Abstract
The cellular microenvironment in follicular lymphoma is of biological and clinical importance. Studies on the clinical significance of non-malignant cell populations have generated conflicting results, which may partly be influenced by poor reproducibility in immunohistochemical marker quantification. In this study, the reproducibility of manual scoring and automated microscopy based on a tissue microarray of 25 follicular lymphomas as compared to flow cytometry is evaluated. The agreement between manual scoring and flow cytometry was moderate for CD3, low for CD4, and moderate to high for CD8, with some laboratories scoring closer to the flow cytometry results. Agreement in manual quantification across the 7 laboratories was low to moderate for CD3, CD4, CD8 and FOXP3 frequencies, moderate for CD21, low for MIB1 and CD68, and high for CD10. Manual scoring of the architectural distribution resulted in moderate agreement for CD3, CD4 and CD8, and low agreement for FOXP3 and CD68. Comparing manual scoring to automated microscopy demonstrated that manual scoring increased the variability in the low and high frequency interval with some laboratories showing a better agreement with automated scores. Manual scoring reliably identified rare architectural patterns of T-cell infiltrates. Automated microscopy analyses for T-cell markers by two different instruments were highly reproducible and provided acceptable agreement with flow cytometry. These validation results provide explanations for the heterogeneous findings on the prognostic value of the microenvironment in follicular lymphoma. We recommend a more objective measurement, such as computer-assisted scoring, in future studies of the prognostic impact of microenvironment in follicular lymphoma patients.
Introduction
Follicular lymphoma (FL) is the second most frequent non-Hodgkin lymphoma in the western world. FL is generally an indolent disease with a 7–12 year median survival.1 The survival distribution is relatively wide and the outcome has improved over recent years.2 In approximately 20% of the patients, long-lasting remissions may be achieved,3 while others die early, often due to transformation to a high-grade lymphoma.4,5 Therefore, determining factors associated with response to treatment and survival are particularly important to guide treatment selection.6 Today, prognostic factors are limited to clinical prognostic indices, such as the Follicular Lymphoma International Prognostic Index (FLIPI).7,8 However, the clinical behavior is also influenced by genetic aberrations present in the tumor cells, by the tumor grade, and by immune cell infiltrates in the tumor microenvironment.
Gene expression analyses have demonstrated that FL biology may be influenced by the non-malignant tumor microenvironment,9,10 stimulating numerous subsequent studies based on immunohistochemical analyses of various cellular components in the microenvironment. Such studies have further explored the prognostic role of specific T-cell and accessory cell populations, such as helper and cytotoxic T cells, regulatory T cells, macrophages, follicular dendritic cells and microvascularity. The results, however, are often contradictory, with specific cell populations significantly correlating with poor prognosis in some series, but with good prognosis or without any significant impact in others.11 A possible explanation for the discrepant results is patient selection bias, with a wide variation in age groups and risk factors. It is also likely that specific types of treatment influence or modify the prognostic impact of certain parameters in the microenvironment.6,11
Another possible explanation for the contradictory results is variation in the immunohistochemical staining and scoring across laboratories. We have previously shown that the reproducibility of scoring immunohistochemical stainings is poor and often inadequate for several frequently reported markers expressed on lymphoma cells in diffuse large B-cell lymphoma (DLBCL).12 Similar issues with reproducibility were reported in solid malignancies for HER2 expression in breast cancer13 and for EGFR in lung carcinoma.14
The studies mentioned above for various markers in DLBCL, and on HER2 and EGFR in carcinomas, focused on analyzing protein markers expressed by tumor cells. In contrast, immunohistochemical studies in FL mostly report on densities and distribution patterns of non-malignant immune cell populations present in the microenvironment. The reproducibility of scoring such parameters across laboratories has not yet been studied. Therefore, we performed a validation study focusing on the reproducibility of scoring cells and cellular distribution patterns in the tumor microenvironment before launching a large study on the impact of the tumor microenvironment in FL. The markers were chosen to be representative of frequent and less frequent cell populations (CD3 vs. CD8 and FOXP3), non-malignant lymphoid populations with membranous and nuclear staining patterns (CD3, CD4, CD8 vs. FOXP3), membranous and nuclear staining patterns on tumor cells (CD10 and MIB1), stromal cell components (CD21), and microvessel density (CD34). Importantly, immunohistochemistry scores (referred to as “manual” scores) are compared to flow cytometry data for CD3, CD4 and CD8 available from the same biopsy in all cases. A computerized system with an automated scanning microscope and computerized image analysis were also used for comparison to the manual scoring by pathologists.
Methods
Flow cytometry
A representative portion of the FL lymph node biopsy was processed by the BC Cancer Agency’s core clinical flow laboratory and dissociated into a single cell suspension by mincing the tissue using a sterile scalpel and phosphate buffered saline (Invitrogen Canada Inc., Burlington, ON, Canada) in a sterile Petri dish. Cells were counted, then co-stained using 6 panels of antibodies (all from Beckman Coulter, Fullerton, CA, USA). Information on antibody panels is provided in the Online Supplementary Appendix. Flow cytometry was performed on a Beckman Coulter FC500 flow cytometer.
Tissue microarray construction
Tissue microarrays (TMAs) were prepared at the Dept. of Pathology of the British Columbia Cancer Agency (BCCA) from 25 representative, newly diagnosed cases of FL and 2 tonsil samples with adequate archival formalin-fixed and paraffin-embedded material for which also routine flow cytometry data for CD3, CD4 and CD8 from the same FL lymph nodes were available. Representative 1.0 mm cores were taken and re-embedded in duplicate according to standard procedures. Five μm sections were stained with antibodies to 9 markers (Table 1) according to standard protocols at the Dept. of Pathology, The Netherlands Cancer Institute, The Netherlands.
Table 1.
Antibodies used for immunohistochemistry.
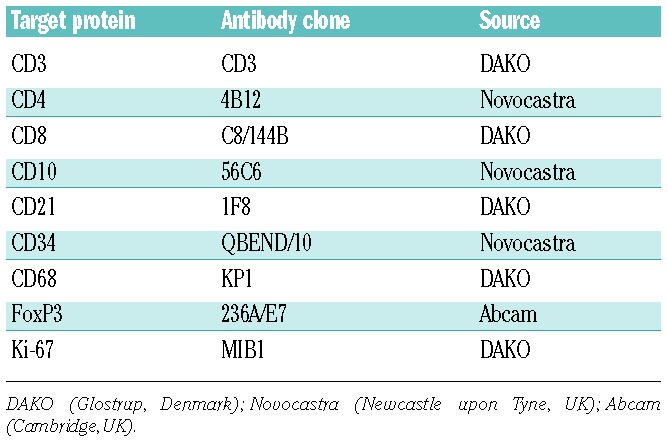
Criteria and scoring methods for immunohistochemistry
Scoring criteria and cut-off points for densities and patterns of non-malignant cells in FL (Table 2) were based on currently used methods published by various research groups.15–18 The scoring method was designed to allow comparison with the flow cytometry data expressed as positive cells for a given marker/total number of cells analyzed. A “scoring manual” was constructed as an additional guideline.
Table 2.
Manual scoring of immunohistochemical markers.
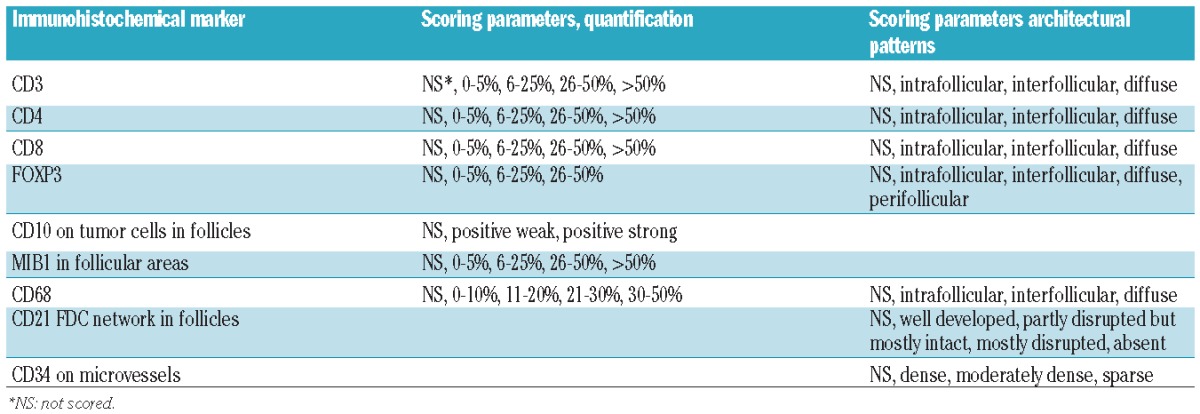
The same slide set was rotated among laboratories and independently analyzed by 7 pathologists/teams using the scoring manual as a guideline. The score for each core was recorded separately on an Excel worksheet. If the core could not be scored, the reason was recorded (Online Supplementary Table S5). Scores were reported to a central laboratory for analysis. Further information on the scoring is provided in the Online Supplementary Appendix. Additionally, all slides were scored using a computerized system with an automated scanning microscope and computerized image analysis (“Ariol Cambridge”) (Ariol SL-50, Genetix Ltd., Queensway, New Milton, Hampshire, UK) in combination with the Multistain assay in the Ariol software for quantification, as previously described.19 For CD3, CD4 and CD8, the automated scoring was repeated using a different operator and a different scanning and image analysis instrument of the same type, scanning microscope Olympus BX61, Ariol SL-50, v.3.4, Genetix Corp. 1998–2009 and the Multistain assay in the Ariol software for quantification (“Ariol Madrid”).20
Further information on the scoring methods is provided in the Online Supplementary Appendix.
Statistical analysis
All information on the statistical analysis is provided in the Online Supplementary Methods.
Ethical Committee approval
Approval to review, analyze and publish the data in this study was given by the University of British Columbia–British Columbia Cancer Agency Research Ethics Board.
Results
Analysis of CD3, CD4 and CD8 positive cells Densities of T-cell infiltration by flow cytometry versus manual or automated microscopy scoring of immunohistochemical stainings
Using flow cytometry, CD3 positive (CD3+) T cells ranged from 7–58% (median 32%), CD4+ T cells ranged from 5–41% (median 20%) and CD8+ T cells ranged from 2–14% (median 7%) of all viable cells in the sample. Thus there was a marked variation in the number of T cells and T-cell subsets, and the values observed are similar to previously reported results.21
Similar distributions of manual scores for T-cell markers in the two cores were observed (Figure 1 and Online Supplementary Tables S1 and S2). As shown in Table 3 and Figure 2A, there was significant variation (F-test; P<0.0001) in the scoring results between the 7 laboratories with 1–2 laboratories with significantly different scores from others (Lab 1 scored higher and Lab 5 scored lower for CD3, Labs 5 and 7 scored lower for CD4, and Lab 1 scored higher and Lab 4 scored lower for CD8, as compared to other laboratories). Agreement among all 7 laboratories was observed in 8–17% of the cases and among all but one laboratory in 38–46% of the cases (Table 4). The average pairwise agreement of 54–63% resulted in a low to moderate reproducibility with free marginal kappa statistic values of 0.43–0.54 (Table 4 and Figure 3).
Figure 1.
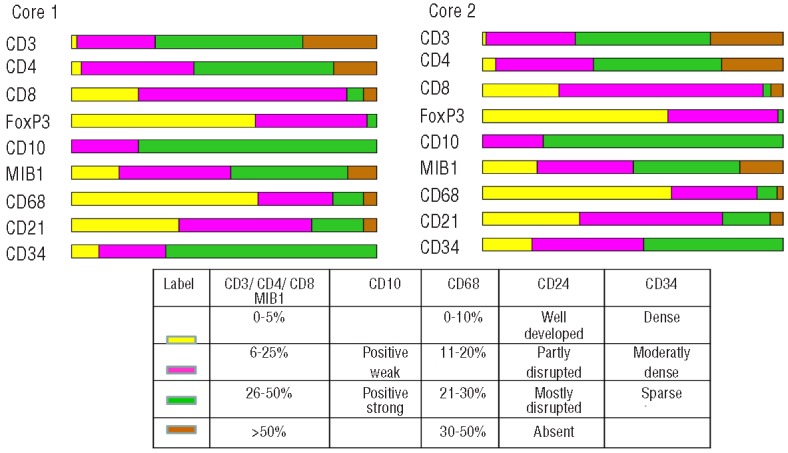
The distribution of manual scores of all investigated markers for core 1 and core 2 across the 7 laboratories, excluding the cores not scored. The color-coded labels for the scoring categories for each marker are explained in the lower part of the figure. Note that the results obtained in the two different cores are very similar for all markers except CD34.
Table 3.
Comparison of T-cell marker scores from flow cytometry, image analysis and manual scoring (Core 1).
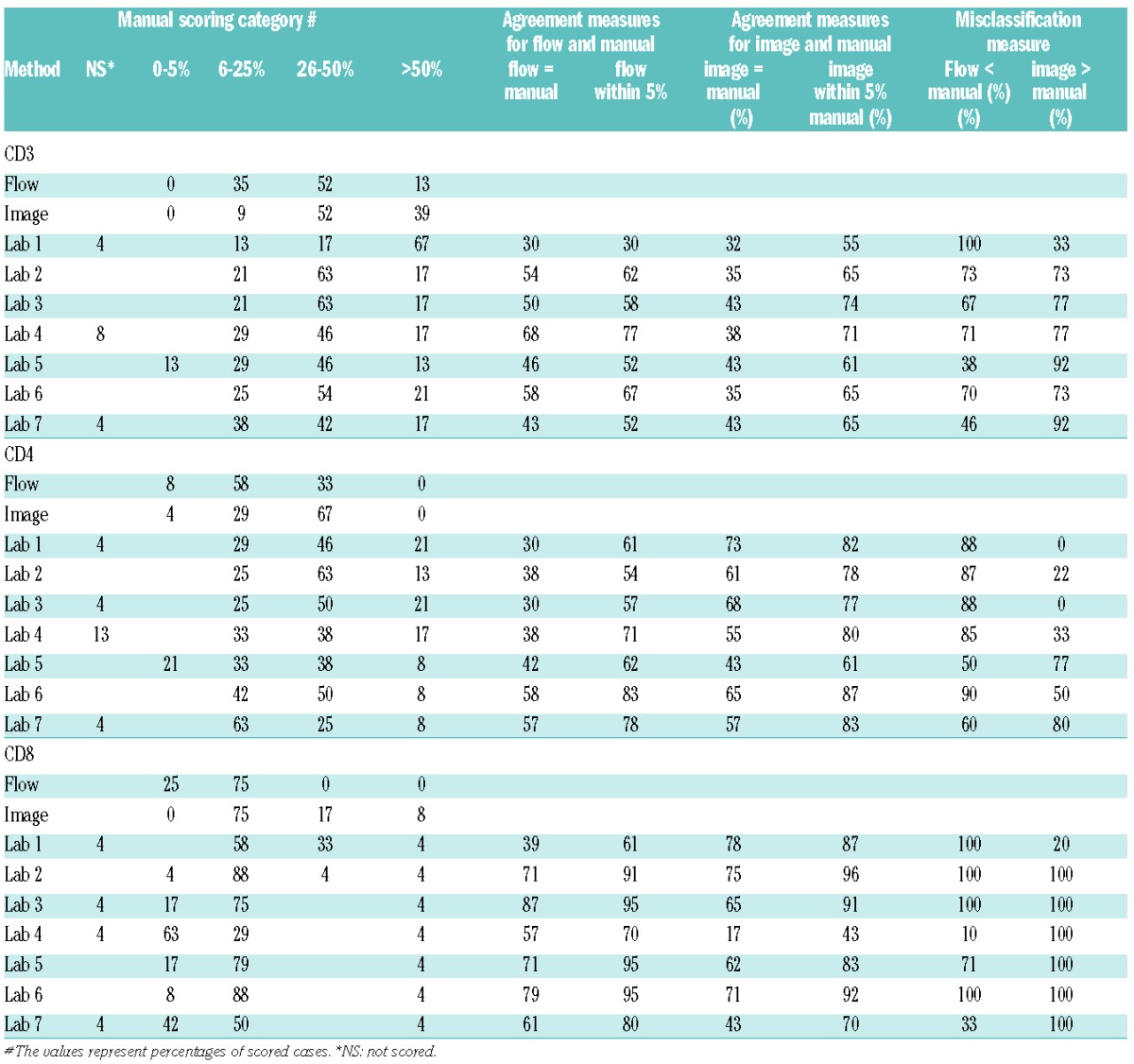
Figure 2.
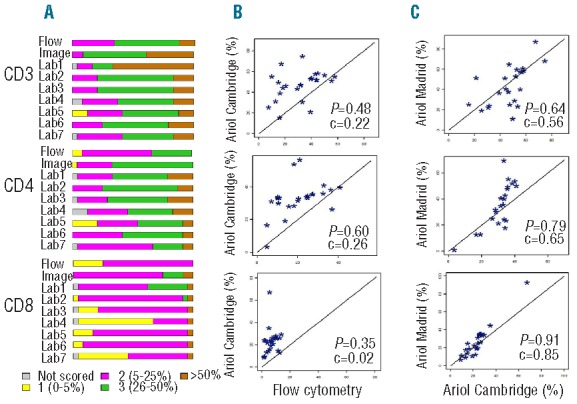
T-cell frequencies as analyzed by flow cytometry, image analysis and manual scoring. (A) Quantification of CD3, CD4 and CD8 by flow cytometry, automated microscopy and image analysis and by manual scoring in the 7 laboratories. Note that certain laboratories scored very different from others, resulting in high variability. However, for some markers these laboratories were in better agreement with flow cytometry and/or image analysis results. (B) Comparison of frequencies of CD3, CD4 and CD8 as analyzed by image analysis and flow cytometry. There is moderate correlation (p) but low concordance (c) between results obtained by the two methods. (C) Comparison of frequencies of CD3, CD4 and CD8 by two separate automated microscopes, Ariol “Cambridge” and Ariol “Madrid” show high correlation between the two instruments.
Table 4.
Agreement of manual scores among the laboratories. Agreement based on the free marginal kappa statistic is categorized as low agreement (<0.40), moderate agreement (0.40–0.75) and high agreement (>0.75).
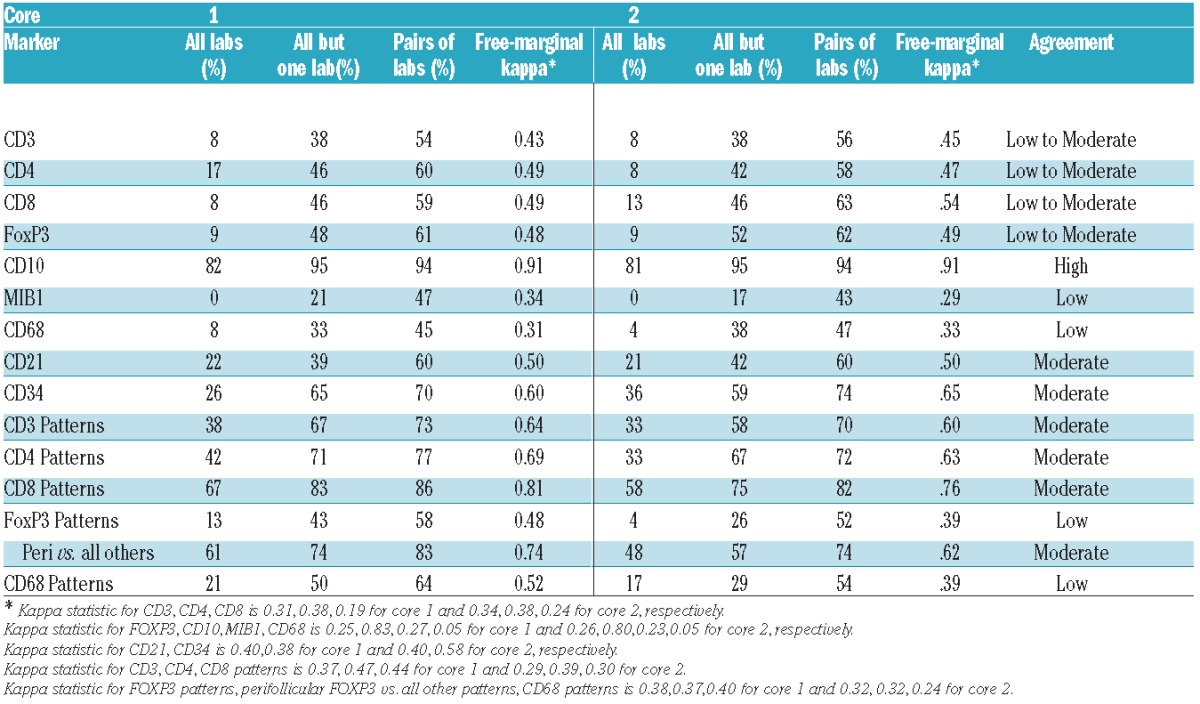
Figure 3.
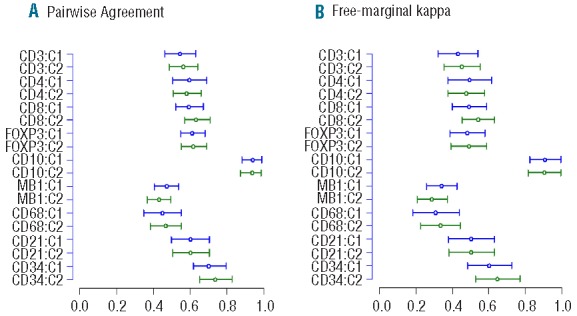
Agreement of manual scoring across labs (A) the pairwise agreement and 95% bootstrap confidence intervals and (B) the free marginal kappa statistic and 95% bootstrap confidence intervals. The cases coded as not scored are included as this indicates disagreement from the labs which scored these cases.
A comparison of manual scoring and flow cytometry (Tables 3 and 5) showed that the agreement on average was moderate for CD3 (a median of 50% of the core scores agree with flow cytometry across laboratories), low for CD4 (a median of 38% of the scores agree), and moderate to high for CD8 (a median of 71% of the scores agree) (Table 5) with variation in agreement observed by laboratory (Table 3). Thus, while certain laboratories agreed well with the flow cytometry results (e.g. Labs 4 and 6 for CD3) other laboratories did not (e.g. Lab 1). Disagreement tended to occur on the high end with manual scoring overestimating the frequencies of cells in the high range for CD3 and CD4 (Table 3). Specifically, among those misclassified, on average 70, 87 and 100% of the cases had manual scores higher than flow for CD3, CD4 and CD8, respectively (Table 5). As an example, for CD4, 0% of cases fell into the category of more than 50% by flow cytometry as compared to between 8% to 21% by manual scoring (Table 3). Thus, the manual scoring on average across laboratories tended to increase the variability and range of T-cell frequencies compared to flow cytometry. However, higher levels of agreement were observed for specific laboratories and markers (e.g. Lab 4 for CD3 and Labs 3, 5 and 6 for CD8), but no systematic trends were observed.
Table 5.
Summary statistics for the comparison of flow, image and manual scoring for T-cell densities for core 1. The median percent of cores and range within each category (as appropriate) is reported.
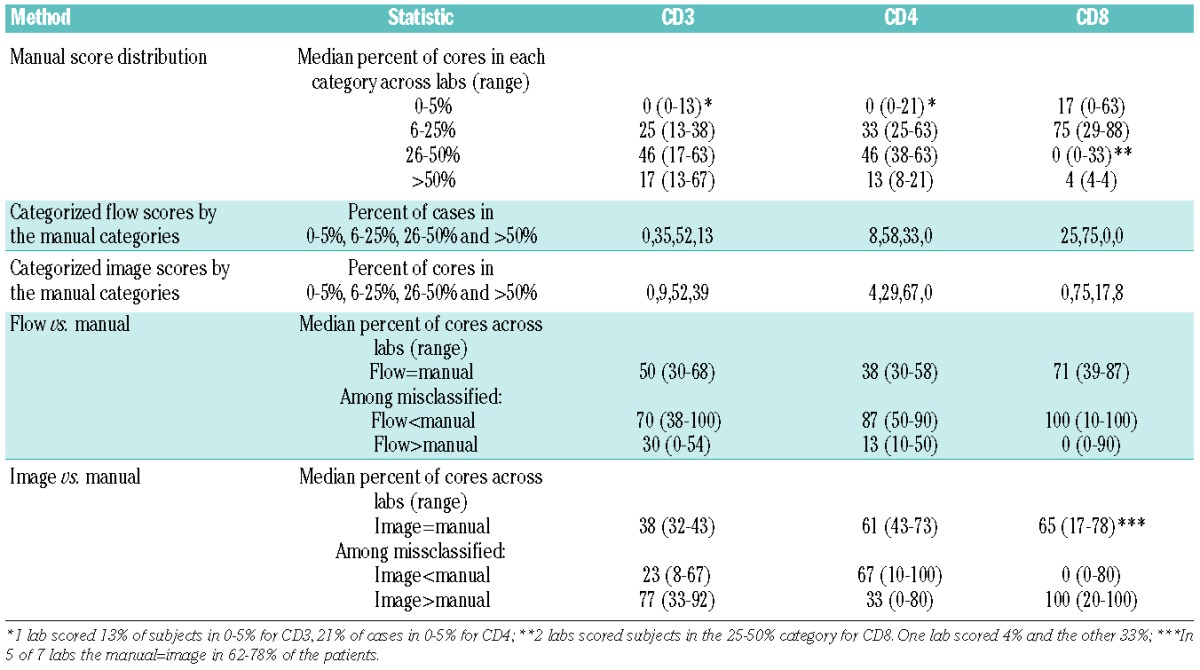
T-cell frequencies analyzed by an automated scanning microscope (Ariol SL-50 “Cambridge”) were expressed as percentages of cells positive out of all cells in the core. For CD3, CD4 and CD8, the median image score was 48, 30 and 20 for core 1 and 50, 34 and 24 for core 2, respectively (Figure 4). Comparison with the flow cytometry data demonstrated a moderate correlation between the results (Spearman correlation of 0.48, 0.60 and 0.35 for CD3, CD4 and CD8, respectively) (Figure 2B). The frequencies of T cells and T-cell subsets were higher by the automated scanning than by flow cytometry (Table 5), resulting in a low concordance between the two methods (<0.26).
Figure 4.
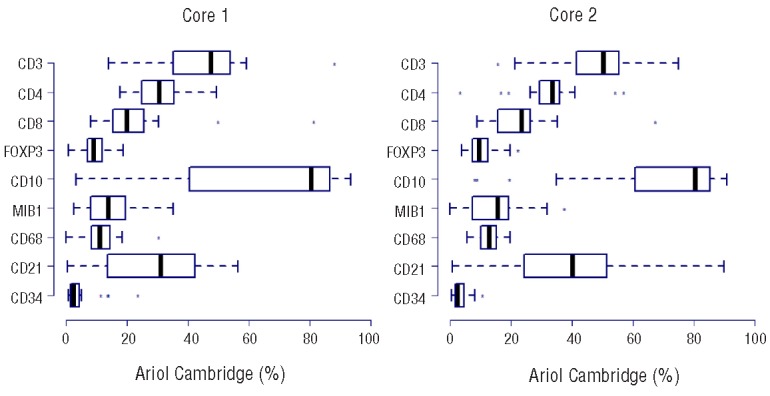
Distribution of the image scores of all investigated markers for core 1 and core 2, excluding the cores not scored.
Comparing manual and image scoring for core 1 resulted in low to moderate agreement on average in 38, 61 and 65% of the cases for CD3, CD4 and CD8, respectively (Table 5). The disagreement tended to occur in the low and high manual categories for CD3 with a higher percent of cases scoring 6–25% and lower percent scoring more than 50% by manual as compared to image (misclassification occurring with image scores higher than manual in 77% of misclassified cases on average), in the high manual categories for CD4 with a higher percent of cases scored more than 50% for manual as compared to image (misclassification occurring with image scores lower than manual in 67% of misclassified cases on average) and in the low manual categories for CD8 with a higher percent of cases scored in 0–5% for manual as compared to image (image scores higher than manual in 100% of the misclassified cases on average) (Table 5 and Figure 5). Similar results were obtained for core 2.
Figure 5.
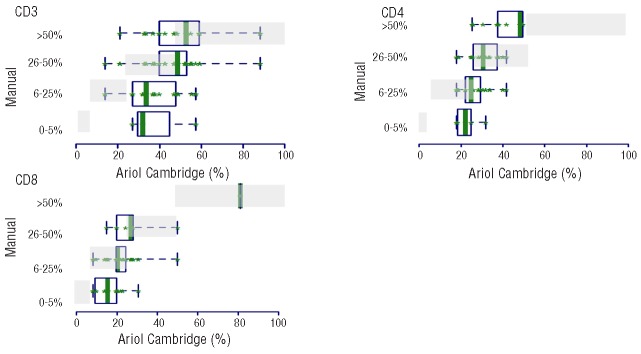
Distribution of manual versus automated microscopy scores for CD3, CD4 and CD8 in core 1. The automated microscopy values are represented on the x-axis and the manual score categories on the y-axis. The green boxes represent the image distribution of the cores within the corresponding categories to which the cores were manually scored. The gray shaded boxes indicate where the automated microscopy scores should have been if there was perfect agreement between the two scoring methods, and thus give us a pictorial view of missclassification. As can be seen, there is a discrepancy between manual scoring and automated microscopy in the low range for CD3 and CD8 and the higher range for CD3 and CD4.
High correlation was observed between the two image analysis instruments (Spearman correlation 0.64, 0.79 and 0.91 for CD3, CD4 and CD8, respectively) (Figure 2C). The concordance was moderate to high with estimates for core 1 of 0.39 (95%CI: 0.01, 0.69), 0.57 (95%CI: 0.30, 0.75), 0.83 (95%CI: 0.67, 0.92) for CD3, CD4 and CD8, respectively. For core 2, respectively, the estimated concordance was 0.56 (95%CI: 0.24, 0.77), 0.65 (95%CI: 0.48, 0.78) and 0.85 (95%CI: 0.75, 0.91).
Architectural patterns of T-cell infiltration
Next we investigated whether manual scoring could identify common and rare distribution patterns of T-cell subsets (Table 2). There was a moderate agreement in the scoring of CD3, CD4 and CD8 architectural patterns (Table 4). The majority of the cases were scored as interfollicular (Online Supplementary Tables S1 and S2) and, therefore, the agreement level indicates that the laboratories consistently identified the interfollicular pattern. The number of cases with diffuse or intrafollicular patterns was relatively low, limiting the ability to evaluate agreement for these patterns. However, one case had a predominantly intrafollicular distribution pattern for CD4 and all scoring laboratories correctly identified this case.
Densities and distribution of FOXP3 cells, representing nuclear staining in a sparse T-cell subset
Considerable variation (P<0.001) was observed in the FOXP3+ scoring across the 7 laboratories with 3 of them (Labs 4, 6 and 7) scoring a higher percentage of cases as 0–5% and Lab 1 scoring a higher percentage of cases as 5–25%, indicating the difficulty distinguishing between these two categories (Online Supplementary Tables S1, S2 and S3). Agreement among all 7 laboratories was 9%, among all but one laboratory was 48–52%, and average pairwise agreement of 61–62% resulting in a low to moderate agreement with free marginal kappa statistic value of 0.48–0.49 (Table 4). In the automated microscopy analysis, most cores were classified with less than 20% FOXP3+ cells (median 9%, range 1–19% for core 1; median 10%, range 4–22% for core 2) (Figure 4). Compared with image analysis, manual scoring misclassified many cases in the lower range resulting in manual scores being lower with 92% of misclassified cases on average (range of 50–100%) (Figure 6). In addition, a few cores in 1–2 laboratories were manually scored in the highest category (26–50%) while no such cores were identified by the automated microscopy analysis (Figure 6). Thus, the variation observed with the manual scores was greater than that observed with the automated microscopy analysis with misclassification mainly occurring in the lower range.
Figure 6.
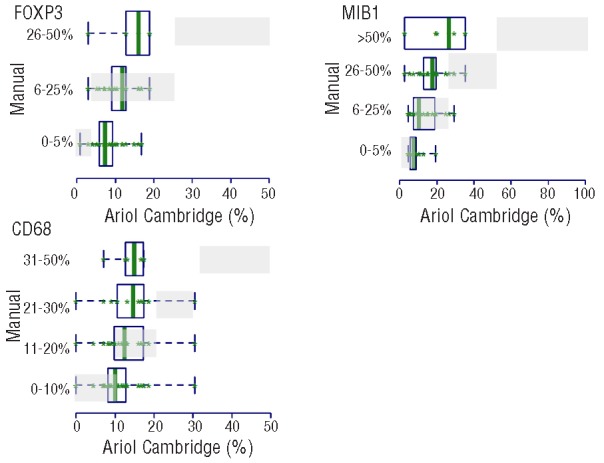
Distribution of manual versus automated microscopy scores for FOXP3, CD68 and MIB1 in core 1. The automated microscopy values are represented on the x-axis and the manual score categories on the y-axis. The green boxes represent the image distribution of the cores within the corresponding categories to which the cores were manually scored. The gray shaded boxes indicate where the automated microscopy scores should have been if there was perfect agreement between the two scoring methods, and thus give us a pictorial view of missclassification. Note that the manual scoring underestimated the frequencies of FOXP3 and MIB1 in the low range and overestimated the frequencies of MIB1 in the higher range.
Low agreement between the laboratories was observed for scoring the architectural pattern of FOXP3+ cells using 4 categories (Tables 2 and 4). Since the perifollicular pattern (Figure 7) is claimed to be discriminative as a prognostic marker,22,23 we evaluated agreement for this specific pattern relative to the three others combined and found that the characteristic perifollicular localization of FOXP3 was identified in all scoring laboratories except one, resulting in a moderate agreement (Table 4).
Figure 7.
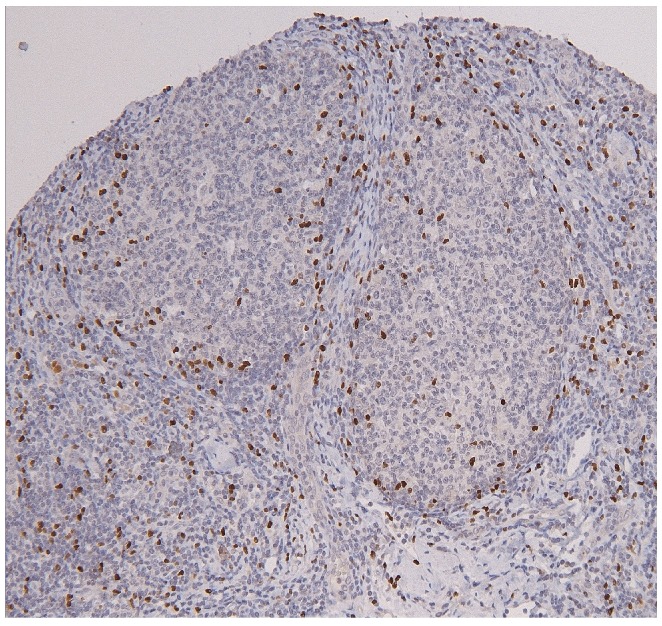
Perifollicular distribution of FOXP3 positive T cells.
Markers expressed on tumor cells: CD10 and MIB1 analyzed by manual scoring and automated microscopy
Most cores were classified as strongly positive and the reproducibility of the CD10 scores was high with no difference between the scoring laboratories from analysis of variance (P=0.9) and agreement in all laboratories of 81–82% (free marginal kappa of 0.91) indicating high agreement (Table 4 and Figure 3). For MIB1, the reproducibility of the manual scoring was suboptimal with no agreement between all 7 laboratories and 17–21% agreement between 6 of 7 laboratories, and average pairwise agreement of 43–47%, resulting in low agreement (free-marginal kappa of 0.29–0.34) (Table 4 and Figure 3). This result was influenced by two laboratories which systematically scored low (ANOVA, P<0.0001). However, these two laboratories were in better agreement with the automated analysis, while the others had a tendency to overestimate the variation with a higher percent of scores in the 0–5% and 26–50% and more than 50% categories. The distribution of the automated microscopy scores was narrower than appreciated with the manual scoring (Figure 4), with a median of 14% and range of 3–35% for core 1 and a median of 16% and range of 0–38% for core 2. For agreement, the manual scores showed a systematic underestimation of the percentages of MIB1+ cells in the low range as compared to the automated scores (manual scores in the 0–5% interval had image values higher than 5%) and an overestimation in the higher range as reflected by the fact that 4% of cases were scored by image as more than 26% while 51% on average (range 4–71%) were scored manually as more than 26% (Figure 6 and Online Supplementary Table S3).
Scoring of frequencies and architectural patterns of cells present in the microenvironment of follicular lymphoma
In spite of the fact that the majority of CD68+ macrophage scores were in the lowest category (Figure 1), there was variation (ANOVA, P<0.0001) among scoring laboratories with 3 laboratories using at least 3 of the 4 categories and the other laboratories with most cases scored as 0–10% resulting in low agreement with pairwise agreement of 45–47% and free marginal kappa of 0.31–0.33 (Table 4). Automated microscopy analysis showed a narrow distribution with a median of 11% (range 0–30) for core 1 and 6% (range 6–20) for core 2 (Figure 4). The agreement of CD68 patterns between laboratories was low (Table 4).
Stainings of CD21 and CD34 represent a different category of markers highlighting the intricate pattern of the follicular dendritic cell network and the microvessel density, respectively. For both of these markers the manual scoring showed a moderate agreement between laboratories with pairwise agreement of 60–74% and free marginal kappa statistics of 0.50–0.65 (Table 4). For CD34, the distribution differed in the two cores with a higher frequency of scores in the category “sparse” in core 1 compared to core 2 (Figure 1).
Discussion
The FL tumor microenvironment contains a variable proportion of non-malignant cells that contribute to the tumor cell survival and are important contributors to FL biology.24–31 Gene expression studies have demonstrated that patient survival, risk for transformation and response to therapy are associated with activation of genes reflecting non-malignant cell content and activation, rather than genes expressed by the tumor cells.9,10,32 It is hypothesized that the frequency and distribution of non-malignant cells can in part serve as surrogate markers for the clinically relevant gene expression signatures. This issue has recently been the focus of numerous immunohistochemical studies, including evaluating markers for T-helper, cytotoxic and regulatory cell subsets,17–21,23,33–35 for stromal cells including macrophages, mast cells and dendritic cells,15,20,33,36–39 and for endothelial cells assessing microvessel density.40–42 The results of these studies are often contradictory, which in part can be explained by the small number of patients and variations in patient selection and treatment protocol.11,43 Another potential reason is the scoring reproducibility of the immunohistochemical markers. It could well be that evaluating immunohistochemical markers by manual scoring are reproducible within a study where scoring is carried out by one individual but is not reproducible over a series of studies where other individuals score other lymphoma cohorts. In addition, the manual scoring may be more reproducible once a cut-off point of clinical relevance based on a larger series of patients has been established. However, in the development of prognostic and/or predictive markers, which often have the aim of determining the optimal cut-off point to separate populations into low or high risk for failure, these markers are scored in 4–5 categories for most markers. The data in this study show that with 4–5 categories, the manual scoring agreement is low to moderate. Therefore, in the development of prognostic and/or predictive markers (and as stratification for randomized clinical trials) it is important to centralize and standardize marker scoring, or consider alternative approaches to manual scoring, to minimize the variation and increase the power. Once a cut-off point has been established, it is likely that the manual scoring agreement will increase across scoring laboratories. The main purpose of this study was to investigate the feasibility and reproducibility of scoring common FL microenvironment markers by comparing manual scoring, automated microscopy and, for T-cell markers, also flow cytometry. We focused on quantification and architectural localization of cell types considered to be important for FL biology and outcome.
The main conclusions from this study are: 1) T-cell frequencies are highly variable in the lymphoma tissue and the reproducibility of manual quantification of T cells is low to moderate; 2) T-cell frequencies as analyzed by flow cytometry and automated microscopy show moderate agreement but image analysis overestimates frequencies; 3) for many markers, manual scoring increases the variability in the low and high frequency interval as compared to automated microscopy; 4) automated microscopy analyses for T-cell markers by two different instruments shows a high degree of correlation; 5) manual scoring can readily identify rare architectural patterns of T-cell infiltrates in FL tissue.
T-cell frequencies measured by flow cytometry have previously been suggested to predict outcome in FL.21,34 In the present study, we used flow cytometry data for comparison with T-cell frequencies obtained by manual and automated scoring of immunohistochemistry. Flow cytometry was considered to be the “gold standard” since it measures and quantifies a high number of cells as single events and since multiple antigens and CD markers are used to define a population resulting in high accuracy.44 Flow cytometry is thus a very reliable and quantitative method for comparing and enumerating frequencies of numerous and rare cell populations and it can be standardized between laboratories. Flow cytometry can also be used for detecting intracellular markers, such as BCL2 and FOXP3, in combination with surface markers, allowing detailed information on cell subsets to be gathered. The major disadvantages with this method is that it requires fresh tissue and does not provide information on the cellular localization, which is of relevance in FL. Furthermore, in case of fibrosis, or when tumor cells are large and fragile, the cell preparation used for flow cytometry might not fully represent the cell composition in the tissue.
A comparison of flow cytometry with image analysis showed there was a moderate correlation between CD3, CD4 and CD8 positive T-cell frequencies similar to previously published results.20 However, both in the present study and in the study by Wahlin et al. examining T cells in FL,20 image analysis resulted in higher values for the T-cell markers compared to flow cytometry, resulting in a low concordance between the measurements. One possible explanation for this difference could be cell loss during preparation of the single cell suspension used for flow cytometry,44 although this is less likely since preferentially large cells are lost. Another possibility for the discrepancy between flow cytometry and image analysis is the presence of incomplete cells in the tissue sections. It could also be a “calibration effect” caused by a lower detection threshold in the image analysis. The low concordance between flow cytometry and image analysis does not, in our opinion, rule out using image analysis to reduce scoring variability and to obtain objective quantification since the effects will be similar across all cases. An advantage in using automated microscopy or flow cytometry compared to manual scoring is that quantification is in a continuous scale rather than a categorical measurement, providing more information when evaluating biologically relevant cut-off points.
For several markers, the manual scoring overestimated the variation in cell frequencies as compared to flow cytometry and automated microscopy. On the other hand, while the variation across laboratories was considerable, certain laboratories agreed well with each other and with flow cytometry results. However, this was not consistent but differed between markers and laboratories, with no laboratory identified as uniformly reliable. Studies using immunohistochemical scoring should be carefully validated by flow cytometry or image analysis to ensure reproducibility.
The analysis evaluating the reproducibility of T-cell marker quantification using two separate automated microscopes in combination with image analysis performed by 2 different instrument operators, showed a high correlation in scoring CD3, CD4 and CD8 densities. These results suggest that it is possible to reproducibly score T-cell markers in FL using image analysis. Image scoring also enumerates cells as actual values, an obvious advantage over manual scoring according to category. The frequency and localization of FOXP3+ Treg cells has been associated with clinical behavior in FL.20,22,23 For CD4+ T cells, one study has suggested that a high number of intrafollicular CD4+ cells is associated with a poor prognosis.20 In the present validation study, certain architectural patterns were highly reproducible, such as localization of CD8+ T cells, while agreement was moderate for localization of CD3+ and CD4+ T cells, as well of CD68+ macrophages. Importantly, manual scoring did well when it came to recognizing rare patterns, as exemplified by the perifollicular localization of FOXP3+ regulatory T cells and intrafollicular localization of CD4 positive T cells.
This validation study provides explanations for the heterogeneous results of various reports in the literature on the prognostic value of the microenvironment in FL. Based on these results, we recommend refraining from manual scoring for research purposes in this field unless careful validation by other methods is carried out, and recommend the use of a more objective measurement, such as computer-assisted scoring.
Acknowledgments
The Lunenburg Lymphoma Biomarker Consortium is a collaboration of 9 international lymphoma collaborative groups, each represented by a clinical investigator and one or more hematopathologists and supported by a team of statisticians. The project has been initiated and is financially supported by the van Vlissingen Lymphoma Foundation. In addition, unrestricted grants were received from Genentech/Roche, GlaxoSmithKline, Pfizer Pharma, Teva Pharmaceuticals/Cephalon and Millenium Pharmaceuticals Inc. EORTC Lymphoma group: Daphne de Jong, John Raemaekers; HOVON: Daphne de Jong, Marie José Kersten, Anton Hagenbeek; LYSA (GELA): Philippe Gaulard, Thierry Molina, Josette Briere, Gilles Salles; British Columbia Cancer Center: Randy Gascoyne, Laurie Sehn; ECOG: Randy Gascoyne; DSHBHL (German High Grade non-Hodgkin Lymphoma Group): Christoph Thorns, Andreas Rosenwald, Wolfram Klapper, German Ott, Sylvia Hoeller, Heinz-Wolfram Bernd, Michael Pfreundschuh; NLG: Birgitta Sander, Eva Kimby, Björn E. Wahlin; Barts Cancer Institute: Abigail Lee, Andrew Clear, Maria Calaminici, John Gribben, Andrew Lister; Dana-Farber Cancer Institute, Boston, USA: Edie Weller, Wanling Xie; Independent pathology advisor: Elias Campo, Barcelona, Spain.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Pulte D, Jansen L, Gondos A, Emrich K, Holleczek B, Katalinic A, et al. Survival of patients with non-Hodgkin lymphoma in Germany in the early 21st century. Leuk Lymphoma. 2012;54(5):979–85 [DOI] [PubMed] [Google Scholar]
- 2.Pulte D, Gondos A, Brenner H. Expected long-term survival of older patients diagnosed with non-Hodgkin lymphoma in 2008–2012. Cancer Epidemiol. 2012;36(1):e19–25 [DOI] [PubMed] [Google Scholar]
- 3.Horning SJ, Rosenberg SA. The natural history of initially untreated low-grade non-Hodgkin’s lymphomas. N Engl J Med. 1984;311(23):1471–5 [DOI] [PubMed] [Google Scholar]
- 4.Rohatiner AZ, Lister TA. The clinical course of follicular lymphoma. Best Pract Res Clin Haematol. 2005;18(1):1–10 [DOI] [PubMed] [Google Scholar]
- 5.Solal-Celigny P, Cahu X, Cartron G. Follicular lymphoma prognostic factors in the modern era: what is clinically meaningful? Int J Hematol. 2010;92(2):246–54 [DOI] [PubMed] [Google Scholar]
- 6.Relander T, Johnson NA, Farinha P, Connors JM, Sehn LH, Gascoyne RD. Prognostic factors in follicular lymphoma. J Clin Oncol. 2010;28(17):2902–13 [DOI] [PubMed] [Google Scholar]
- 7.Solal-Celigny P, Roy P, Colombat P, White J, Armitage JO, Arranz-Saez R, et al. Follicular lymphoma international prognostic index. Blood. 2004;104(5):1258–65 [DOI] [PubMed] [Google Scholar]
- 8.Federico M, Bellei M, Marcheselli L, Luminari S, Lopez-Guillermo A, Vitolo U, et al. Follicular lymphoma international prognostic index 2: a new prognostic index for follicular lymphoma developed by the international follicular lymphoma prognostic factor project. J Clin Oncol. 2009;27(27):4555–62 [DOI] [PubMed] [Google Scholar]
- 9.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159–69 [DOI] [PubMed] [Google Scholar]
- 10.Glas AM, Knoops L, Delahaye L, Kersten MJ, Kibbelaar RE, Wessels LA, et al. Geneexpression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007;25(4):390–8 [DOI] [PubMed] [Google Scholar]
- 11.de Jong D, Fest T. The microenvironment in follicular lymphoma. Best Pract Res Clin Haematol. 2011;24(2):135–46 [DOI] [PubMed] [Google Scholar]
- 12.de Jong D, Rosenwald A, Chhanabhai M, Gaulard P, Klapper W, Lee A, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications–a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2007;25(7):805–12 [DOI] [PubMed] [Google Scholar]
- 13.Perez EA, Suman VJ, Davidson NE, Martino S, Kaufman PA, Lingle WL, et al. HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol. 2006;24(19):3032–8 [DOI] [PubMed] [Google Scholar]
- 14.Ruschoff J, Kerr KM, Grote HJ, Middel P, von Heydebreck A, Alves VA, et al. Reproducibility of Immunohistochemical Scoring for Epidermal Growth Factor Receptor Expression in Non-Small Cell Lung Cancer. Arch Pathol Lab Med. 2013;137(9):1255–61 [DOI] [PubMed] [Google Scholar]
- 15.Farinha P, Masoudi H, Skinnider BF, Shumansky K, Spinelli JJ, Gill K, et al. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood. 2005;106(6):2169–74 [DOI] [PubMed] [Google Scholar]
- 16.de Jong D, Koster A, Hagenbeek A, Raemaekers J, Veldhuizen D, Heisterkamp S, et al. Impact of the tumor microenvironment on prognosis in follicular lymphoma is dependent on specific treatment protocols. Haematologica. 2009;94(1):70–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreras J, Lopez-Guillermo A, Roncador G, Villamor N, Colomo L, Martinez A, et al. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol. 2009;27(9):1470–6 [DOI] [PubMed] [Google Scholar]
- 18.Alvaro T, Lejeune M, Salvado MT, Lopez C, Jaen J, Bosch R, et al. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J Clin Oncol. 2006;24(34):5350–7 [DOI] [PubMed] [Google Scholar]
- 19.Carreras J, Lopez-Guillermo A, Fox BC, Colomo L, Martinez A, Roncador G, et al. High numbers of tumor-infiltrating FOXP3-positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood. 2006;108(9):2957–64 [DOI] [PubMed] [Google Scholar]
- 20.Wahlin BE, Aggarwal M, Montes-Moreno S, Gonzalez LF, Roncador G, Sanchez-Verde L, et al. A unifying microenvironment model in follicular lymphoma: outcome is predicted by programmed death-1–positive, regulatory, cytotoxic, and helper T cells and macrophages. Clin Cancer Res. 2010;16(2):637–50 [DOI] [PubMed] [Google Scholar]
- 21.Wahlin BE, Sundstrom C, Holte H, Hagberg H, Erlanson M, Nilsson-Ehle H, et al. T cells in tumors and blood predict outcome in follicular lymphoma treated with rituximab. Clin Cancer Res. 2011;17(12):4136–44 [DOI] [PubMed] [Google Scholar]
- 22.Farinha P, Al-Tourah A, Gill K, Klasa R, Connors JM, Gascoyne RD. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood. 2010;115(2):289–95 [DOI] [PubMed] [Google Scholar]
- 23.Lee AM, Clear AJ, Calaminici M, Davies AJ, Jordan S, MacDougall F, et al. Number of CD4+ cells and location of forkhead box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J Clin Oncol. 2006;24(31):5052–9 [DOI] [PubMed] [Google Scholar]
- 24.Carbone A, Gloghini A, Gruss HJ, Pinto A. CD40 ligand is constitutively expressed in a subset of T cell lymphomas and on the microenvironmental reactive T cells of follicular lymphomas and Hodgkin’s disease. Am J Pathol. 1995;147(4):912–22 [PMC free article] [PubMed] [Google Scholar]
- 25.Ghia P, Boussiotis VA, Schultze JL, Cardoso AA, Dorfman DM, Gribben JG, et al. Unbalanced expression of bcl-2 family proteins in follicular lymphoma: contribution of CD40 signaling in promoting survival. Blood. 1998;91(1):244–51 [PubMed] [Google Scholar]
- 26.Buske C, Twiling A, Gogowski G, Schreiber K, Feuring-Buske M, Wulf GG, et al. In vitro activation of low-grade non-Hodgkin’s lymphoma by murine fibroblasts, IL-4, anti-CD40 antibodies and the soluble CD40 ligand. Leukemia 1997;11(11):1862–7 [DOI] [PubMed] [Google Scholar]
- 27.Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK, et al. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci USA. 2010;107(29):12747–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epron G, Ame-Thomas P, Le Priol J, Pangault C, Dulong J, Lamy T, et al. Monocytes and T cells cooperate to favor normal and follicular lymphoma B-cell growth: role of IL-15 and CD40L signaling. Leukemia 2012;26(1):139–48 [DOI] [PubMed] [Google Scholar]
- 29.Johnson PW, Watt SM, Betts DR, Davies D, Jordan S, Norton AJ, et al. Isolated follicular lymphoma cells are resistant to apoptosis and can be grown in vitro in the CD40/stromal cell system. Blood. 1993;82(6):1848–57 [PubMed] [Google Scholar]
- 30.Schmitter D, Koss M, Niederer E, Stahel RA, Pichert G. T-cell derived cytokines co-stimulate proliferation of CD40-activated germinal centre as well as follicular lymphoma cells. Hematol Oncol. 1997;15(4):197–207 [DOI] [PubMed] [Google Scholar]
- 31.Travert M, Ame-Thomas P, Pangault C, Morizot A, Micheau O, Semana G, et al. CD40 ligand protects from TRAIL-induced apoptosis in follicular lymphomas through NF-kappaB activation and up-regulation of c-FLIP and Bcl-xL. J Immunol. 2008;181(2):1001–11 [DOI] [PubMed] [Google Scholar]
- 32.Bohen SP, Troyanskaya OG, Alter O, Warnke R, Botstein D, Brown PO, et al. Variation in gene expression patterns in follicular lymphoma and the response to rituximab. Proc Natl Acad Sci USA. 2003;100(4):1926–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley T, Beck R, Absi A, Jin T, Pohlman B, Hsi E. Biologic predictors in follicular lymphoma: importance of markers of immune response. Leuk Lymphoma. 2007;48(12):2403–11 [DOI] [PubMed] [Google Scholar]
- 34.Wahlin BE, Sander B, Christensson B, Kimby E. CD8+ T-cell content in diagnostic lymph nodes measured by flow cytometry is a predictor of survival in follicular lymphoma. Clin Cancer Res. 2007;13(2 Pt 1):388–97 [DOI] [PubMed] [Google Scholar]
- 35.Sweetenham JW, Goldman B, LeBlanc ML, Cook JR, Tubbs RR, Press OW, et al. Prognostic value of regulatory T cells, lymphoma-associated macrophages, and MUM-1 expression in follicular lymphoma treated before and after the introduction of monoclonal antibody therapy: a Southwest Oncology Group Study. Ann Oncol. 2010;21(6):1196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klapper W, Hoster E, Rolver L, Schrader C, Janssen D, Tiemann M, et al. Tumor sclerosis but not cell proliferation or malignancy grade is a prognostic marker in advanced-stage follicular lymphoma: the German Low Grade Lymphoma Study Group. J Clin Oncol. 2007;25(22):3330–6 [DOI] [PubMed] [Google Scholar]
- 37.Taskinen M, Karjalainen-Lindsberg ML, Leppa S. Prognostic influence of tumor-infiltrating mast cells in patients with follicular lymphoma treated with rituximab and CHOP. Blood. 2008;111(9):4664–7 [DOI] [PubMed] [Google Scholar]
- 38.Taskinen M, Karjalainen-Lindsberg ML, Nyman H, Eerola LM, Leppa S. A high tumor-associated macrophage content predicts favorable outcome in follicular lymphoma patients treated with rituximab and cyclophosphamide-doxorubicin-vincristine-prednisone. Clin Cancer Res. 2007;13(19):5784–9 [DOI] [PubMed] [Google Scholar]
- 39.Alvaro T, Lejeune M, Camacho FI, Salvado MT, Sanchez L, Garcia JF, et al. The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma. Haematologica. 2006;91(12):1605–12 [PubMed] [Google Scholar]
- 40.Farinha P, Kyle AH, Minchinton AI, Connors JM, Karsan A, Gascoyne RD. Vascularization predicts overall survival and risk of transformation in follicular lymphoma. Haematologica. 2010;95(12):2157–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taskinen M, Jantunen E, Kosma VM, Bono P, Karjalainen-Lindsberg ML, Leppa S. Prognostic impact of CD31-positive microvessel density in follicular lymphoma patients treated with immunochemotherapy. Eur J Cancer. 2010;46(13):2506–12 [DOI] [PubMed] [Google Scholar]
- 42.Clear AJ, Lee AM, Calaminici M, Ramsay AG, Morris KJ, Hallam S, et al. Increased angiogenic sprouting in poor prognosis FL is associated with elevated numbers of CD163+ macrophages within the immediate sprouting microenvironment. Blood. 2010;115(24):5053–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leich E, Ott G, Rosenwald A. Pathology, pathogenesis and molecular genetics of follicular NHL. Best Pract Res Clin Haematol. 2011;24(2):95–109 [DOI] [PubMed] [Google Scholar]
- 44.Craig FE, Foon KA. Flow cytometric immunophenotyping for hematologic neoplasms. Blood. 2008;111(8):3941–67 [DOI] [PubMed] [Google Scholar]


