Abstract
IL-6 a multi-functional cytokine with important effects in both inflammation and haematopoiesis. SOCS3 is the primary inhibitor of IL-6 signalling, interacting with gp130, the common shared chain of the IL-6 family of cytokines, and JAK1, JAK2 and TYK2 to control both the duration of signalling and the biological response. Recent biochemical and structural studies have shown SOCS3 binds to only these three JAKs, all of which are associated with IL-6 signalling, and not JAK3. This specificity is determined by a three residue “GQM” motif in the kinase domain of JAK1, JAK2 and TYK2. SOCS3 binds to JAK and gp130 simultaneously, and inhibits JAK activity in an ATP-independent manner by partially occluding the kinase’s substrate binding groove with its kinase inhibitory region. We therefore propose a model in which each of gp130, JAK and SOCS3 are directly bound to the other two, allowing SOCS3 to inhibit IL6 signalling with high potency and specificity
Keywords: IL-6, Janus Kinases, SOCS, cytokine signalling, JAK/STAT
Introduction: IL-6 Signalling
IL-6 is a pleiotropic cytokine that exerts both inflammatory and anti-inflammatory effects depending upon its cellular context and is an important differentiation factor during haematopoiesis (reviewed in [1]). IL-6 belongs to a family of cytokines that also include IL-11, IL-27, LIF, OSM, CT-1 and CNTF. These cytokines are structurally similar[2] and signal via association with cell-surface trans-membrane receptors that each consist of a dimer (or higher-order oligomer) of the common shared chain, gp130 and a cytokine-specific alpha chain[3, 4].
In classical IL-6 signalling, IL-6 first associates with its specific receptor alpha chain, IL-6Rα, and this dimer then associates with gp130 to form a hexameric signalling competent complex with 2:2:2 stoichiometry[5, 6]. Whilst gp130 is expressed on the surface of most cell-types, IL-6Rα expression is more restricted. However, many cells which do not express IL-6Rα still respond to IL-6 by virtue of circulating soluble IL-6Rα (sIL-6Rα). This is termed trans-signalling and is often associated with the pro-inflammatory effects of IL-6[7, 8]
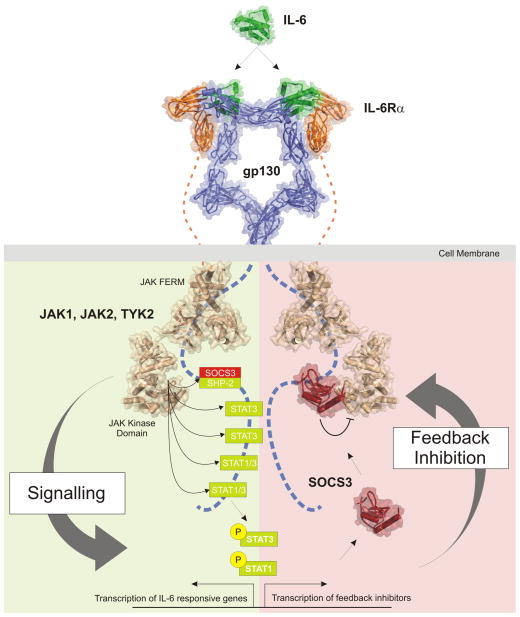
In both classical and trans-signalling, once the gp130 dimer is occupied there is an autoactivation of associated JAKs (Janus Kinases) which are found in an inactive state prior to cytokine exposure [9]. Based on similarity to receptor tyrosine kinase (RTK) signalling (for example insulin signalling) [10], activation is thought to occur by auto-phosphorylation in trans. In more detail, according to this model one JAK molecule bound to one cytokine receptor chain is phosphorylated by the JAK molecule bound to the other receptor chain (and vice-versa) within the receptor homo- or hetero-dimer. Activation involves phosphorylation of specific tyrosine(s) within the activation loop of the kinase [9] which causes the activation loop to translocate out of the active site in order to allow ATP and substrate to bind [11]. JAK1, JAK2 and TYK2 have all been found associated with gp130[12] in certain contexts however genetic deletion of these kinases has implicated JAK1 as the most important member of the family for gp130 induced signalling[13]. Upon activation, JAKs then phosphorylate five specific tyrosines on the cytoplasmic domain of gp130. Four of these phosphotyrosines are recruitment sites for STAT1 and/or STAT3 (Signal Transducer and Activator of Transcription-1 and −3) which are then activated by phosphorylation, again through the kinase activity of JAK1, JAK2 or TYK2[14]. STAT1 and STAT3 are latent transcription factors and once activated, they translocate into the nucleus and induce the transcription of appropriate IL-6-responsive genes. Thus STATs are the primary drivers of the biological response (See Figure 1, left). However, in addition to the JAK/STAT cascade, the MAP kinase and PI3 kinase pathways are also activated. This is via the fifth tyrosine, Y759, which, once phosphorylated, is a docking site for SHP2. SHP2 is activated by phosphorylation after binding and this leads to stimulation of both the MAPK/ERK and PI3 kinase pathways[15].
Figure 1. IL-6 signalling and its inhibition.
Schematic diagram illustrating activation (left) of the JAK/STAT signalling cascade in response to IL-6 and the termination of signalling (right) catalysed by SOCS3. IL-6 signals via a cell-surface receptor that consists of a 2:2 gp130(blue):IL-6Rα (orange) tetramer. Interaction between the cytokine and its receptor induces the autoactivation (in trans) of Janus Kinases (JAKs,: JAK1, JAK2, TYK2; shown in beige) bound to the cytoplasmic domain of gp130. Activated JAK then phosphorylates five tyrosines within gp130cyt. Four of these phosphotyrosines recruit STAT3 or STAT1/STAT3 which are then themselves phosphorylated, and thereby activated, by JAK, translocate to the nucleus and begin inducing the transcription of IL-6-responsive genes. STATs also upregulate the transcription of SOCS3 (red) which binds to the fifth phosphotyrosine in gp130cyt (pY759) and shuts down the JAK/STAT signalling cascade by binding to JAK and directly inhibiting its catalytic activity, forming a negative feedback loop. This phosphotyrosine also recruits SHP-2, which leads to activation of the MAPK/ERK and PI3K pathways (not shown here) and therefore SOCS3, which competes for this site, is also capable of inhibiting those signalling cascades. Signalling and inhibition is symmetric with respect to both gp130 chains and is shown here divided into left and right for ease of illustration. The structures shown are those solved and/or modelled for components of the signalling cascade, Note that the pseudokinase and SH2-like domains of JAK are omitted for clarity in this figure.
In addition to driving the biological response, activated STAT3 also induces expression of SOCS3 (Suppressor of Cytokine Signalling-3). SOCS3 in turn terminates the JAK/STAT signalling cascade, forming a negative feedback loop that allows the cell to return to its basal (unstimulated) state (Figure 1, right). This action of SOCS3 appears to be the primary mechanism by which IL-6 signalling is regulated within the organism. This review will focus on the mechanism by which SOCS3 inhibits IL-6 (and IL-6 family) signalling.
Discovery of the SOCS proteins
In 1997 the SOCS family of proteins were discovered concurrently by the groups of Hilton (Walter and Eliza Hall Institute, Australia), Yoshimura (Kurume University, Japan) and Kishimoto (Osaka University, Japan)[16–18]. Each group used a different approach. Hilton et al., used an expression cloning methodology to identify proteins capable of inhibiting the IL-6-induced differentiation of the mouse M1 myelomonocytic cell-line and discovered, and named, SOCS1 (Suppressor of Cytokine Signalling 1). Yoshimura’s group discovered the same entity via a yeast two-hybrid screen aimed at identifying proteins that bind to JAK and termed the protein JAB (JAK-binding protein). Finally, Kishimoto et al isolated a protein (SSI-1) on the basis of a short region of sequence similarity with STAT3. SSI-1 was found to be related to the SH2 domain-containing protein CIS (Cytokine inducible SH2 domain containing protein) and identical in sequence to SOCS1/JAB identified by the other two groups. Collectively, these three manuscripts described the major attributes of the SOCS1 protein: (A) That its expression is induced by a variety of cytokines; (B) it then inhibits the signalling cascade initiated by those same cytokines, forming a negative feedback loop; (C) it acts by binding to, and directly inhibiting, JAK with (D) the end result that STAT activation is curtailed.
At the same time as they discovered SOCS1, the group of Hilton et al., cloned two other proteins with similar domain architectures, termed SOCS2 and SOCS3. These three proteins, in addition to the already identified protein CIS[19–21], all contained an SH2 domain (responsible for binding phosphotyrosine residues) and a short, approximately 40 amino acid domain at their C-terminus that the authors termed the SOCS box. Subsequently, an extensive search of the genome databases discovered a further four proteins that shared this common domain structure (SH2 domain-SOCS box domain) and these were termed SOCS4-7[22].
The SOCS family
Evolutionarily, SOCS proteins are first seen in animals having bilateral symmetry[23]. Recent analyses suggest the existence of three SOCS proteins in these organisms: orthologues of CIS/SOCS1/SOCS2/SOCS3 as well as SOCS4/5 and SOCS6/7[23, 24]. Whilst certain species, most notably the fruit fly D. melanogaster have lost one or more of these three subgroups, they are all represented in vertebrates where they have expanded to form the eight family members seen in mammals. Of these three subgroups it is the CIS/SOCS1/SOCS2/SOCS3 class which are strictly associated with the control of cytokine signalling whereas the function of the SOCS4-7 homologues may also include regulation of RTK (Receptor Tyrosine Kinase) signalling such as that initiated by epidermal growth factor [25, 26], [27, 28].
The most notable feature of the SOCS family is their SOCS box domain[22]. The SOCS box is responsible, via an interaction with elonginsB/C and Cullin5, for promoting the ubiquitination of target proteins[29–31]. In general, these target proteins are cytokine receptors and thus SOCS proteins mostly function to control cytokine action by inducing the degradation of specific cytokine receptors[29, 31–34].
However the two most potent SOCS family members, SOCS1 and SOCS3 act primarily via a different mechanism[35, 36], distinct from that common to other SOCS proteins[26, 37–42]. They function by directly inhibiting the enzymatic activity of the JAKs, the initiators of the intracellular signalling cascade induced upon cytokine exposure and this mode-of-action is the major subject of this review.
SOCS3 is the primary regulator of IL-6 signalling
Despite SOCS1 being discovered on the basis of its ability to inhibit IL-6 action when overexpressed[17, 18], genetic deletion studies have surprisingly shown that SOCS1 plays little, if any, role in inhibiting IL-6 in vivo[43]. Rather it is SOCS3 that is the family member responsible for inhibiting IL-6 under physiological conditions[44–46]. This is a cautionary tale regarding the interpretation of the effects of individual SOCS proteins on various cytokines; whilst many SOCS proteins inhibit a number of different cytokines when artificially over-expressed, under normal conditions their activity is usually highly specific for only a few cytokines. This has been made clear by genetic deletion of SOCS1 and SOCS3 in mice which has highlighted their true role as regulators of signalling by interferon-α/γ36,40–43 and IL-6/G-CSF/Leptin/LIF[44, 46–50] respectively.
SOCS3 controls the duration of IL-6 signalling
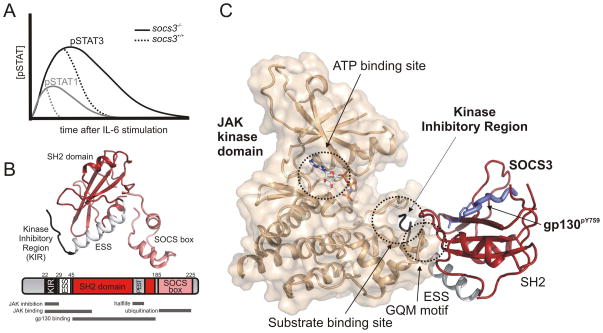
Genetic deletion of SOCS3 in mice is lethal due to placental insufficiency as a result of dysregulated signalling by LIF[48]. Therefore, confirmation of the important role that SOCS3 plays in regulating signalling by other IL-6 family members, including IL-6 itself, has been via conditional knockout of the Socs3 gene. The first such studies were knockouts of SOCS3 in hepatocytes[46] and macrophages[44] (using cre recombinase under control of the Albumin or LysM promoters respectively) and the use of Socs3−/− fetal liver cells to repopulate wild-type mice[51]. These experiments showed that the loss of SOCS3 had a profound effect on the duration of signalling induced by exposure of these cells to IL-6. For example, when wild type mice are injected with IL-6, activated (phosphorylated) STAT1 and STAT3 are detectable in liver cells from approximately 15 minutes after IL-6 exposure, but return to basal (undetectable) levels after approximately 30 minutes and 2 hours respectively[46] (Figure 2a). Loss of SOCS3 has no effect on the magnitude or time of initiation of JAK/STAT signalling after IL-6 exposure but led to a four- and two-fold increase in the persistence of activated STAT1 and STAT3 respectively. Whilst a 2–4 fold increase may seem like only a mild molecular defect it has drastic consequences for the animal. Mice lacking SOCS3 in their haematopoietic system (vavCre) develop a lethal inflammatory disease, largely due to dysregulated IL-6 signalling[45].
Figure 2. SOCS3 inhibits the duration of IL-6 signalling by direct inhibition of JAK1, JAK2 and TYK2 but not JAK3.
(A) Schematic diagram showing the effect of SOCS3 on STAT1 and STAT3 activation post IL-6 stimulation. Shown is a representation of the data from[46] (B). The structure of SOCS3 (PDB 4GL9) with an explanation of the major functional motifs shown as a schematic below. Note that the PEST motif is absent from the structure and that the SOCS box has been modelled based on the structure of the SOCS2 SOCS box (PDB 2C9W). (C) SOCS3 (red) binds the kinase domains of JAK1, JAK2 and TYK2 and inhibits its catalytic activity by blocking the substrate binding site with its kinase inhibitory region (black). Note that SOCS3 remains bound to gp130 (blue) whilst in complex with JAK (beige) and that ATP binding is unaffected.
SOCS3 shapes the cells response to IL-6
As well as controlling the duration of IL-6 signalling, SOCS3 also helps shape the cell’s response to IL-6. For example, the transcriptional output of Socs3−/− macrophages stimulated with IL-6 differs not just quantitatively but also qualitatively from that of wild-type cells. In particular, loss of SOCS3 leads to an IL-6-induced transcriptional response that in part resembles that for interferon-γ with a number of interferon-inducible genes being switched on by IL-6 in these cells[51]. Likewise, IL-6 stimulation of Socs3−/− haematopoietic progenitor cells skewed differentiation toward the macrophage lineage rather than neutrophil lineage seen with wild-type cells, again indicating that it shapes the response to IL-6 rather than simply inhibiting it[52]. One explanation for this phenomenon is that, in addition to inducing the phosphorylation of STAT3, IL-6 also induces low level STAT1 activation. In the presence of SOCS3, this activation of STAT1 is even more effectively curtailed than is the activation of STAT3[46, 53] thus preventing induction of a STAT1 (interferon-γ-like) transcriptional response. In the absence of SOCS3 therefore, STAT1 is “on” for long enough to induce the transcription of interferon-inducible genes leading to a qualitatively different cellular response.
SOCS3 interacts with gp130, the shared receptor for IL-6 family cytokines
STAT3 is activated by a number of different cytokines and is a powerful inducer of SOCS3 expression[54] . However, SOCS3 only feeds back to inhibit STAT3 that is activated in response to particular cytokines (for example IL-6) and not others (for example IL-10 or interferon-γ)[44, 51, 55]. The key to this specificity is that SOCS3 directly interacts with gp130, the co-receptor for IL-6 family cytokines[56–58]. This allows SOCS3 to specifically target the IL-6 signalling cascade and not those induced by other cytokines.
SOCS3 binds gp130 with high affinity [56, 57, 59, 60]. The interaction occurs via the SH2 domain of SOCS3 that binds to a motif on gp130 (centered upon pTyr759) only once it has been phosphorylated. As Tyr759 is only phosphorylated by JAK after IL-6 stimulation, SOCS3 cannot bind to an unstimulated IL-6 receptor and this ensures that a cell can still respond to the first wave of IL-6 stimulation, even if SOCS3 is present in the cytoplasm (for example if a different cytokine has already induced its expression). The interaction between the SOCS3 SH2 domain and gp130 has been well described both biochemically[56, 57, 59] and structurally[61] (see Figure 2b) and there are a number of features of the interaction responsible for its high affinity that are worth noting.
SH2 domains bind phosphotyrosine residues in peptides and proteins[62–66] but usually only when embedded within a particular sequence motif. This specificity is generally achieved by an interaction with the so-called BG loop of the SH2 domain (sometimes termed the specificity-determining loop) and amino-acids located 2–4 residues downstream (i.e. C-terminal) of the phosphotyrosine on the target molecule. For the vast majority of SH2 domains therefore, specificity and affinity for a target sequence are encoded only by the pTyr residue and the amino-acids immediately downstream of it. However, SOCS3 is unusual in that it also has a requirement for specific residues upstream of the phosphotyrosine on its target molecules[56]. In particular, SOCS3 contacts a hydrophobic residue at the pY-2 position on gp130, a valine. The interaction with this valine adds 10-fold to the affinity with which SOCS3 targets gp130[56] (KD=100nM) compared to a typical SH2-target interaction which is usually of micromolar affinity[66]. The SH2 domain of SHP-2 is similar in this regard and SHP-2 is known to bind a number of SOCS3 targets[56, 57, 67]. In addition to the pY-2 interaction, the BG loop of SOCS3 makes extensive contacts with the Val-Val-His sequence in the pY +3 to +5 region of gp130[61] which also contributes to the high affinity. In addition to gp130, SOCS3 also interacts with the receptors for leptin[58], G-CSF[68] and potentially EPO[69] although all of these interactions are at least 10-fold weaker than that seen for gp130. These interactions reveal a minimal consensus motif of V/L-X-pY-X-X-V/L-V/L-X.
The other major feature of the SOCS3 SH2 domain is that it contains a large (35 amino acid) unstructured loop inserted immediately prior to the specificity determining BG loop[60]. This loop is a PEST motif (Pro, Glu, Ser, Thr rich motif), a motif first described by Rogers on the basis of their being found in a number of intra-cellular proteins with very short half-lives[70]. The PEST motif does not effect the structure of the SH2 domain, as a comparison between the NMR structure of wild-type SOCS3[71] and the crystal structure of a PEST-deleted construct[61] clearly shows. Neither does it interfere with the SH2 domain function as both wild-type and PEST-deleted SOCS3 bind a gp130 phosphopeptide with similar affinties[71]. Rather it reduces the stability of SOCS3 inside the cell and leads to its proteolytic degradation in a mostly non-proteasome dependent fashion when tested in 293T cells[71]. Several other SOCS proteins are predicted to contain a PEST motif although this has not yet been verified by half-life studies. The most obvious is CIS, which contains a predicted PEST motif in the same position within its SH2 domain as SOCS3[60]. SOCS3 is known to have an extremely short half-life in certain cell-types[72]. This is likely the result of several mechanisms, including ubiquitination of lysine6[72], the presence of the SOCS box[73, 74] and PEST motif itself. SOCS3 turnover appears necessary for the cell to be able to respond to subsequent rounds of cytokine stimulation and therefore its half-life is strictly regulated.
The interaction of SOCS3 with gp130 at pTyr759 is competitive with the binding of SHP2. It has been shown that SOCS3 deletion (but not just deletion of its SOCS box) leads to hyper-phosphorylation of SHP2, presumably via increased accessibility to its binding site on gp130, in response to LIF stimulation of embryonic stem (ES) cells32,[75]. Phosphorylation of SHP2 appears to be the major mechanism of activating the MAPK/ERK pathway in IL-6/LIF signalling[76]. Consequently, Socs3−/− ES cells display extended activation of pERK1/2 in response to LIF signalling32,[75]. Socs3−/− ES cells, unlike wild-type ES cells, display reduced self-renewal and spontaneous differentiation into primitive endoderm in the presence of LIF and this differentiation could be prevented by the use of MAPK/ERK pathway inhibitors[75]. This indicates that this mode of inhibition by SOCS3 (attenuation of MAPK signalling) is independent of its E3 ligase activity and has an important role in the biological outcome of IL-6/LIF signalling.
The interaction of SOCS3 with gp130 is a key molecular determinant of its specificity and its ability to inhibit cytokine signalling. However, whilst the SH2 domain-gp130 interaction is sufficient to inhibit the MAPK/ERK pathway post IL-6 stimulation it is not sufficient to inhibit the JAK/STAT signalling cascade. Its ability to inhibit the JAK/STAT pathway relies upon an interaction with, and inhibition of, JAK whilst both entities are scaffolded on gp130. This is largely due to the “kinase inhibitory region” (KIR) of SOCS3 and this mechanism will now be discussed in detail.
The Kinase Inhibitory Region (KIR) of SOCS3 allows it to directly inhibit JAK’s catalytic activity
SOCS3 in mice and humans is a 225 amino acid protein that, like all SOCS proteins, contains an SH2 domain (residues 45–185) and a SOCS box domain (residues 186–225)[18, 22]. SOCS3 also contains a short N-terminal segment (residues 1–44), the most notable feature of which is the so-called Kinase Inhibitory Region (KIR)[77–80], an 8–12 amino acid sequence that allows it to directly inhibit JAK’s catalytic domain and is absolutely required for function (Figure 2b).
The existence of the KIR was first identified in both SOCS3 and SOCS1 in two seminal papers by Yoshimura’s group in 1999[78, 79]. These manuscripts defined the KIR as a 12 amino-acid sequence (residues 22–33), upstream of the SH2 domain. The KIR is only found in SOCS1 and SOCS3 and has been shown to be required for both interaction with, and inhibition of, JAK[78]. Although they are unstructured in the absence of JAK[60, 61, 71], the first eight residues of the KIR adopt an extended conformation that occupies the interface between the JAK kinase and the SOCS3 SH2 domain when the two proteins are in complex[81]. The four C-terminal residues of the KIR are also structured upon JAK binding, forming an extra, N-terminal, turn on the ESS (extended SH2 subdomain) helix. The ESS helix is a 14-residue alpha-helix immediately prior to the N-terminus of the SH2 domain and is a feature that is shared by all eight SOCS proteins. This helix is integral to the stability of the SOCS3 SH2 domain as, when deleted, the protein becomes unstable. The ESS covers a large hydrophobic surface on the under-side of the central β-sheet of the SH2 domain which gives it a very fixed geometry relative to the rest of the domain. This geometry may be important for positioning KIR in the case of SOCS1 and SOCS3. Now that the structure of SOCS3 in complex with JAK has been solved, we favour a redefinition of the KIR as consisting of residues 22–29 of SOCS3 and the ESS helix as residues 30–44 (Figure 2b).
The KIR inhibits JAK by partially blocking the substrate binding groove on the surface of the kinase (Figure 2c). This prevents substrates (for example STAT3) from accessing the active site. Tyrosine kinases catalyse the transfer of the terminal (γ) phosphate from ATP to a tyrosine hydroxyl moiety and are therefore two-substrate enzymes: ATP (the phosphate donor) and the tyrosine-containing protein/peptide (the phosphate acceptor). Whilst the tyrosine-containing protein/peptide is occluded from its binding site when SOCS3 is bound to JAK, ATP binding remains unperturbed (Figure 2c). This makes SOCS3 a non-competitive inhibitor (with regards to ATP) of JAK and may be an important aspect to its function as it does not need to compete with the high concentrations of ATP found in the cytoplasm[82].
SOCS3 inhibits JAK1, JAK2 and TYK2 but not JAK3
Whilst the KIR is required to inhibit JAK, it is not sufficient. There is no detectable inhibition of JAK using a SOCS3 KIR peptide[82]. The structure of SOCS3 bound to JAK2 shows that only approximately 20% of the buried surface area within the complex involves the KIR. The majority of the SOCS3:JAK affinity is derived from an interaction between the SH2 domain of SOCS3 and JAK. It is important to note that this does not involve the classical phosphotyrosine binding groove on the SH2 domain, which remains accessible for binding to gp130. Rather it is on the opposing face of the domain and also involves residues on the ESS. This surface wraps around helix αG of JAK, in particular a three residue “GQM” (Gly-Gln-Met) motif at its N-terminal end.
Interestingly, this GQM motif is only found in JAK1, JAK2 and TYK2 but not JAK3. In vitro inhibition assays have shown that SOCS3 can only directly inhibit JAK1, JAK2 and TYK2 and that the GQM motif is responsible for this specificity[82]. Thus all three JAKs found associated with gp130 are susceptible to SOCS3 inhibition whilst JAK3 is not. The GQM motif is conserved in JAK1, JAK2 and TYK2 throughout vertebrate evolution and is always absent in JAK3 suggesting an important distinction.
A model of SOCS3 inhibition of IL-6 signalling
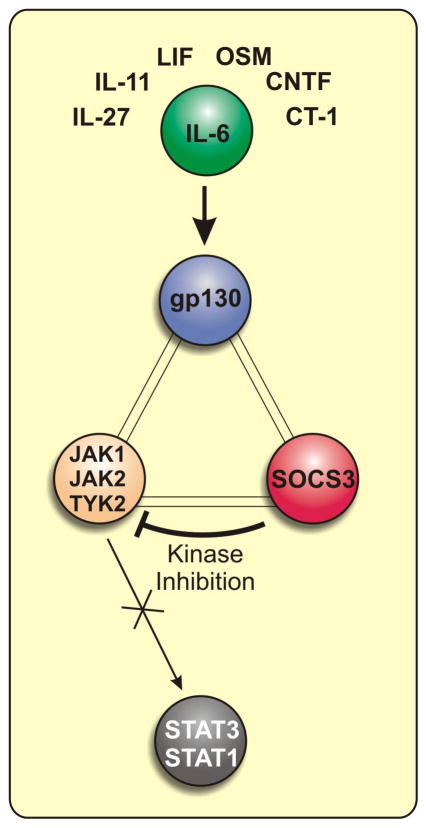
Knockout studies have shown that SOCS3 is a highly potent and specific inhibitor of IL-6 family cytokines, G-CSF and leptin, despite the fact that its expression is induced by a much larger number of cytokines. Any model of SOCS3 action must explain this specificity. Our model is centred upon the fact that SOCS3 binds JAK and the IL-6 receptor simultaneously via two opposing surfaces. Thus it is a particular JAK/Receptor complex that is the true target of SOCS3, rather than an individual JAK or receptor alone. This allows SOCS3 to inhibit IL-6 signalling (in addition to G-CSF and leptin) with both (A) high specificity and (B) high potency (affinity).
Specificity is derived from the fact that only particular JAK/receptor pairs are targeted, overcoming the redundancy caused by the fact that all cytokines signal through only four different JAK kinases and six different STAT transcription factors. SOCS3 can inhibit JAK1, JAK2 and TYK2 however it does so effectively only when they are already bound to a cytokine receptor that contains a SOCS3 binding site such as gp130, G-CSFR and lepR.
High affinity is derived from the formation of an unusual three-way complex (JAK/gp130/SOCS3) in which each member is directly bound to the other two (see figure 3). JAK binds gp130 through its FERM domain and SOCS3 through its kinase domain. gp130 binds JAK via its Box1 motif and SOCS3 via pY759. Finally, SOCS3 binds gp130 via its phosphotyrosine binding groove and JAK via the KIR and a surface on the opposing face of its SH2 domain. This creates a SOCS3:JAK/gp130 avidity that is higher than the mere sum of the individual SOCS:JAK and SOCS:gp130 affinities, reminiscent of certain antibody-antigen interactions. This avidity arises from SOCS3 containing two independent binding sites for the JAK/gp130 complex (see Figure 3).
Figure 3.
SOCS3 inhibits IL-6 family signalling by targeting a gp130:JAK dimer.
There are two predictions from this model of SOCS3 action that can be made: (1) it will inhibit a broad range of cytokines, in fact any cytokine that uses JAK1, JAK2 or TYK2, when overexpressed and (2) when present at physiological levels it will only inhibit of cytokines whose receptors contain SOCS3 binding sites. The former phenomenon has been well described elsewhere[83–85], whilst the latter phenomenon correlates well with the known specificity of SOCS3 for IL-6, LIF, G-CSF and leptin (all of which use receptors with SOCS3 binding sites) signalling. To date, the highest affinity SOCS3 binding site is found on gp130 (pY759). SOCS3 binds gp130 with a >10-fold higher affinity than it does G-CSFR and LepR. This explains why SOCS3 is such an effective inhibitor of all IL-6 family cytokines.
The mechanism of SOCS3 is reminiscent of the inhibition of insulin signalling by Grb14[86]. Like SOCS3, Grb14 inhibits insulin signalling by binding to a specific phosphotyrosine on the insulin receptor cytoplasmic domain and then inhibiting its associated kinase activity by blocking the kinase’s substrate binding site with a short inhibitory region. The difference is that rather than being bound to a kinase, the insulin receptor is the kinase and thus both interactions made by Grb14 (scaffolding via its SH2 domain and kinase inhibition by it kinase inhibitory sequence) are with the same molecular entity, rather than a dimeric kinase/receptor complex consisting of two separate chains. Given this conservation of mechanism between these major inhibitors of cytokine and RTK (receptor tyrosine kinase) signalling pathways it will be interesting to determine whether other kinase-based signalling pathways are similarly controlled.
SOCS3 is the primary feedback inhibitor of IL-6 family signalling
SOCS3 controls the duration of IL-6 signalling and shapes the cells response to it.
SOCS3 binds to gp130, the shared IL-6 family co-receptor
SOCS3 directly inhibits JAK1, JAK2 and TYK2 but not JAK3
SOCS3 targets gp130/JAK complexes
Acknowledgments
The original research described in this review was supported by the National Health and Medical Research Council of Australia (program grant nos. 461219 and 487922, 1011804), the U.S. National Institutes of Health (grant no. CA22556), the Victorian State Government Operational Infrastructure Support Grant, and the NHMRC Independent Research Institutes Infrastructure Support Scheme (361646). N.A.N. acknowledges fellowship support from the National Health and Medical Research Council, L.N.V. from the Leukaemia Foundation of Australia and the Australian Stem Cell Centre and J.J.B. from the Australian Research Council.
Abbreviations
- JAKs
Janus Kinases
- SOCS
Suppressor of Cytokine Signalling)
- STAT
Signal Transducers and Activators of Transcription
- IL-6
Interleukin-6
- IL-10
Interleukin-10
- IL-11
Interleukin-11
- IL-27
Interleukin-27
- gp130
glycoprotein 130
- OSM
Oncostatin M
- LIF
Leukemia Inhibitory Factor
- CNTF
cillary neurotrophic factor
- CT-1
Cardiotrophin 1
- IL-6Rα
Interleukin-6 Receptor alpha-chain
- GCSF
Granulocyte colony-stimulating factor
- KIR
kinase inhibitory region
- SH2
Src homology 2
- SHP2
SH2 domain containing phosphatase
- PI3K
Phosphoinositide 3-kinase
- MAPK
Mitogen-activated protein kinase
- ERK
extracellular-signal-regulated kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. 2011;1813:878–88. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 2.Nicola NA, editor. Guidebook to Cytokines and Their Receptors. Oxford University Press; 1994. [Google Scholar]
- 3.Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Kershaw NJ, Luo CS, Soo P, Pocock MJ, Czabotar PE, Hilton DJ, Nicola NA, Garrett TP, Zhang JG. Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J Biol Chem. 2010;285:21214–8. doi: 10.1074/jbc.C110.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–4. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 7.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J. 1994;300 ( Pt 2):281–90. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabe B, Chalaris A, May U, Waetzig GH, Seegert D, Williams AS, Jones SA, Rose-John S, Scheller J. Transgenic blockade of interleukin 6 transsignaling abrogates inflammation. Blood. 2008;111:1021–8. doi: 10.1182/blood-2007-07-102137. [DOI] [PubMed] [Google Scholar]
- 9.Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol. 1997;17:2497–501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–12. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 11.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. Embo J. 1997;16:5572–81. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, Quelle FW, Silvennoinen O, Barbieri G, Pellegrini S, et al. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science. 1994;263:92–5. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 13.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–83. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 14.Hemmann U, Gerhartz C, Heesel B, Sasse J, Kurapkat G, Grotzinger J, Wollmer A, Zhong Z, Darnell JE, Jr, Graeve L, Heinrich PC, Horn F. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. II. Src homology SH2 domains define the specificity of stat factor activation. J Biol Chem. 1996;271:12999–3007. doi: 10.1074/jbc.271.22.12999. [DOI] [PubMed] [Google Scholar]
- 15.Eulenfeld R, Dittrich A, Khouri C, Muller PJ, Mutze B, Wolf A, Schaper F. Interleukin-6 signalling: more than Jaks and STATs. Eur J Cell Biol. 2012;91:486–95. doi: 10.1016/j.ejcb.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–4. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 17.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–9. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 18.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–21. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins NA, Gilbert DJ, Copeland NG, Hara T, Miyajima A. A novel cytokine-inducible gene CIS encodes an SH2- containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. Embo J. 1995;14:2816–26. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, Yokouchi M, Ohtsubo M, Yoshimura A. Cloning and characterization of novel CIS family genes. Biochem Biophys Res Commun. 1997;239:439–46. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–54. [PubMed] [Google Scholar]
- 22.Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liongue C, O’Sullivan LA, Trengove MC, Ward AC. Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS One. 2012;7:e32777. doi: 10.1371/journal.pone.0032777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liongue C, Ward AC. Evolution of the JAK-STAT pathway. JAKSTAT. 2013;2:e22756. doi: 10.4161/jkst.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linossi EM, Chandrashekaran IR, Kolesnik TB, Murphy JM, Webb AI, Willson TA, Kedzierski L, Bullock AN, Babon JJ, Norton RS, Nicola NA, Nicholson SE. Suppressor of Cytokine Signaling (SOCS) 5 Utilises Distinct Domains for Regulation of JAK1 and Interaction with the Adaptor Protein Shc-1. PLoS One. 2013;8:e70536. doi: 10.1371/journal.pone.0070536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholson SE, Metcalf D, Sprigg NS, Columbus R, Walker F, Silva A, Cary D, Willson TA, Zhang JG, Hilton DJ, Alexander WS, Nicola NA. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc Natl Acad Sci U S A. 2005;102:2328–33. doi: 10.1073/pnas.0409675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stec WJ, Zeidler MP. Drosophila SOCS Proteins. J Signal Transduct. 2011;2011:894510. doi: 10.1155/2011/894510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–21. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- 29.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 30.Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–13. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- 31.Muniz JR, Guo K, Kershaw NJ, Ayinampudi V, von Delft F, Babon JJ, Bullock AN. Molecular Architecture of the Ankyrin SOCS Box Family of Cul5-Dependent E3 Ubiquitin Ligases. J Mol Biol. 2013;425:3166–77. doi: 10.1016/j.jmb.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci U S A. 2006;103:7637–42. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C Complex Reveals a Distinct SOCS Box Interface and the Molecular Basis for SOCS-Dependent EGFR Degradation. Structure. 2007;15:1493–504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babon JJ, Sabo JK, Soetopo A, Yao SG, Bailey MF, Zhang JG, Nicola NA, Norton RS. The SOCS box domain of SOCS3: Structure and interaction with the elonginBC-cullin5 ubiquitin ligase. Journal of Molecular Biology. 2008;381:928–940. doi: 10.1016/j.jmb.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle K, Egan P, Rakar S, Willson TA, Wicks IP, Metcalf D, Hilton DJ, Nicola NA, Alexander WS, Roberts AW, Robb L. The SOCS box of suppressor of cytokine signaling-3 contributes to the control of G-CSF responsiveness in vivo. Blood. 2007;110:1466–1474. doi: 10.1182/blood-2007-03-079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle K, Zhang JG, Nicholson SE, Trounson E, Babon JJ, McManus EJ, Nicola NA, Robb L. Deletion of the SOCS box of suppressor of cytokine signaling 3 (SOCS3) in embryonic stem cells reveals SOCS box-dependent regulation of JAK but not STAT phosphorylation. Cellular Signalling. 2009;21:394–404. doi: 10.1016/j.cellsig.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenhalgh CJ, Metcalf D, Thaus AL, Corbin JE, Uren R, Morgan PO, Fabri LJ, Zhang JG, Martin HM, Willson TA, Billestrup N, Nicola NA, Baca M, Alexander WS, Hilton DJ. Biological evidence that SOCS-2 can act either as an enhancer or suppressor of growth hormone signaling. J Biol Chem. 2002;277:40181–4. doi: 10.1074/jbc.C200450200. [DOI] [PubMed] [Google Scholar]
- 38.Banks AS, Li J, McKeag L, Hribal ML, Kashiwada M, Accili D, Rothman PB. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J Clin Invest. 2005;115:2462–71. doi: 10.1172/JCI23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pazienza V, Clement S, Pugnale P, Conzelman S, Foti M, Mangia A, Negro F. The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology. 2007;45:1164–71. doi: 10.1002/hep.21634. [DOI] [PubMed] [Google Scholar]
- 40.Kario E, Marmor MD, Adamsky K, Citri A, Amit I, Amariglio N, Rechavi G, Yarden Y. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J Biol Chem. 2005;280:7038–48. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- 41.Verdier F, Chretien S, Muller O, Varlet P, Yoshimura A, Gisselbrecht S, Lacombe C, Mayeux P. Proteasomes regulate erythropoietin receptor and signal transducer and activator of transcription 5 (STAT5) activation. Possible involvement of the ubiquitinated Cis protein. J Biol Chem. 1998;273:28185–90. doi: 10.1074/jbc.273.43.28185. [DOI] [PubMed] [Google Scholar]
- 42.Ram PA, Waxman DJ. Role of the cytokine-inducible SH2 protein CIS in desensitization of STAT5b signaling by continuous growth hormone. J Biol Chem. 2000;275:39487–96. doi: 10.1074/jbc.M004755200. [DOI] [PubMed] [Google Scholar]
- 43.Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, Kay TW, Nicola NA, Hertzog PJ, Metcalf D, Hilton DJ. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 44.Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4:551–6. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 45.Croker BA, Kiu H, Pellegrini M, Toe J, Preston S, Metcalf D, O’Donnell JA, Cengia LH, McArthur K, Nicola NA, Alexander WS, Roberts AW. IL-6 promotes acute and chronic inflammatory disease in the absence of SOCS3. Immunol Cell Biol. 2012;90:124–9. doi: 10.1038/icb.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS. SOCS3 negatively regulates IL-6 signaling in vivo. Nature Immunology. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 47.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 48.Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, Metcalf D, Hilton DJ, Alexander WS. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nature Medicine. 2004;10:739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 50.Kievit P, Howard JK, Badman MK, Balthasar N, Coppari R, Mori H, Lee CE, Elmquist JK, Yoshimura A, Flier JS. Enhanced leptin sensitivity and improved glucose homeostasis in mice lacking suppressor of cytokine signaling-3 in POMC-expressing cells. Cell Metabolism. 2006;4:123–132. doi: 10.1016/j.cmet.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Lang R, Pauleau AL, Parganas E, Takahashi Y, Mages J, Ihle JN, Rutschman R, Murray PJ. SOCS3 regulates the plasticity of gp130 signaling. Nature Immunology. 2003;4:546–550. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- 52.Croker BA, Mielke LA, Wormald S, Metcalf D, Kiu H, Alexander WS, Hilton DJ, Roberts AW. Socs3 maintains the specificity of biological responses to cytokine signals during granulocyte and macrophage differentiation. Exp Hematol. 2008;36:786–98. doi: 10.1016/j.exphem.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wormald S, Zhang JG, Krebs DL, Mielke LA, Silver J, Alexander WS, Speed TP, Nicola NA, Hilton DJ. The comparative roles of suppressor of cytokine signaling-1 and −3 in the inhibition and desensitization of cytokine signaling. J Biol Chem. 2006;281:11135–43. doi: 10.1074/jbc.M509595200. [DOI] [PubMed] [Google Scholar]
- 54.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc Natl Acad Sci U S A. 1999;96:6964–9. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, Heinrich PC, Muller-Newen G. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially cytokine signaling 3. Journal of Immunology. 2003;170:3263–3272. doi: 10.4049/jimmunol.170.6.3263. [DOI] [PubMed] [Google Scholar]
- 56.Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, Metcalf D, Hilton DJ, Nicola NA, Baca M. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6493–6498. doi: 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmitz J, Weissenbach M, Haan S, Heinrich PC, Schaper F. SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. Journal of Biological Chemistry. 2000;275:12848–12856. doi: 10.1074/jbc.275.17.12848. [DOI] [PubMed] [Google Scholar]
- 58.De Souza D, Fabri LJ, Nash A, Hilton DJ, Nicola NA, Baca M. SH2 domains from suppressor of cytokine signaling-3 and protein tyrosine phosphatase SHP-2 have similar binding specificities. Biochemistry. 2002;41:9229–9236. doi: 10.1021/bi0259507. [DOI] [PubMed] [Google Scholar]
- 59.Lehmann U, Schmitz J, Weissenbach M, Sobota RM, Hortner M, Friederichs K, Behrmann I, Tsiaris W, Sasaki A, Schneider-Mergener J, Yoshimura A, Neel BG, Heinrich PC, Schaper F. SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130. Journal of Biological Chemistry. 2003;278:661–671. doi: 10.1074/jbc.M210552200. [DOI] [PubMed] [Google Scholar]
- 60.Babon JJ, Yao S, DeSouza DP, Harrison CF, Fabri LJ, Liepinsh E, Scrofani SD, Baca M, Norton RS. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. Febs J. 2005;272:6120–30. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 61.Bergamin E, Wu J, Hubbard SR. Structural basis for phosphotyrosine recognition by suppressor of cytokine signaling-3. Structure. 2006;14:1285–92. doi: 10.1016/j.str.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 62.Pawson T, Gish GD. SH2 and SH3 domains: from structure to function. Cell. 1992;71:359–62. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- 63.Koch CA, Moran M, Sadowski I, Pawson T. The common src homology region 2 domain of cytoplasmic signaling proteins is a positive effector of v-fps tyrosine kinase function. Mol Cell Biol. 1989;9:4131–40. doi: 10.1128/mcb.9.10.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeClue JE, Sadowski I, Martin GS, Pawson T. A conserved domain regulates interactions of the v-fps protein-tyrosine kinase with the host cell. Proc Natl Acad Sci U S A. 1987;84:9064–8. doi: 10.1073/pnas.84.24.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grucza RA, Bradshaw JM, Futterer K, Waksman G. SH2 domains: from structure to energetics, a dual approach to the study of structure-function relationships. Med Res Rev. 1999;19:273–93. doi: 10.1002/(sici)1098-1128(199907)19:4<273::aid-med2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 66.Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC, Schlessinger J. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc Natl Acad Sci U S A. 1995;92:3199–203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fairlie WD, De Souza D, Nicola NA, Baca M. Negative regulation of gp130 signalling mediated through tyrosine-757 is not dependent on the recruitment of SHP2. Biochemical Journal. 2003;372:495–502. doi: 10.1042/BJ20030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hortner M, Nielsch U, Mayr LM, Johnston JA, Heinrich PC, Haan S. Suppressor of cytokine signaling-3 is recruited to the activated granulocyte-colony stimulating factor receptor and modulates its signal transduction. Journal of Immunology. 2002;169:1219–1227. doi: 10.4049/jimmunol.169.3.1219. [DOI] [PubMed] [Google Scholar]
- 69.Hortner M, Nielsch U, Mayr LM, Heinrich PC, Haan S. A new high affinity binding site for suppressor of cytokine signaling-3 on the erythropoietin receptor. European Journal of Biochemistry. 2002;269:2516–2526. doi: 10.1046/j.1432-1033.2002.02916.x. [DOI] [PubMed] [Google Scholar]
- 70.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–8. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 71.Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, Nicholson SE, Norton RS. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell. 2006;22:205–16. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 72.Sasaki A, Inagaki-Ohara K, Yoshida T, Yamanaka A, Sasaki M, Yasukawa H, Koromilas AE, Yoshimura A. The N-terminal truncated isoform of SOCS3 translated from an alternative initiation AUG codon under stress conditions is stable due to the lack of a major ubiquitination site, Lys-6. Journal of Biological Chemistry. 2003;278:2432–2436. doi: 10.1074/jbc.C200608200. [DOI] [PubMed] [Google Scholar]
- 73.Haan S, Ferguson P, Sommer U, Hiremath M, McVicar DW, Heinrich PC, Johnston JA, Cacalano NA. Tyrosine phosphorylation disrupts elongin interaction and accelerates SOCS3 degradation. Journal of Biological Chemistry. 2003;278:31972–31979. doi: 10.1074/jbc.M303170200. [DOI] [PubMed] [Google Scholar]
- 74.Chen XP, Losman JA, Cowan S, Donahue E, Fay S, Vuong BQ, Nawijn MC, Capece D, Cohan VL, Rothman P. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc Natl Acad Sci U S A. 2002;99:2175–80. doi: 10.1073/pnas.042035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forrai A, Boyle K, Hart AH, Hartley L, Rakar S, Willson TA, Simpson KM, Roberts AW, Alexander WS, Voss AK, Robb L. Absence of suppressor of cytokine signalling 3 reduces self-renewal and promotes differentiation in murine embryonic stem cells. Stem Cells. 2006;24:604–614. doi: 10.1634/stemcells.2005-0323. [DOI] [PubMed] [Google Scholar]
- 76.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–60. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 77.Narazaki M, Fujimoto M, Matsumoto T, Morita Y, Saito H, Kajita T, Yoshizaki K, Naka T, Kishimoto T. Three distinct domains of SSI-1/SOCS-1/JAB protein are required for its suppression of interleukin 6 signaling. Proc Natl Acad Sci U S A. 1998;95:13130–4. doi: 10.1073/pnas.95.22.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes to Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 79.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. Embo J. 1999;18:1309–20. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sasaki A, Yasukawa H, Shouda T, Kitamura T, Dikic I, Yoshimura A. CIS3/SOCS-3 suppresses erythropoietin (EPO) signaling by binding the EPO receptor and JAK2. Journal of Biological Chemistry. 2000;275:29338–29347. doi: 10.1074/jbc.M003456200. [DOI] [PubMed] [Google Scholar]
- 81.Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA. Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–50. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. Journal of Biological Chemistry. 1998;273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 84.Ram PA, Waxman DJ. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. Journal of Biological Chemistry. 1999;274:35553–35561. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 85.Cohney SJ, Sanden D, Cacalano NA, Yoshimura A, Mui A, Migone TS, Johnston JA. SOCS-3 is tyrosine phosphorylated in response to interleukin-2 and suppresses STAT5 phosphorylation and lymphocyte proliferation. Mol Cell Biol. 1999;19:4980–8. doi: 10.1128/mcb.19.7.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Depetris RS, Hu J, Gimpelevich I, Holt LJ, Daly RJ, Hubbard SR. Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell. 2005;20:325–33. doi: 10.1016/j.molcel.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]