Abstract
Almiramide C is a marine natural product with low micromolar activity against Leishmania donovani, the causative agent of leishmaniasis. We have now shown that almiramide C is also active against the related parasite Trypanosoma brucei, the causative agent of human African trypanosomiasis. A series of activity-based probes have been synthesized to explore both the molecular target of this compound series in T. brucei lysates, and site localization through epifluorescence microscopy. These target identification studies indicate that the almiramides likely disrupt glycosomal function through disruption of membrane assembly machinery. Glycosomes, which are organelles specific to kinetoplastid parasites, house the first seven steps of glycolysis and have been shown to be essential for parasite survival in the bloodstream stage. There are currently no reported small molecule disruptors of glycosome function, making the almiramides unique molecular probes for this understudied parasite-specific organelle. Additionally, examination of toxicity in an in vivo zebrafish model has shown that these compounds have little effect on organism development even at high concentrations, and has uncovered a potential side effect through localization of fluorescent derivatives to zebrafish neuromast cells. Combined, these results further our understanding of the potential value of this lead series as development candidates against T. brucei.
Human African trypanosomiasis (HAT) is a neglected tropical disease (NTD) endemic to sub-Saharan Africa caused by the parasite Trypanosoma brucei. There are an estimated 60 million people at risk, and 45,000 deaths a year from this infectious parasite.1 HAT is transmitted when an infected tsetse fly takes a blood meal from a host, transmitting metacyclic trypomastigotes from the salivary gland of the tsetse fly to the host bloodstream. These injected metacyclic trypomastigotes then transform into bloodstream trypomastigotes and are dispersed throughout the host circulatory system, where they go on to cause persistent systemic infection.
The disease manifests itself in two stages; stage one is typically characterized by nonspecific symptoms that occur as a result of the parasites multiplying by binary fission throughout the haemolymphatic system. During this transformation, the host can experience a variety of symptoms which can include skin lesions, cardiac, endocrine, or gastrointestinal problems causing HAT to be misdiagnosed or, more frequently, undiagnosed. In stage two, trypomastigotes cross into the central nervous system (CNS) and cause a wide range of neurological dysfunctions; the disease gets its name (African sleeping sickness) due to the fact that patients often experience disturbances in the sleep-wake cycle in the late stage of the disease.
Currently there are only four licensed drugs to treat the two stages of HAT (Figure 1), most of which have been in use for more than 50 years.2 Stage one infections are treated with the polysulphonated napthyl urea, suramin (1) and the diamidine, pentamidine (2). Suramin was developed in 1921 and is strongly negatively charged at physiological pH, precluding transfer across the blood brain barrier (BBB) into the CNS, and limiting its use to stage one infections. Treatments for stage two infections include eflornithine (3) and melarsolprol (4). Eflornithine, the only new option for therapy, was developed in the 1980s and has been used as combination therapy with nifurtimox (used to treat Chagas disease) to decrease both daily dose and treatment duration in the hope of curbing the development of resistance. Unfortunately, 3 is expensive to produce and is only effective against T.b. gambiense. The most commonly administered drug for stage two infections is melarsoprol, which is an arsenic-based drug, which suffers from issues of both toxicity and resistance. It is estimated that between three and ten percent of patients suffer drug-related mortality due to the effects of melarsoprol.3 While the molecular targets of these therapeutics remain elusive, our understanding of the mechanisms of antitrypanosomal drug action has recently been advanced using a genome-scale RNA interference target sequencing (RIT-seq) screen.4 This screen has yielded a broad picture of the metabolic pathways and proteins affected by the therapeutics, and offers insight into the putative mechanisms of action for several of these compound classes.
Figure 1.
Current therapeutics for HAT.
Although a number of new treatments are currently under development, including fexinidazole (5)5,6 and SCYX-7158,3 the drug pipeline for HAT remains comparatively weak. In order to overcome this issue there has been a focused effort to identify trypanosome-specific enzymes or pathways that could serve as targets for screening campaigns. In particular, folate metabolism, glycolysis, and protein N-myristoylation have been identified as promising targets for T. brucei.7 Along these lines, glycolysis is recognized as a promising target because it is essential for survival in the bloodstream form of the parasite, and is the sole source of ATP production for the parasite once it has been transmitted to the human host. Typically, glycolysis is a cytosolic process in eukaryotes, but kinetoplastid parasites house this process in an unique organelle termed the glycosome.8 This unique compartmentalization has inspired several research programs to pursue targets in this pathway for potential drug leads.9 However, no compound from these studies has yet progressed to clinical trials.
We now report the discovery of the first small molecules that are candidates for disruption of glycosomal function in trypanosomes. Building upon previous work to develop the almiramide class of natural products as lead compounds against Leishmania donovani, we have shown that these compounds are also active against the related kinetoplastid parasite T. brucei, and have employed a derivatization strategy to generate a suite of chemical probes for target identification. Using these probes we have shown that the almiramides bind to two related glycosomal membrane proteins, GIM5A and PEX11, and have examined the site localization of fluorescent almiramide derivatives in fixed procyclic parasites. Finally, we have examined the vertebrate toxicity of these compounds in zebrafish using fluorescence site localization and have shown that these compounds have little effect on development. In addition, all embryos survived up to 168 µM, the determined maximum tolerated dose (MTD).
RESULTS AND DISCUSSION
Creation of Natural Product-Based Chemical Probes
Almiramides A – C are marine-derived N-methylated lipopentapeptide natural products, originally isolated from a Panamanian collection of Lyngbya majuscula in conjunction with the Panama International Cooperative Biodiversity Group (ICBG).10 Almiramides A – C (7–9) and 12 related synthetic analogues (10–21) have previously been shown to possess antiparasitic activities against L. donovani, the causative agent of leishmaniasis. The close structural relationship between L. donovani and other kinetoplastid parasites motivated us to extend the scope of this program to include other global health targets from this class.
In vitro screening against T. b. brucei identified several library members with activities that were comparable to values previously obtained for L. donovani (Table 1). Overall, variation in functional groups at the C- terminus of the amino acid portion had the largest effect on activity, with compounds 14–17, which contained carboxylic acid termini, being weakly active compared to the compounds containing either tertiary amides (10–13) or methyl esters (18–21). This result suggested that, while free acids were not well tolerated at this position, it may be possible to substitute the methyl group in either the amide or ester series for other alkyl side chains, and that this could be a viable handle for chemical probe derivatization.
Table 1.
Screening Results for Almiramides A – C (7 – 9) and Related Synthetic Compounds 10 – 23.
| Compound | R1 | R2 | R3 | R4 | IC50 Leishmania sp. (µM) |
IC50 T.b.brucei (µM) |
LD50 Vero Cells (µM) |
SI T.b. brucei |
|---|---|---|---|---|---|---|---|---|
| Almiramide A (7) | NH2 | H | CH3 |  |
>13.5 | nt | 113.1 | - |
| Almiramide B (8) | NH2 | H | CH3 | 2.4 | 6.0 | 52.3 | 8.7 | |
| Almiramide C (9) | NH2 | H | CH3 | 1.9 | 3.0 | 33.1 | 11 | |
| 10 | N(CH3)2 | CH3 | H | 3.1 | 1.3 | 155.7 | 120 | |
| 11 | N(CH3)2 | CH3 | H | 6.7 | 0.6 | 93.1 | 160 | |
| 12 | N(CH3)2 | CH3 | H | 5.9 | 0.6 | 159.4 | 270 | |
| 13 | N(CH3)2 | CH3 | H | 2.7 | 0.6 | 23.8 | 40 | |
| 14 | OH | CH3 | H | 5.6 | >13.7 | 281.1 | - | |
| 15 | OH | CH3 | H | >14.0 | 2.6 | >70.0 | >27 | |
| 16 | OH | CH3 | H | >14.0 | 10.3 | >68.9 | >6.7 | |
| 17 | OH | CH3 | H | 4.1 | >13.7 | 192.4 | - | |
| 18 | OCH3 | CH3 | H | >14.0 | 0.6 | 41.4 | 69 | |
| 19 | OCH3 | CH3 | H | >14.0 | 0.6 | 24.7 | 41 | |
| 20 | OCH3 | CH3 | H | >14.0 | 0.6 | 27.0 | 45 | |
| 21 | OCH3 | CH3 | H | >14.0 | 0.4 | 18.8 | 47 | |
| 22 |  |
CH3 | H | - | 0.7 | nt | - | |
| 23 |  |
CH3 | H | - | 3.5 | nt | - |

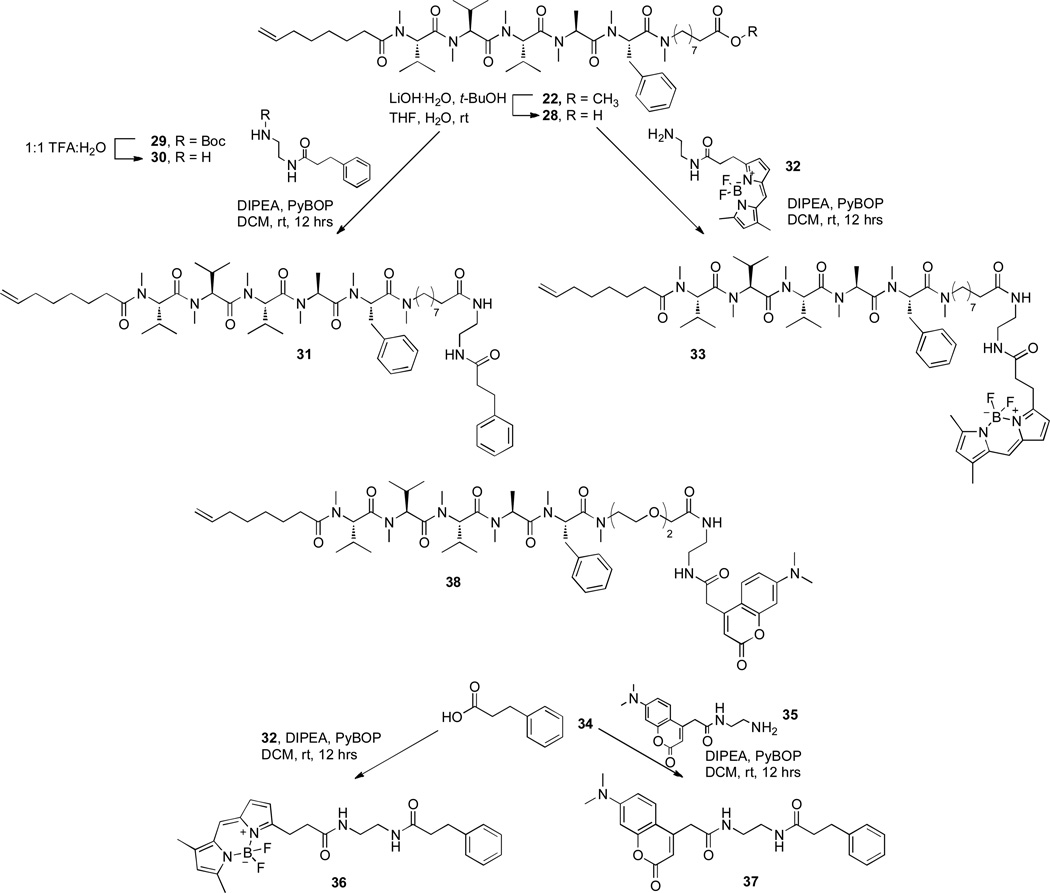
By contrast, variations in the alkyl chain length and terminal moieties at the N-terminus had little effect on the activities of compounds within each series (e.g. 10–13). While this result may appear to suggest this as a viable position for compound derivatization, previous work with L. donovani has shown that the unsaturated terminus of the alkyl side chain is important for activity, and that compounds with saturated alkyl chain termini are inactive against L. donovani. We therefore elected to retain this motif, and to develop the almiramide warhead through derivatization at the C-terminus. To this end, two almiramide – linker conjugates (22 and 23) were synthesized using standard solid phase synthesis methods, followed by solution phase permethylation. Both of these linker derivatives retained low micromolar activities against T. brucei (IC50 = 0.7 and 3.5 µM respectively) comparable to the parent molecule at 1.9 µM, and were therefore deemed suitable for subsequent derivatization as chemical probes.
The almiramide series possesses a chemical scaffold that is distinct from all current and developing therapeutics for HAT. The flexibility of permitted functional groups at the C-terminus, sub-micromolar biological activities, and facile synthetic access to the core scaffold indicated that this series was well suited to a target identification study against T. brucei. An affinity-capture study for target identification was therefore initiated against bloodstream form trypanosomes, using compound 21 as the core structure for affinity probe design and synthesis.
Target Identification from Whole Cell Lysates
A dual approach using either biotinylated- or affinity resin-derivatized almiramide C was pursued for affinity purification of candidate target proteins in T. brucei.11 Biotinylated probes have been successfully applied in the Activity Based Protein Profiling (ABPP) targeting active site reactivity,12 such as in the cases of the marine natural product didemnin B, recently shown to target the elongation factor 1-α (EF1-α),13 or the siderophore petrobactin found to interact with the siderophore-binding membrane-bound protein ferric petrobactin import A (FpiA).14 Our first approach was therefore to design biotinylated almiramide derivatives to explore the molecular targets of the almiramides in T. brucei.
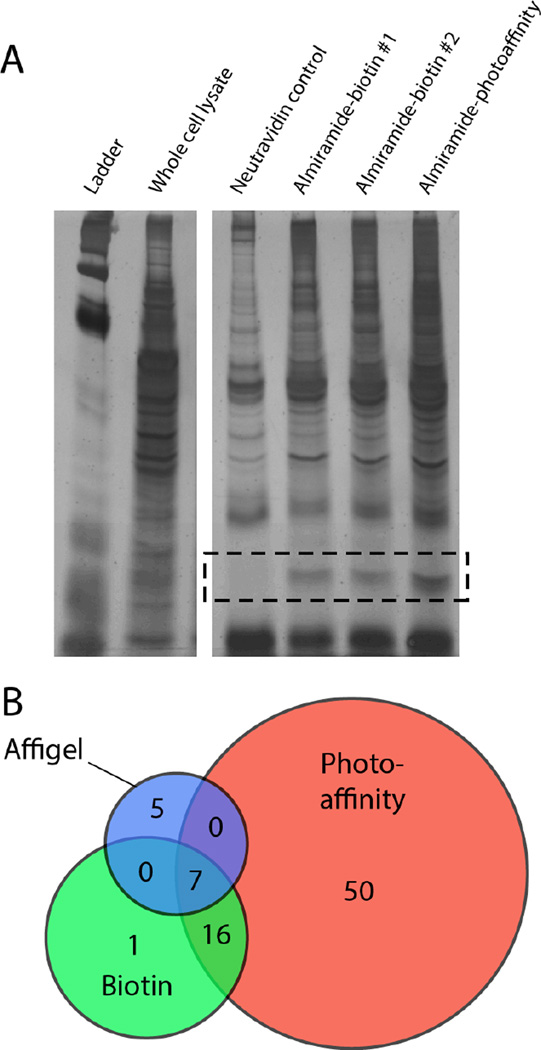
Synthesis of our biotinylated affinity capture probe began with a commercially available biotin reporter tag containing a primary amine terminus. This reporter was coupled to the almiramide moiety (17) using standard coupling reagents (Supporting Information) to generate biotinylated probe 24 (IC50 = 9.9 µM). This log unit drop in activity is common for biotinylated probes, due to the significant alterations in physicochemical properties imparted by the addition of biotin-linker motifs.15 Briefly, the workflow for protein affinity capture using biotinylated probe 24 was as follows (Figure 2); firstly, biotinylated probe 24 was incubated with the preclarified whole cell lysate of T. b. brucei at room temperature for two hours to allow the almiramide moiety to bind with target protein(s). Secondly, NeutrAvidin resin was added to capture the probe-protein complex, and the resin was washed three times with detergent-free buffer. Finally, SDS-page analysis was performed on the biotinylated-almiramide-protein-resin complex, followed by excision, tryptic digest and LCMS/MS analysis of bands of interest from the SDS-page gel to identify candidate target proteins. In order to reduce the abundance of non-specific, endogenously biotinylated proteins as well as high abundance background proteins that may have some resin affinity,16 whole cell lysates were preclarified by incubation with NeutrAvidin resin prior to biotin affinity capture. Affinity capture experiments with biotinylated almiramide 24 were repeated in four independent biological replicates, and all major and minor bands from gel lanes for both the negative control and biotinylated probe 24 affinity capture samples were examined in all experiments.
Figure 2.
Parallel target identification strategies for almiramide-based affinity purification probes. Orange triangle = biotin; Yellow star = photoaffinity benzophenone moiety; Grey sphere = Affigel resin
Cumulative analysis of the affinity capture results for four independent replicate experiments using biotinylated almiramide 24 identified 38 unique proteins that were present in two or more replicates. Of these, 24 were significantly enriched in the affinity capture condition over the control, including a strong gel band enhancement in the 29 kDa range (Figure 3A). This band contained both GIM5A (Tbg972.9.6900) and PEX11 (Tbg972.11.12940), which are integral membrane proteins found in glycosomes. These two proteins were consistently identified with high sequence coverage in replicate biotin affinity capture experiments (Table 2), with strong visual gel band enhancement in all cases. However, a number of additional candidate target proteins were also identified in at least three replicates using this approach (Table 2), precluding unambiguous assignment of either GIM5A or PEX11 as the unique target proteins of the almiramides. This result is common for protein affinity capture experiments, which often result in the identification of lists of putative target proteins for further study. GIM5A and PEX11 are normally present in low titer in whole cell lysates, so the identification of these two proteins using the biotin affinity strategy suggests that they have strong affinity for the almiramide probe, either individually or in complex.
Figure 3.
A) Representative SDS-page gel for protein affinity capture results. PEX11/GIM5A band enhancement outlined by dashed box. B) Venn diagram showing relative number of candidate target proteins pulled down using each approach. Relative circle areas correspond to the total number of proteins pulled down using each approach.
Table 2.
Candidate Proteins from All Three Orthogonal Protein Affinity Capture Approaches.
| Experiment, # of n replicates | ||||
|---|---|---|---|---|
| Name | TriTryp ID | Biotin | Affi | Benzo |
| glycosomal membrane protein (PEX11) | Tbg972.11.12940 | 3 | 1 | 2 |
| glycerol kinase, glycosomal | Tbg972.9.7700 | 3 | 2 | 1 |
| elongation factor 1-alpha | Tbg972.10.2530 | 3 | 2 | 1 |
| beta tubulin | Tbg972.1.1350 | 3 | 1 | 1 |
| glyceraldehyde 3-phosphate dehydrogenase, glycosomal | Tbg972.6.4080 | 3 | 1 | 1 |
| Gim5A protein | Tbg972.9.6900 | 2 | 1 | 2 |
| orotidine-5-phosphate decarboxylase | Tbg972.5.5300 | 2 | 1 | 1 |
To address the shortcomings of the biotin reporter tag approach, we also employed an orthogonal affinity chromatographic method. This approach identifies target proteins by the covalent attachment of the ligand to the surface of an aqueous-compatible affinity matrix via a long linker, followed by incubation of the whole cell lysate with the derivatized affinity gel and removal of the unbound protein fraction. This approach does not suffer from the non-specific enrichment of endogenous biotinylated proteins observed with avidin capture, but relies on the ligand being able to bind to its molecular target(s) while anchored to the surface of the affinity resin. Affinity matrices have been used successfully in the past for parasite lead compounds to discover the target for 2,4-diaminopyrimidines against T. brucei as MAPKs and CRKs.16
To generate the affinity resin-derivatized almiramide, compound 17 was first coupled to a 15Å PEG linker, to provide sufficient physical separation between the almiramide reporter and the surface of the affinity gel. Linker-almiramide conjugate 25 (Figure 2) was then anchored to the surface of an aqueous-compatible agarose resin (Affi-Gel) using standard amide coupling conditions (Supporting Information). Although the Affi-Gel resin has a binding capacity of 15 µmoles/mL, the resin was loaded to just 8 µmoles/mL in order to allow the almiramide warhead sufficient flexibility and steric space to bind to target proteins. The remaining open positions on the surface of the resin were capped with 6-amino hexanol, which was shown in a control experiment not to retain proteins from the whole cell lysate (Supporting Information). For comparative analysis, an inactive analog of almiramide affinity resin 26 was prepared at the same loading capacity, using a molecule containing the same methylation pattern and length of lipophilic terminus as compound 21, but containing a poly-alanine peptide portion in place of the Phe-Ala-Val-Val-Val almiramide core (Supporting Information).
The workflow for the affinity resin capture experiments was as follows (Figure 2); the parasite cell lysate was preclarified with a control resin capped with 6-amino hexanol, and incubated with either almiramide affinity resin 25 or inactive analogue resin 26. Following incubation, the affinity resins were washed with detergent-free buffer, and analyzed by SDS-PAGE gel electrophoresis. The affinity capture was conducted twice (n = 2) and compared with control resin-captured proteins to include only those proteins that were present in at least one replicate experiment, but not present in control lanes. This analysis revealed 12 proteins that were unique to the almiramide-affinity capture resin. These included both PEX11 and GIM5A, which were originally identified as candidates in the previous biotin capture experiments, lending further credence to the hypothesis that the almiramides target glycosomal function. However, a further six proteins were also common to both the biotin and affigel approaches (Figure 3 and Table 2), which precluded the identification of a unique target protein using these data alone.
Neither the biotin nor the affinity resin capture methods reported above were able to address the reversibility of binding to candidate targets, although competitive binding was unsuccessful using either almiramide addition at either the affinity capture or the complex elution steps, indicating that almiramide binding to target proteins must have an extremely slow off rate if it was not irreversible. An inherent flaw with both affinity gel and biotin reporters is their reliance on irreversible binding or slow turnover of the warhead to target protein(s). To force the equilibrium toward complete binding, a bifunctional probe was designed which contained both a photo-affinity motif and a biotin functional group on the same probe. This was accomplished by replacement of the phenylalanine residue with a benzophenone substituent (27). Bifunctional probes can be valuable for the examination of systems involving non-covalent protein interactions, because they can simultaneously capture and enrich target proteins with weak binding that would not be observed under standard biotin affinity capture conditions. The disadvantage of benzophenone photo-reactive probes is that the lifetime of photo-activated reactive species is long, which can allow diffusion-mediated non-specific covalent bond formation, meaning that photo-affinity probes are often somewhat more promiscuous than other methods. Despite this limitation, bifunctional photoaffinity biotin probes have been used successfully in a number of recent target identification studies.17, 18
Bifunctional biotinylated probe 27 was synthesized under standard Fmoc solid phase peptide synthesis (SPPS) conditions in an analogous fashion to biotinylated probe 24, differing only in the replacement of the initial phenylalanine residue with a commercial benzophenone-derived amino acid mimic. The probe was completed using standard SPPS and methylation conditions as previously described (Supporting Information) followed by coupling to a commercially available biotin-linker conjugate to yield probe 27.
The workflow for 27 is shown in Figure 2, and closely resembles the methodology used for biotinylated probe 24, with the exception of one additional key step. Following incubation of 27 with the preclarified cell lysate, the photoaffinity probe was activated by irradiation with UV light (366 nm, 3 min) to generate a short-lived carbene species (1–2 µsec in benzene).19 This highly reactive carbene will rapidly form a covalent attachment to neighboring proteins, creating a permanent bond between target proteins and the biotin reporter tag. The photoaffinity capture experiment was repeated twice (n = 2), and identified 73 proteins that were present in at least one replicate, but were not present in the streptavidin-only control lane. Identification of a larger number of candidates compared with the biotin-only affinity capture experiments is typical for photoaffinity experiments with benzophenone probes, however this list did include a number of candidate proteins that were common to the other two strategies used for protein affinity capture in this study.
To integrate the results of all three orthogonal affinity capture approaches we sorted the results by frequency of observation across all methods, and ordered this list based on the frequency of appearance in replicate experiments for each method (Figure 3 and Table 2). Using this approach seven candidate proteins were identified that were present in all three methods. Of these, five are associated with the glycosome, while the remaining two (β-tubulin and EF1-α) are highly abundant proteins that are likely not relevant to the mechanism of action of the almiramides. All of the glycosomal proteins were present in at least one replicate from all three affinity capture methods, and at least two of three replicates in the biotin affinity capture experiments.
Glycosomes are globular organelles roughly 0.2–0.3 µM in diameter, surrounded by a single phospholipid bilayer,20 and have been found in all kinetoplastid parasites so far examined. Uniquely, glycosomes compartmentalize the first seven steps of glucose metabolism, and two of glycerol metabolism:21 due to the unusual regulatory characteristics of the enzymes concerned, this compartmentation prevents runaway ‘turbo-explosion’ glycolysis and cell death.22 Other glycosomal pathways include ether lipid metabolism, the pentose phosphate pathway, and purine salvage; they vary somewhat depending on the organism and life-cycle stage.23,24 Each T. brucei parasite contains roughly 60 glycosomes, which in the bloodstream form of the parasite represents the sole source of energy production.20 By contrast, the insect procyclic form of the parasite is able to utilize more substrates for energy production, such as the ability to catabolize amino acids.25 Inhibition of glycosomal function has been proposed as an attractive target for therapeutic intervention because the target has no mammalian homolog and is essential for parasite survival. However, to date no compounds have been identified that directly disrupt the viability of glycosome structures.26–29 The proteins and signals involved in glycosomal protein import and membrane biogenesis are clearly related to those of peroxisomes and glyoxysomes, allowing glycosomes to be classified as microbodies.30,27 Approximately 32 “PEX” proteins (or peroxins), are involved in microbody biogenesis: some are found in all microbodies so far examined, whereas others are specific to subgroups of organisms. The three most abundant integral membrane proteins in the glycosomal membrane are the glycosomal integral membrane proteins GIM5A and GIM5B, and PEX11.31 All three of these proteins are members of the PEX11 family, although sequence identity with mammalian and yeast PEX11 proteins is low. Both GIM5 and PEX11 have been shown to be essential to the bloodstream form of T. brucei.31,32 GIM5A and GIM5B have also been shown to form heterodimers in vivo;32 each protein has two trans-membrane domains of 24–32 amino acids and the orientation is such that the C and N termini, which are much better conserved within Kinetoplastidae than the trans-membrane domains, protrude into the cytosol.33 Over-expression of either GIM5 or PEX11 altered glycosome morphology and numbers, suggesting roles in maintaining glycosomal membrane structure. More recent results clearly indicate that PEX11-family proteins are required for elongation34 and proliferation35 of peroxisomes, including the generation of membrane curvature during peroxisomal fission.36 It is also possible that PEX11 proteins might form a pore to allow transit of low molecular weight metabolites.37 In procyclic forms, depletion of GIM5A in the presence of GIM5B results in depletion of ether lipids,32 supporting a specific role of GIM5A for this particular pathway.
Remarkably, all five candidate target proteins identified in the protein pull-down experiments are glycosome-associated: GIM5A, PEX11 and three matrix enzymes, GAPDH, glycerol kinase and orotidine-5-phosphate decarboxylase.31 This is strongly indicative of glycosomal function as the target of the almiramides, and we speculated that they may target glycosome biogenesis by disrupting the functions of GIM5 and PEX11. To test this hypothesis we explored a number of approaches to validate this mechanism of almiramide function. We stained almiramide-treated trypanosomes with anti-aldolase antibody, but it rapidly became clear that all drug concentrations that affected growth caused such drastic morphological alterations that there was no chance of detecting a specific effect on glycosomes.
Next, we created a suite of fluorescent probes for imaging. The BODIPY fluorophore was selected because BODIPY’s spectral properties surpass those of fluorescein, tetramethylrhodamine, Texas Red and longer-wavelength dyes,38 and BODIPY dyes are relatively nonpolar with an electrically neutral chromophore. Together, these properties minimize dye-induced perturbations of parent molecule properties, and reduce the biological impact of introducing this chromophore to test compounds.39 The synthesis of these probes was accomplished by extension of the methodology described above. Briefly, almiramide-linker compound 22 was de-protected with LiOH to give the corresponding free acid 28 (Figure 4), and coupled to commercially available BODIPY FL EDA (32) under standard coupling conditions to give fluorescent derivative 33. Non-fluorescent control compound 31 was synthesized using a similar approach by coupling 28 to N-(3-phenylpropionyl)-1,2-ethanediamine 30. Finally, a BODIPY control compound lacking the almiramide warhead was prepared by coupling BODIPY FL EDA (32) to commercially available hydrocinnamic acid (34) under standard coupling conditions (Figure 4).
Figure 4.
Synthetic route to fluorescent almiramide derivatives and control compounds for fluorescence imaging.
Localization of Fluorescent Almiramides in Trypanosomes
As a prelude to fluorescence imaging, all compounds were screened for activity against T. b. brucei. The BODIPY control 36 was inactive against the parasite up to the highest tested concentration (50 µM), while fluorescent almiramide derivative 33 retained activity of 3.1 µM, indicating both that the fluorophore possessed no intrinsic activity against the parasite, and that derivatization of the almiramide moiety with the fluorescent tag did not eliminate its biological activity. To visualize the localization of the almiramide derivative within the parasite, we tested a multitude of different conditions. After brief in vivo labeling with high drug amounts, live parasites were fluorescent but the signal was lost following fixation, which is necessary to detect subcellular structures. In vivo labeling with lower amounts, for longer times, was problematic because of toxic effects; and the drug did not bind to anything in formaldehyde-fixed preparations. The only successful protocol involved EtOH fixation of dried procyclic trypanosomes, followed by permeabilization using T×100, and incubation with either 5 or 50 µM of 33. DAPI (nuclear stain) and anti-aldolase (glycosome stain) were used for co-localization. Figure 5A shows that compound 33 (5 µM) was distributed throughout the cytosol with localized brighter regions. Compound 33 did not localize to the nucleus, nor to the kinetoplast DNA, which can be seen as a small bright region below the nucleus in the DAPI images. At the higher concentration (50 µM, Figure 5B) the almiramide probe localized preferentially to some non-glycosome structures that lay in the parasite posterior, between the nucleus and flagellar pocket. This location is suggestive of structures of the secretory pathway, but we have not tested this in more detail. Glycosomal membranes are, like those of peroxisomes, probably initially formed by the ER, and some PEX11 family proteins are involved.35 A preferential association of almiramide with a minor portion of GIM5 or PEX11, within the ER membrane, therefore cannot be ruled out. Overall, however, the results of these experiments were inconclusive, since it is possible that under the conditions used, the almiramide derivative was binding in a non-specific fashion.
Figure 5.
Fluorescence site localization results for BODIPY-labeled almiramide probe 33. A) Uptake of probe 33 at 5 µM, and distribution of staining with respect to the nucleus and kinetoplast (large and small staining regions respectively stained in DAPI channel). B) Distribution of probe 33 with respect to glycosome distributions at 50 µM. Glycosomes stained with red anti-aldolase antibody.
Genetic Validation of Protein Targets
Unfortunately, previous attempts to express GIM5A and GIM5B in Escherichia coli were unsuccessful,26 precluding in vitro binding experiments. To complement the results from the pull-down, we therefore attempted to test the effect of the almiramides on GIM5 function in vivo. If the almiramides target GIM5, over-expression of GIM5 ought to confer almiramide resistance. Over-expression of GIM5 or PEX11 in procyclic trypanosomes is, however, toxic by itself.31,33 We transfected bloodstream-form trypanosomes with a vector containing a tetracycline-inducible GIM5B gene,27 hoping that perhaps we could counter this toxicity using almiramide. Several clones were obtained but in these forms, no over-expression could be detected by Western blotting (data not shown), and no effect on almiramide toxicity was observed. It is possible that GIM5B expression is already too high for additional expression to have any impact. Alternatively, it is possible that even ‘leaky’ expression of extra GIM5B, in the absence of inducer, is lethal in bloodstream forms, precluding selection of appropriate clones. Although the fact that both over-and under-expression of GIM5 and PEX11 severely inhibit parasite growth makes these proteins attractive potential drug targets, it also represents a serious technical limitation on genetic approaches to verify them as the targets of potential inhibitors.
Zebrafish Toxicity and Site Localization Imaging
Current front-line therapeutics for HAT are antiquated, and suffer from issues of toxicity and development of resistance. The almiramides and their analogues represent a unique chemical scaffold for the treatment of HAT that is distinct from any of the current therapeutic options. However, to be of value as candidates for further development, a large number of potential liabilities must be examined, including toxicity in whole animal models. Zebrafish are gaining prominence for exploring ADMETox properties of early-stage leads in hit-to-lead drug development projects40 because of their straightforward husbandry requirements, their small size, which allows testing with small amounts of test compounds, and their ability to report on a wide variety of developmental and neurological characteristics.
Both the unlabeled compound 21 and fluorescent derivative 38 were tested against zebrafish with the objectives of identifying toxic side effects of the compound series, and determining whether the compounds localized to any organs or specific regions of the fish. To this end, compound 21 was incubated with zebrafish embryos (48 hr post fertilization) for 24 hr and embryo viability was then assessed to establish the maximum tolerated dose (MTD). After 24 hr of incubation with 21, the MTD was 168 µM, giving a wide therapeutic index considering that this compound series was active against T. b. brucei in the low micromolar range. These doses also agreed well with the previously reported LD50 values against Vero cells.10
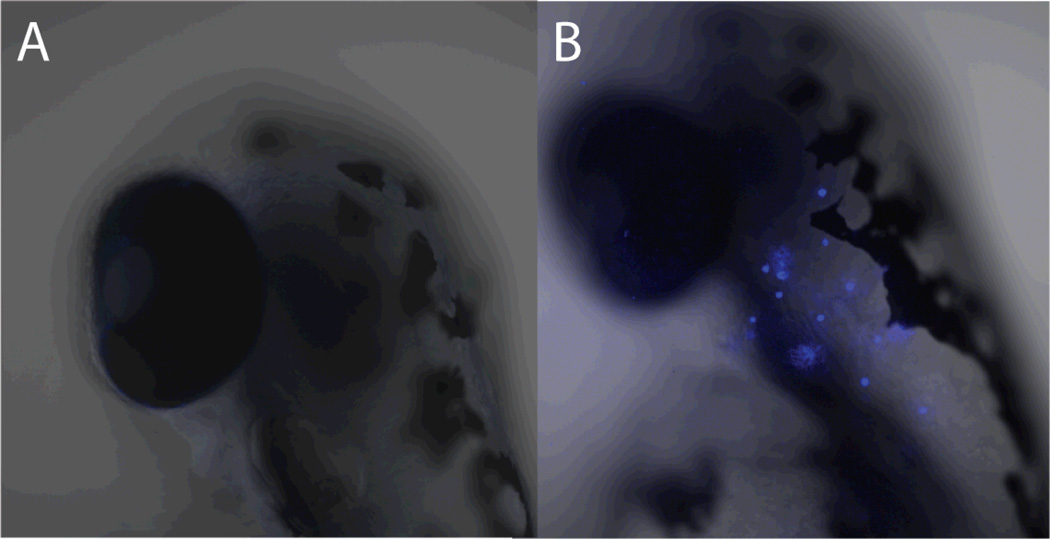
Using these results to determine appropriate concentrations for imaging probes, we tested two different almiramide-fluorophore conjugates for fluorescence staining and site localization. Imaging using 48 hr post fertilization (hpf) embryos revealed high levels of fluorescence to rosette structures along the lateral line of the zebrafish. Changing the protocol to use 120 hpf embryos, revealed strong fluorescence in the intestinal tract of the zebrafish, likely the result of fish ‘gulping’ water during compound treatment. With fluorescent probe 33 no fluorescence was observed after 3 hr of exposure. However at high doses of probe 38 (250 and 500 µM), bright blue rosettes were observed along the lateral line of the fish (Figure 6). This patterning was not observed with the other fluorescent analogue, or with fluorophore control compounds alone. From the structure and patterning of the observed fluorescence, it appeared that 38 may have affinity for zebrafish neuromasts at very high doses. Neuromasts are sensory organs located on the epithelial of the skin of the fish and are composed of different cell types, including hair cells, which allow the fish to sense water vibrations. In humans, neuromasts are analogous to the hair cells found in the cochlea, which is located in the inner ear. The hair cells play a role in auditory function. Human hair cells and neuromasts have a well documented sensitivity to the amino glycoside antibiotics, for example neomycin, tobramycin and gentamycin.41,42 To test the affinity of probe 38 for neuromasts in zebrafish, we examined the effect of pre-treatment with neomycin on neuromast staining. From this study embryos which were untreated with neomycin, but treated with 250 µM 38 displayed the blue rosette patterning along the lateral line. Embryos which were untreated with neomycin, but treated with fluorescent control compound 37 displayed no rosette patterning, and embryos that were pretreated with neomycin (500 µM) followed by treatment with 250 µM 38 displayed no fluorescent uptake. These results indicate that 38 has affinity for neuromasts at high concentrations. Additional experimentation is necessary to determine whether this uptake is specific to 38 or the nonspecific result of the exposure of these cells to the external environment, as the high probe concentration might suggest. Regardless, the almiramides should be closely monitored for potential ototoxicity as the scaffold is further developed. If such a side effect is observed, ototoxicity can be prevented by blocking the c-Jun-N-terminal kinase (JNK) with the peptide D-JKNI-1.43
Figure 6.
Fluorescence imaging for A) control compound 37 and B) almiramide probe 38 in juvenile zebrafish. Images display lateral region at the intersection of the head and the upper spine. Zebrafish eye visible as a large dark region in the center-left of each image. The fluorescent staining indicate binding of almiramide to neuromast cells. These data are also presented as a z-stack video in the supporting information (Video S1)
The relatively high concentration of 21 at which toxicity is observed in zebrafish embryos is in stark contrast to the low concentrations necessary to cause T. b. brucei death. This differential activity bodes well for the development of the almiramides as antiparisitics. However, evaluation in higher vertebrates will be necessary to ensure that toxic effects to the host from these molecules remain minimal.
In conclusion, this work represents the discovery and characterization of the first example of small molecules that may disrupt glycosomal function in T. brucei. The combined results from the affinity capture experiments and fluorescent microscopy experiments strongly support the conclusion that the almiramides target the glycosomal membrane. Although we were unable to confirm this by immunofluorescent staining or reverse genetics, both approaches suffered from serious technical limitations, rendering the results inconclusive. Glycosomes are an encouraging potential target for kinetoplastid drug discovery because glycolysis is not compartmentalized in mammalian cells, making this target parasite-specific. More work is still needed to uncover the precise molecular target of the almiramides, but given that GIM5A and GIM5B have not been successfully expressed, and in vitro binding experiments are therefore precluded, alternative methods for target identification will be required. Along with GIM5A, PEX11 is involved in glycosomal biogenesis and proper fission of budding glycosomes. Affinity capture of PEX11 and GIM5A is therefore consistent with the almiramides affecting glycosomal biogenesis. The almiramides represent a promising molecular tool with which to further probe glycosomal function during parasite development. Given that the almiramides display low vertebrate toxicity, they are encouraging candidates for further lead development. This scaffold is synthetically tractable and derivatizable, and amenable to large-scale production for evaluation in murine parasitemia models. In conclusion the almiramide scaffold represents the first potential molecular probe for the disruption of glycosomal membranes in Trypanosoma brucei, and offers a new avenue for future drug development against African sleeping sickness.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Jasco P-2000 polarimeter using a 10 mm path length cell at 589 nm. UV spectra were acquired on an Agilent 8453 UV-vis spectrophotometer. NMR spectra were acquired on a Varian Inova 600 MHz spectrometer equipped with a 5 mm HCN triple resonance cryoprobe, and referenced to residual solvent proton and carbon signals (δH 7.26, δC 77.1 for CDCl3 and δH 1.94, δC 1.4 for CD3CN). Unless otherwise stated, reactions were performed under an argon atmosphere using freshly dried solvents. Methylene chloride (DCM) was dried by passing through an activated alumina column. Solvents used for HPLC chromatography were HPLC grade and were used without further purification.
Trypanosomes
Trypanosoma brucei brucei strain 427–221 was grown at 37º C, 5% CO2 in HMI-9 medium containing 10% fetal bovine serum, 10% Serum Plus (JRH Inc.) and penicillin/streptomycin.
Trypanosome screening assay
T.b.b. 427–221 cells were diluted to 2×104 per mL in complete HMI-9 medium and aliquoted in Greiner sterile 384-well flat white opaque culture plates using a WellMate cell dispenser from Matrix Tech. Test compounds were serially diluted in dimethyl sulfoxide (DMSO) and added to the assay plates with the robotic dispenser BiomekFXp liquid handler (Beckman Coulter). Thimerosal (2 µM final concentration) was added as a positive control and DMSO as a negative control (1% final concentration). Trypanosomes were incubated with compounds for 48 hr at 37 ºC with 5% CO2 before monitoring cell viability as previously described.44 Briefly, the trypanosomes were lysed in the wells by adding 25 µL of CellTiter-Glo (Promega), and the lysed trypanosomes placed on an orbital shaker at room temperature (rt) for 2 min. After lysis, the resulting ATP-bioluminescence was measured at rt using an Analyst HT plate reader (Molecular Devices). Percentage inhibition of parasite growth was calculated for each well as [1-(RLU×-RLU+)/(RLU−-RLU+)]*100 where RLU×, RLU+ and RLU− are respectively the Relative Light Units for each well, positive (thimerosal) and negative (DMSO) controls. A screening window coefficient, denoted Z’ factor, was used to evaluate the performance of the assay. The Z’ factor, calculated as 1−(3σc++3σc−)/(µc+ − µc−) where σc+, σc−, µc+ and µc− are respectively the standard deviation and mean values of positive and negative controls, is reflective of the assay signal dynamic range and the data variation associated with signal measurement.45 IC50s were calculated using Prism (GraphPad).
Target Protein Affinity Capture
Preparation of Trypanosome Lysate
Cells were resuspended (approximately 200 µL 100 million cell pellet−1) in a buffer containing 50 mM HEPES (pH 7.4), 100 mM NaCl, 0.5% NP40, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1% Tween20 and supplemented with Halt protease inhibitor cocktail (ThermoFisher Scientific) and lysed using a dounce homogenizer, with pestle B, 70 strokes. DNA was pelleted by centrifugation.
Affinity Chromatography
The cleared lysate was incubated with either control Affigel resin or neutravidin resin for 1 h at 4 ºC to remove endogenously biotinylated proteins. The lysate was then divided equally between Falcon tubes to provide replicate material for the different affinity approaches. For the biotinylated capture, 20 µM of compound 24 dissolved in DMSO was added to the preclarified lysate and inverted at rt for 2 hr. Neutravidin beads were prepared as follows: 400 µL of slurry was transferred to a 15 mL Falcon tube and 10 mL of cold detergent-free buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, Halt protease inhibitor cocktail was added to wash the resin, followed by centrifugation for 3 min at 2500 rpm to pellet. The wash was removed, the pellet resuspended with 10 mL of cold detergent free buffer, and the process repeated. Upon completion of affinity binding step, the lysate-24 mixture was transferred to a 1.5 mL Eppendorf tube containing the prepared neutravidin beads, and inverted for 2 h at rt to capture the protein-compound complexes. Following affinity capture, the resin was centrifuged, and the supernatant was removed. The resin-protein-24 complex was carefully washed with 1 mL of cold detergent-free buffer, spun down, the wash removed, and the washing protocol repeated and all liquid removed from the resin. Samples were stored overnight at −20 ºC until gel analysis could be performed.
For the biotinylated-benzophenone affinity chromatography (27), the same procedure was utilized, with the addition of UV exposure to induce crosslinking. At the end of incubation of 20 µM of 27 with the preclarified lysate, the sample was irradiated with a UV lamp for 3 min to induce crosslinking.46 Then capture procedure was performed as described above.
For the Affigel-10 affinity chromatography, the preclarified lysate was divided equally and incubated with either the 26 or 25 for 5 h at 4 ºC to capture the protein-compound complexes. The Affigel samples were then washed and prepared for gel analysis as previously described.
Protein Identification by Mass Spectrometry
Protein identification from affinity-purified samples was performed using peptide sequencing by mass spectrometry. Affinity purified samples were eluted from capture resin, then resolved by SDS-PAGE using 10% Tris-glycine gels (Bio-Rad) and stained by silver stain.47 Gel bands were excised and subjected to in-gel trypsin digestion using standard protocols (in-gel digestion protocol, UCSF). Extracted peptides were sequenced using a QStar Pulsar quadrupole-orthogonal-acceleration-time-of-flight hybrid mass spectrometer (Applied Biosystems), coupled to an Ultimate HPLC with Famosautoinjector (LC Packings), and a self-packed C18 column (New Objective Inc., 5 µm bead size, 100 µm × 150 mm). The LC was operated at 350 nL/min flow rate, and peptides were separated using a linear gradient over 42 min from 5% to 50% ACN in 0.1% formic acid. Data were acquired in information-dependent acquisition mode; 1 s survey scans over 310–1400 m/z range were followed by 3 s CID fragmentation spectra over a 60–1400 m/z range.
Data were analyzed in Analyst 2.0 software (Applied Biosystems) with the Mascot script 1.6b20 (Matrix Science). Analyst processing options for peak finding in a spectrum were 0.5% default threshold, 400 Gaussian filter, and a Gaussian filter limit of 10; for TOF auto-centroiding: 20 ppm merge distance, 10 ppm minimum width, 50% percentage height, and 100 ppm maximum width. Default parameters were used except that “no deisotoping” was selected and precursor mass tolerance for grouping was set to 0.2. Database searches were performed using ProteinProspector v. 5.10.1 (http://prospector.ucsf.edu) using the Batch-Tag and Search Compare modules.48 Searches were performed on the SwissProt databank, downloaded March 21, 2012 and containing 535,248 entries, as well as the T. brucei sequences in the TriTrypDB v. 4.1 (http://www.tritrypdb.org), downloaded April 23, 2012 and containing 28,250 entries. For estimation of false discovery rate, these databases were concatenated with a fully randomized set of entries.49 Data were searched with a parent mass tolerance of 200 ppm and fragment mass tolerance of 300 ppm.
For database searching, peptide sequences were matched as tryptic peptides with no missed cleavages, and carbamidomethylated cysteines as a fixed modification. Variable modifications included oxidation of methionine, N-terminal pyroglutamate from glutamine, loss of methionine and N-terminal acetylation. For reporting of protein identifications from this database search, score thresholds were selected that resulted in a protein false discovery rate of approximately 1%. The specific Protein Prospector parameters were: minimum protein score of 22, minimum peptide score of 15, and maximum expectation values of 0.02 for protein and 0.05 for peptide matches. Protein identification results from specific affinity purification experiments are reported with a spectral count as an approximation of protein abundance, along with percent sequence coverage and an expectation value for the probability of the protein identification.50,51
Trypanosome Fluorescence Imaging
2 × 106 procyclic trypanosomes were resuspend in 2 µL of modified Eagle’s medium for procyclics (MEM-PROS) medium supplemented with 10% fetal calf serum,52 spread on a glass slide, allowed to dry, then fixed with 70% EtOH. The cells were rehydrated in 1 × PBS/0.2% Triton for 20 minutes, washed three times with PBS, incubated for 20 min with PBS/0.5% gelatin, and then with anti-aldolase (1h, 1:500) in the same PBS-gelatin mix. After two washes with PBS, the cells were incubated for 1 h with 5 µM 33 or 36, plus 1:500 Alexa 594 α-rabbit, and 100 ng/mL DAPI. The stained spreads were washed three times with 1 × PBS, dried, then mounted in 90% glycerol, 10% 10× PBS and imaged with a Leica DMRXA microscope at 100× oil magnification.
Zebrafish Studies
Animals
Adult zebrafish (Danio rerio) obtained from Aquatica Tropicals were maintained at 27 ± 2 °C in plastic recirculating aquaria (Aquaneering Inc.) on a 12 h light:12 h dark cycle. Fish were fed two to three times daily with a flake mix (57% Aquatox Flake, 19% Spirulina Flake, 8% Hikari Micropellet, 8% Cyclop-eeze, 4% Golden Pearl 300–500 and 4% Golden Pearl 500–800) and live brine shrimp. All animal experimentation was conducted according to NIH and University of California, San Diego guidelines under IACUC-approved protocol S05560.
Breeding and Embryo Collection
Two adult male and two adult female zebrafish were introduced into a two-way fish breeder (Petsmart, Inc.) without tank flow the evening prior to breeding and separated by a plastic divider overnight. The divider was removed the following morning and after 1 h embryos were transferred to petri dishes containing egg water medium consisting of filtered water supplemented with 60 µg/mL ‘Instant Ocean’ salts and 0.001% methylene blue.53
Almiramide Toxicity
Embryos at 48 and 120 h post fertilization were used to determine the maximum tolerated dose of almiramide. A 10 mM stock of 22 in DMSO was diluted to a concentration of 500 µM in egg water lacking methylene blue. Two-fold serial dilutions were performed in a 96-well plate and final concentrations tested were 500, 250, 125, 62.5, 31.25, 15.6, 7.8, 3.9, 1.95, and 0 µM. Two embryos were exposed to each concentration. Survival, defined as the presence of a heartbeat, was assessed using an inverted microscope at 3 h and 24 h following compound addition. Survival of 22-exposed animals was compared to animals exposed to the benzopyrone coumarin prepared in an identical manner.
Localization of Fluorescent Almiramide
Embryos (48 and 120 h.p.f.) were incubated with compounds 37 or 38 diluted in methylene blue-free egg water to concentrations of 270, 90, 30, and 0 µM. Three embryos were exposed per treatment. After 3 h of incubation a Zeiss LSM-700 confocal microscope was used to visualize compound localization.
Localization of Fluorescent Almiramide in Neomycin-Exposed Embryos
Embryos were exposed to the aminoglycoside antibiotic neomycin at 500 µM for 1 h. A control group of animals was incubated in medium only. Embryos from both groups were then divided into 37 (250 µM), 38 (250 µM), or compound-free treatments. After 3 h of incubation a Zeiss LSM-700 confocal microscope was used to visualize compound localization.
Supplementary Material
Acknowledgement
We thank K. Sharaf for assistance with synthetic preparation of some starting materials, A. Hamdoun for access to confocal microscope facilities, and J. Campanale for technical assistance. Mass spectrometry was performed in the UCSF MS facility (A.L. Burlingame, Director) supported by 8P41GM103481. This work was funded by the International Cooperative Biodiversity Group (ICBG) program in Panama (Grant ICBG TW006634, RGL and LG), NIH1R56AI085177-01A1 (RGL), the Sandler Family foundation (JHM), and an NSF graduate research fellowship (LMS).
Footnotes
Supporting Information. Experimental procedures and 1H and 13C NMR spectra for all synthetic compounds. Z-stack video of rosette staining pattern in zebrafish, and spectral count files for all protein identifications. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fevre EM, von Wissmann B, Welburn SC, Lutumba P. PLoS Negl. Trop. Dis. 2008;2:e333. doi: 10.1371/journal.pntd.0000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalli A, Bolognesi ML. J. Med. Chem. 2009;52:7339–7359. doi: 10.1021/jm9004835. [DOI] [PubMed] [Google Scholar]
- 3.Barrett MP. Curr. Opin. Infect. Dis. 2010;23:603–608. doi: 10.1097/QCO.0b013e32833f9fd0. [DOI] [PubMed] [Google Scholar]
- 4.Alsford S, Eckert S, Baker N, Glover L, Sanchez-Flores A, Leung KF, Turner DJ, Field MC, Berriman M, Horn D. Nature. 2012;482:U232–U125. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burri C. Parasitology. 2010;137:1987–1994. doi: 10.1017/S0031182010001137. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser M, Bray MA, Cal M, Trunz BB, Torreele E, Brun R. Antimicrob. Agents Chemother. 2011;55:5602–5608. doi: 10.1128/AAC.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowther GJ, Shanmugam D, Carmona SJ, Doyle MA, Hertz-Fowler C, Berriman M, Nwaka S, Ralph SA, Roos DS, Van Voorhis WC, Aguero F. PLoS Negl. Trop. Dis. 2010:4. doi: 10.1371/journal.pntd.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra-Giraidez C, Quijada L, Clayton CE. J. Cell Sci. 2002;115:2651–2658. doi: 10.1242/jcs.115.13.2651. [DOI] [PubMed] [Google Scholar]
- 9.Nowicki MW, Tulloch LB, Worralll L, McNae IW, Hannaert V, Michels PAM, Fothergill-Gilmore LA, Walkinshaw MD, Turner NJ. Bioorg. Med. Chem. 2008;16:5050–5061. doi: 10.1016/j.bmc.2008.03.045. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez LM, Lopez D, Vesely BA, Della Togna G, Gerwick WH, Kyle DE, Linington RG. J. Med. Chem. 2010;53:4187–4197. doi: 10.1021/jm100265s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leslie BJ, Hergenrother PJ. Chem. Soc. Rev. 2008;37:1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 12.Cravatt BF, Wright AT, Kozarich JW. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 13.Lomenick B, Olsen RW, Huang J. ACS Chem. Biol. 2011;6:34–46. doi: 10.1021/cb100294v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugdahn N, Peuckert F, Albrecht AG, Miethke M, Marahiel MA, Oberthur M. Angew. Chem. Int. Ed. 2010;49:10210–10213. doi: 10.1002/anie.201005527. [DOI] [PubMed] [Google Scholar]
- 15.Barton V, Ward SA, Chadwick J, Hill A, O'Neill PM. J. Med. Chem. 2010;53:4555–4559. doi: 10.1021/jm100201j. [DOI] [PubMed] [Google Scholar]
- 16.Mercer L, Bowling T, Perales J, Freeman J, Tien N, Bacchi C, Yarlett N, Don R, Jacobs R, Nare B. PLoS Negl. Trop. Dis. 2011;5:e956. doi: 10.1371/journal.pntd.0000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa M, Miura T, Kuzuya K, Inoue A, Ki SW, Horinouchi S, Yoshida T, Kunoh T, Koseki K, Mino K, Sasaki R, Yoshida M, Mizukami T. ACS Chem. Biol. 2011;6:229–233. doi: 10.1021/cb100248e. [DOI] [PubMed] [Google Scholar]
- 18.Barglow KT, Cravatt BF. Nat. Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- 19.Hirai K, Itoh T, Tomioka H. Chem. Rev. 2009;109:3275–3332. doi: 10.1021/cr800518t. [DOI] [PubMed] [Google Scholar]
- 20.Sommer JM, Wang CC. Annu. Rev. Microbiol. 1994;48:105–138. doi: 10.1146/annurev.mi.48.100194.000541. [DOI] [PubMed] [Google Scholar]
- 21.Michels PAM, Moyersoen J, Krazy H, Galland N, Herman M, Hannaert V. Mol. Membr. Biol. 2005;22:133–145. doi: 10.1080/09687860400024186. [DOI] [PubMed] [Google Scholar]
- 22.Michels PAM, Hannaert V, Bringaud F. Parasitol. Today. 2000;16:482–489. doi: 10.1016/s0169-4758(00)01810-x. [DOI] [PubMed] [Google Scholar]
- 23.Colasante C, Ellis M, Ruppert T, Voncken F. Proteomics. 2006;6:3275–3293. doi: 10.1002/pmic.200500668. [DOI] [PubMed] [Google Scholar]
- 24.Vertommen D, Van Roy J, Szikora JP, Rider MH, Michels PAM, Opperdoes FR. Mol. Biochem. Parasitol. 2008;158:189–201. doi: 10.1016/j.molbiopara.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Furuya T, Kessler P, Jardim A, Schnaufer A, Crudder C, Parsons M. Proc. Natl. Acad. Sci. USA. 2002;99:14177–14182. doi: 10.1073/pnas.222454899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra S, Ruhela D, Deb A, Vishwakarma RA. Expert Opin. Ther. Targets. 2010;14:739–757. doi: 10.1517/14728222.2010.495125. [DOI] [PubMed] [Google Scholar]
- 27.Moyersoen J, Choe J, Fan E, Hol WGJ, Michels PAM. FEMS Microbiol. Rev. 2004;28:603–643. doi: 10.1016/j.femsre.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Opperdoes FR, Michels PAM. Caister Academic Press; 2008. pp. 123–158. [Google Scholar]
- 29.Schliebs W. Biochim. Biophys. Acta, Mol. Cell Res. 2006;1763:4–5. doi: 10.1016/j.bbamcr.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Parsons M. Mol. Microbiol. 2004;53:717–724. doi: 10.1111/j.1365-2958.2004.04203.x. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz P, Maier AG, Baumgart E, Erdmann R, Clayton C. EMBO J. 1998;17:3542–3555. doi: 10.1093/emboj/17.13.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voncken F, van Hellemond JJ, Pfisterer I, Maier A, Hillmer S, Clayton C. J. Biol. Chem. 2003;278:35299–35310. doi: 10.1074/jbc.M301811200. [DOI] [PubMed] [Google Scholar]
- 33.Maier A, Lorenz P, Voncken F, Clayton C. Mol. Microbiol. 2001;39:1443–1451. doi: 10.1046/j.1365-2958.2001.02333.x. [DOI] [PubMed] [Google Scholar]
- 34.Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, Brocard C. J. Cell Sci. 2010;123:3389–3400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]
- 35.Huber A, Koch J, Kragler F, Brocard C, Hartig A. Traffic. 2012;13:157–167. doi: 10.1111/j.1600-0854.2011.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opalinski L, Kiel J, Williams C, Veenhuis M, van der Klei IJ. EMBO J. 2011;30:5–16. doi: 10.1038/emboj.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gualdron-Lopez M, Brennand A, Avilan L, Michels PA. Parasitology. 2013;140:1–20. doi: 10.1017/S0031182012001278. [DOI] [PubMed] [Google Scholar]
- 38.Ulrich G, Ziessel R, Harriman A. Angew. Chem. Int. Ed. 2008;47:1184–1201. doi: 10.1002/anie.200702070. [DOI] [PubMed] [Google Scholar]
- 39.Daly CJ, McGrath JC. Pharmacol. Ther. 2003;100:101–118. doi: 10.1016/j.pharmthera.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Govind P. Int. Res. J. Pharm. 2011;2:33–36. [Google Scholar]
- 41.Froehlicher M, Liedtke A, Groh KJ, Neuhauss SCF, Segner H, Eggen RIL. Aquat. Toxicol. 2009;95:307–319. doi: 10.1016/j.aquatox.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Schacht J, Talaska AE, Rybak LP. Anat. Rec. (Hoboken) 2012;295:1837–1850. doi: 10.1002/ar.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eshraghi AA, Wang J, Adil E, He J, Zine A, Bublik M, Bonny C, Puel J-L, Balkany TJ, Van DWTR. Hear. Res. 2007;226:168–177. doi: 10.1016/j.heares.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Mackey ZB, Baca AM, Mallari JP, Apsel B, Shelat A, Hansell EJ, Chiang PK, Wolff B, Guy KR, Williams J, McKerrow JH. Chem. Biol. Drug Des. 2006;67:355–363. doi: 10.1111/j.1747-0285.2006.00389.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang JH, Chung TDY, Oldenburg KR. J. Biomol. Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 46.Bi XH, Schmitz A, Hayallah AM, Song JN, Famulok M. Angew. Chem. Int. Ed. 2008;47:9565–9568. doi: 10.1002/anie.200803962. [DOI] [PubMed] [Google Scholar]
- 47.Shevchenko A, Wilm M, Vorm O, Mann M. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 48.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. Mol. Cell. Proteomics. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elias JE, Gygi SP. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 50.Choi H, Fermin D, Nesvizhskii AI. Mol. Cell. Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu HB, Sadygov RG, Yates JR. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 52.Overath P, Czichos J, Haas C. European Journal of Biochemistry. 1986;160:175–182. doi: 10.1111/j.1432-1033.1986.tb09955.x. [DOI] [PubMed] [Google Scholar]
- 53.Westerfield M. The zebrafish book. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.