOI] [PubMed] [Google Scholar]
- Laposky AD, Bass J, Kohsaka A, Turek FW. Sleep and circadian rhythms: Key components in the regulation of energy metabolism. FEBS Lett. 2008a;582:142–151. doi: 10.1016/j.febslet.2007.06.079. [DOI] [PubMed] [Google Scholar]
- Laposky AD, Bradley MA, Williams DL, Bass J, Turek FW. Sleep - wake regulation is altered in leptin-resistant (db/db) genetically obese and diabetic mice. Am J Physiol Regul Integr Comp Physiol. 2008b;295:R2059–R2066. doi: 10.1152/ajpregu.00026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, III, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant EG, Mistlberger RE. Anticipation and entrainment to feeding time in intact and SCN-ablated C57BL/6j mice. Brain Res. 1997;765:273–282. doi: 10.1016/s0006-8993(97)00571-4. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Graff C, Dardente H, Pevet P, Challet E. Feeding cues alter clock gene oscillations and photic responses in the suprachiasmatic nuclei of mice exposed to a light/dark cycle. J Neurosci. 2005;25:1514–1522. doi: 10.1523/JNEUROSCI.4397-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza J, Pevet P, Challet E. Circadian and photic regulation of clock and clock-controlled proteins in the suprachiasmatic nuclei of calorie-restricted mice. Eur J Neurosci. 2007;25:3691–3701. doi: 10.1111/j.1460-9568.2007.05626.x. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Pevet P, Challet E. High-fat feeding alters the clock synchronization to light. J Physiol. 2008;586:5901–5910. doi: 10.1113/jphysiol.2008.159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci. 2011;34:349–358. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nelson W, Scheving L, Halberg F. Circadian rhythms in mice fed a single daily meal at different stages of lighting regimen. J Nutr. 1975;105:171–184. doi: 10.1093/jn/105.2.171. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50:1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Nakamura W, Friday RC, Hatanaka F, Takumi T, Yamazaki S. Robust food anticipatory activity in BMAL1-deficient mice. PLoS One. 2009;4:e4860. doi: 10.1371/journal.pone.0004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Ptacek LJ, Jones CR, Fu YH. Novel insights from genetic and molecular characterization of the human clock. Cold Spring Harbor Symp Quant Biol. 2007;72:273–277. doi: 10.1101/sqb.2007.72.017. [DOI] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resuehr D, Olcese J. Caloric restriction and melatonin substitution: Effects on murine circadian parameters. Brain Res. 2005;1048:146–152. doi: 10.1016/j.brainres.2005.04.063. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, et al. Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- Saper CB. Staying awake for dinner: Hypothalamic integration of sleep, feeding, and circadian rhythms. Prog Brain Res. 2006;153:243–252. doi: 10.1016/S0079-6123(06)53014-6. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, Takahashi JS. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci. 2009;106:17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanath PR, Podrez EA, Chen J, Ma Y, Marchant K, Antoch M, Byzova TV. Deficiency in core circadian protein Bmal1 is associated with a prothrombotic and vascular phenotype. J Cell Physiol. 2011;226:132–140. doi: 10.1002/jcp.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–850. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- Spiegel K, Tasali E, Leproult R, Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol. 2009;5:253–261. doi: 10.1038/nrendo.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch KF, Weitz CJ. Daily rhythms of food-anticipatory behavioral activity do not require the known circadian clock. Proc Natl Acad Sci. 2009;106:6808–6813. doi: 10.1073/pnas.0902063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Strunk KE, Amann V, Threadgill DW. Phenotypic variation resulting from a deficiency of epidermal growth factor receptor in mice is caused by extensive genetic heterogeneity that can be genetically and molecularly partitioned. Genetics. 2004;167:1821–1832. doi: 10.1534/genetics.103.020495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard A, Lu XY. Rapid changes in night eating: Considering mechanisms. Eat Weight Disord. 2010;15:e2–e8. doi: 10.1007/BF03325274. [DOI] [PubMed] [Google Scholar]
- Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:e62. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 50-AMP-activated kinase with diabetes drug metformin induces casein kinase I1 (CKI1)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Honma S, Mitome M, Honma K. Independence of feeding-associated circadian rhythm from light conditions and meal intervals in SCN lesioned rats. Neurosci Lett. 1997;222:95–98. doi: 10.1016/s0304-3940(97)13353-5. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]