Erythropoietin (Epo) is the main humoral regulator of erythropoiesis. During development, Epo production switches in a species-specific manner from the liver to the kidneys which account for approximately 90% of total Epo synthesis in the adult. Renal Epo is produced by peritubular interstitial cells with fibroblastic and neuronal features, located in the juxtamedullary cortex.1–4 These cells respond to a decrease in tissue oxygen partial pressure by hypoxia-inducible transcription factor-2 (HIF-2)-dependent induction of Epo synthesis.5 DNA sequences required for oxygen-dependent regulation of the EPO gene are different in liver and kidney. Under physiological conditions, EPO gene expression is controlled by elements located within 0.4 kb of the 5′ and 0.7 kb of the 3′-flanking region in the liver, while an essential regulatory element for renal Epo expression resides between −14 and −6 kb in the distal 5′-region.6,7 The liver-inducible element consists of a proximal downstream enhancer which synergizes with the minimal promoter region to achieve up to 100-fold transcriptional induction.8 Detailed analysis of this conserved 50 bp 3′-enhancer revealed a tripartite cis-regulatory structure with a consensus HIF binding site (HBS, ACGTG) and a CACA repeat downstream of the HBS.9 The latter element is necessary, but not sufficient, for hypoxia inducibility of the 3′-enhancer. However, so far no protein binding to this element has been defined. While the Epo 3′-enhancer is well characterized and has been confirmed to be both essential and sufficient for liver-specific Epo expression in mice beyond embryonic day 14.5,10 the kidney-inducible element (KIE) remains unexplored. In the present study, we functionally analyzed a distal upstream hypoxia response element (HRE) which confers oxygen-regulated Epo transcription and presumably represents the hitherto uncharacterized KIE.
We used the ENCODE data integrated in the UCSC Genome Browser (http://genome.ucsc.edu/) to identify a distal conserved putative HBS in the Epo 5′-region, located within a DNase I hypersensitivity site (DSS) observed in 9 different non-Epo-producing cell lines and strongly conserved in multiple vertebrate species (Figure 1A). This distant locus, 9248 bp upstream of the Epo transcriptional start site, contains a putative HBS and is flanked by the most frequently neighboring residues observed around the consensus HBS11 and a CACA repeat 8 bp upstream, very similar to the established 3′-HRE. The striking overlap of this region with a previously reported anemia-inducible DSS in liver and kidney of anemic Epo transgenic mice6 prompted us to perform a detailed functional analysis. Two Epo-expressing (Hep3B and Kelly, Figure 1B) and two non-expressing (HeLa and HK-2) human cell lines were transfected with firefly luciferase reporter gene constructs under the control of the exogenous SV40 promoter and a 100 bp fragment encompassing the novel candidate 5′-HRE, containing either wild-type or mutant HBS and CACA elements. The isolated distal 5′-HRE conferred robust hypoxic reporter gene induction in all cell lines tested (Figure 1C), which was abrogated upon mutation of either the HBS or the CACA sequence (Figure 1D).
Figure 1.
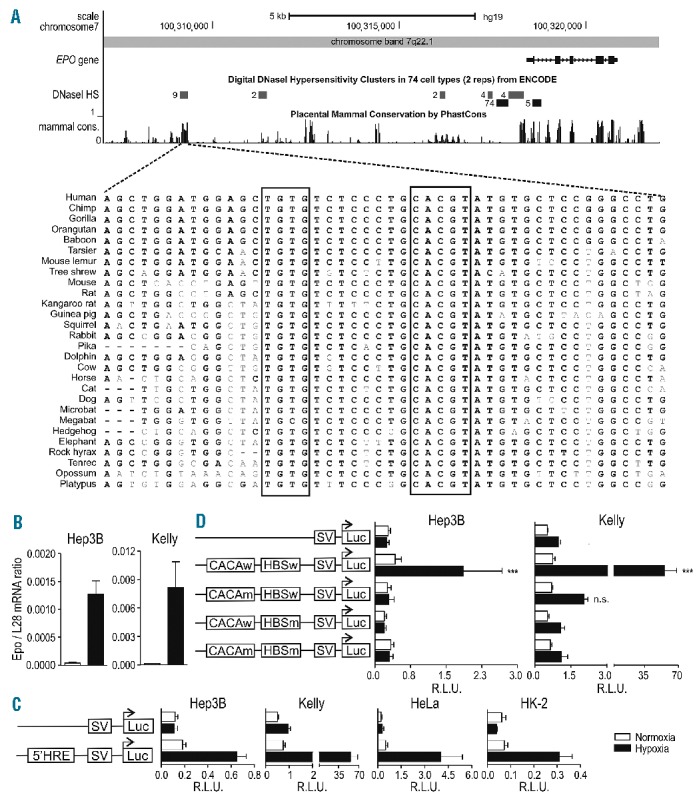
Location of a functional distal HRE in the Epo 5′ regulatory region. (A) UCSC Genome Browser output (hg19) of the Epo genomic and 5′ upstream region. Shown are the ENCODE DSS clusters and mammalian PhastCons conservation tracks with a closer view of the region in 28 vertebrates extracted using the 46-MULTIZ whole-genome multiple alignment algorithm. (B) Epo mRNA levels in human Hep3B and Kelly cells were measured by RT-qPCR and normalized to ribosomal protein L28 mRNA levels. (C) Both cell types (3×105 cells) were co-transfected with the indicated SV40-driven firefly luciferase reporter gene plasmids (500 ng) and a Renilla luciferase control plasmid (5 ng, Promega) in a 6-well format by polyethylenimine (PEI). 24 h post-transfection, cells were incubated for another 24 h under normoxic or hypoxic (0.2% O2) conditions. Luciferase activities of triplicate wells were determined using the Dual Luciferase Reporter Assay System according to the manufacturer’s protocol (Promega). All results are displayed as ratios of firefly to Renilla relative light units (R.L.U.). (D) Cells were co-transfected with pGL3p Epo 5′-HRE wild-type (w), pGL3p Epo HBS mutated (m) (ACGTG to AAAAG), pGL3p Epo CACA mutated (CACA to AAAA) or pGL3p Epo HBSm, CACAm together with a Renilla luciferase control plasmid, and reporter gene experiments performed as in D. (B–D) All data are expressed as mean ± SEM of 3 independent experiments and statistical analyses were performed with one-way ANOVA and Tukey correction for multiple comparisons (*P<0.05; **P<0.01; ***P<0.001).
We next combined the endogenous Epo minimal promoter (117 bp) with the distal 5′-HRE of either 100 bp, 300 bp (covering the entire conserved DSS region), or 3 kb length as indicated in Figure 2A. All three constructs, but not the Epo minimal promoter alone, increased reporter gene activity in hypoxic Hep3B cells (Figure 2B). No further induction was observed by inclusion of the 3′-HRE (126 bp), suggesting that, at least under our experimental conditions, the 5′-HRE and the 3′-HRE do not cooperate. Because the 3 kb containing fragment did not further increase the hypoxic response, it was excluded from later analyses.
Figure 2.
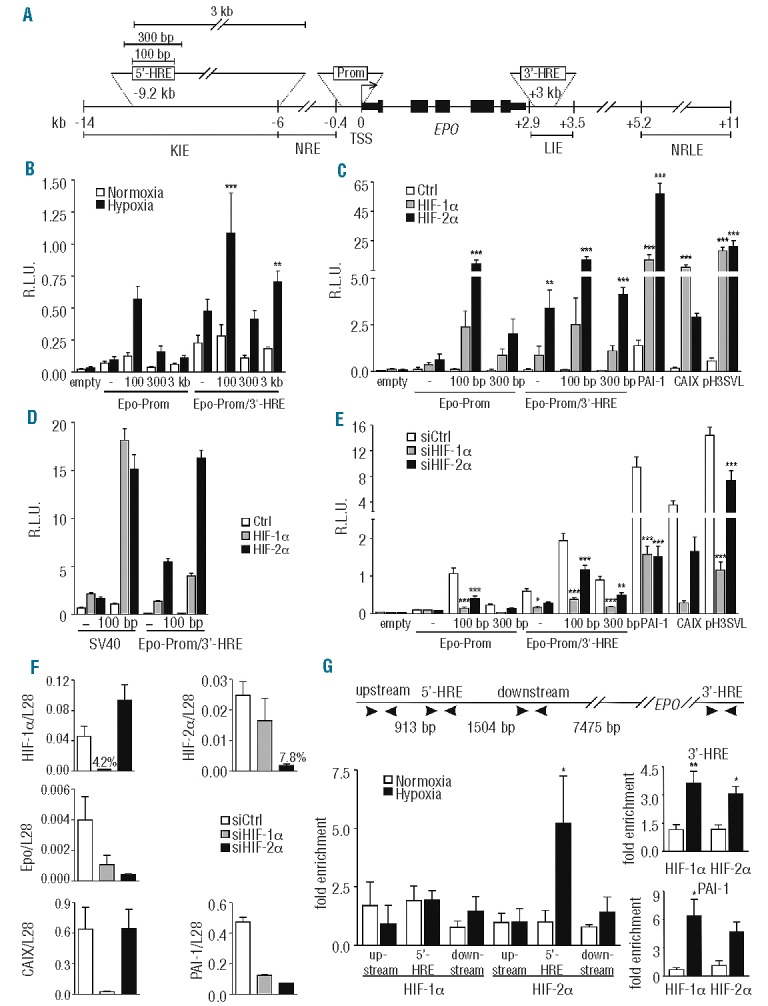
HIF-2α dependent hypoxic regulation of the Epo distal enhancer element. (A) Scheme depicting the constructs used. The numbers are relative to the transcriptional start site (TSS). KIE: kidney-inducible element; NRE: negative regulatory element; LIE: liver-inducible element; NRLE: negative regulatory liver element. (B) Dual luciferase analyses using the Epo minimal promoter (117 bp) and several indicated extensions of the distal 5′-HRE alone or in combination with the 3′-HRE. All reporter gene experiments were performed as in Figure 1. (C) The indicated vectors were co-transfected with HIF-α or empty expression vectors into Hep3B cells. CAIX (carbonic anhydrase IX) and PAI-1 (plasminogen activator inhibitor 1) promoter constructs were used as controls for HIF-1α and HIF-2α specificity, respectively. pH3SVL is controlled by a concatamerized non-HIFα isoform specific HRE and an SV40 promoter. (D) Comparison between SV40-driven and Epo promoter-driven luciferase expression, following HIFα overexpression in Hep3B cells. (E) Effect of siRNA-mediated HIFα knockdown on the indicated vectors in Hep3B cells. (F) HIF-1α, HIF-2α, Epo, PAI-1 and CAIX mRNA levels were measured by RT-qPCR and normalized to ribosomal protein L28 mRNA levels. (G) Scheme depicting the different primer regions used for ChIP experiments. ChIP of normoxic or hypoxic (0.2% O2, 24 h) Hep3B cells using antibodies directed against HIF-1α or HIF-2α. The amount of co-precipitated chromatin derived from the human Epo 5′ regulatory region, its upstream and downstream regions, and the Epo 3′-HRE was determined by qPCR. The PAI-1 promoter was used as an additional positive control. Mean values ± SEM of 3 (B–F) or 4 (G) independent experiments are shown and statistical analyses were performed with one-way ANOVA and Tukey correction for multiple comparisons (*P<0.05; **P<0.01; ***P<0.001).
To analyze HIFα isoform transcriptional selectivity, reporter gene constructs were co-transfected with constitutively active (double proline mutant) HIF-1α or HIF-2α over-expressing plasmids. Reporter assays using the PAI-1 and CAIX promoters, as well as pH3SVL, served as controls for HIFα overexpression. HIF-2α enhanced transactivation of the Epo promoter substantially better than HIF-1α (Figure 2C), and this is in line with the previously reported dominant role of HIF-2α in hypoxia-inducible Epo transcription in vitro.12 Interestingly, no HIFα isoform specificity was observed in reporter genes driven by the heterologous SV40 promoter (Figure 2D), indicating a crucial role of the endogenous Epo minimal promoter in HIFα isoform selectivity. Reporter gene assays were repeated in siRNA-mediated HIFα-deficient Hep3B cells, as described before.13 Both HIF-1α and HIF-2α siRNA reduced 5′-HRE dependent luciferase activity in hypoxic Hep3B cells (Figure 2E). Rather unexpectedly, both Epo 5′ and 3′-HRE dependent luciferase activity was more affected by depletion of HIF-1α than HIF-2α. This might be partially explained by less efficient knockdown of HIF-2α, as reflected by the PAI-1 and pH3SVL reporter activity and HIF-2α mRNA expression levels (Figure 2E and F). Alternatively, knockdown of HIF-1α might decrease hypoxic PHD3 induction, which has been shown to preferentially target HIF-2α.14 Notably, hypoxia-inducible Epo mRNA expression was not entirely HIF-2-dependent in hypoxic Hep3B cells (Figure 2F).
To analyze HIFα DNA binding to the endogenous sites, ChIP assays were performed using either anti-HIF-1α (5 μg, Ab2185; Abcam) or anti-HIF-2α (5 μg, Ab199; Abcam) antibodies in hypoxically-treated Hep3B cells, as described before.13 The PAI-1 and Epo 3′-HRE were analyzed as positive controls, and regions 1 kb upstream and downstream of the 5′-HRE served as negative controls. ChIP analysis revealed hypoxia-inducible HIF-2α, but not HIF-1α enrichment at the 5′-HRE or at any other of the tested 5′ loci (Figure 2G, left panel). In contrast, both HIF-1α and HIF-2α bound to the Epo 3′-HRE and the known PAI-1 HRE (Figure 2G, right panel).
Collectively, our results demonstrate the identification of a functional HIF-2α dependent distal 5′-HRE regulating Epo transcription. The lack of a renal cell culture system capable of hypoxia-inducible Epo expression precluded further validation of the kidney-specificity of this 5′-HRE. There are likely to be additional distal elements contributing to kidney-specific Epo expression because a BAC covering more than 180 kb flanking Epo regulatory regions is required to fully recapitulate endogenous Epo expression in transgenic mice.2,15 Our study is the first to functionally analyze the enigmatic KIE, and provides the basis for further investigations on the cooperation between other putative distal and proximal regions in regulating kidney-specific inducible Epo expression.
Acknowledgments
The authors would like to thank Clemens Cohen for providing HK-2 cells and Patrick Spielmann for expert technical assistance. RHW and DH acknowledge support from the NCCR Kidney. CH, funded by the SNF and from COST Action TD0901 HypoxiaNet. IAR is supported by EU FP7 grant agreement n° 246539. JF and CD acknowledge support from the DFG (GRK1431/2; DA 484/3-1).
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Maxwell PH, Osmond MK, Pugh CW, Heryet A, Nicholls LG, Tan CC, et al. Identification of the renal erythropoietin-producing cells using transgenic mice. Kidney Int. 1993;44(5):1149–62 [DOI] [PubMed] [Google Scholar]
- 2.Obara N, Suzuki N, Kim K, Nagasawa T, Imagawa S, Yamamoto M. Repression via the GATA box is essential for tissue-specific erythro-poietin gene expression. Blood. 2008;111(10):5223–32 [DOI] [PubMed] [Google Scholar]
- 3.Wenger RH, Hoogewijs D. Regulated oxygen sensing by protein hydroxylation in renal erythropoietin-producing cells. Am J Physiol Renal Physiol. 2010;298(6):F1287–F96 [DOI] [PubMed] [Google Scholar]
- 4.Souma T, Yamazaki S, Moriguchi T, Suzuki N, Hirano I, Pan X, et al. Plasticity of renal erythropoietin-producing cells governs fibrosis. J Am Soc Nephrol. 2013;24(10):1599–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapitsinou PP, Liu Q, Unger TL, Rha J, Davidoff O, Keith B, et al. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood. 2010;116(16):3039–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köchling J, Curtin PT, Madan A. Regulation of human erythropoietin gene induction by upstream flanking sequences in transgenic mice. Br J Haematol. 1998;103(4):960–8 [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Human erythropoietin gene expression in transgenic mice: multiple transcription initiation sites and cis-acting regulatory elements. Mol Cell Biol. 1990;10(3):930–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard KL, Acquaviva AM, Galson DL, Bunn HF. Hypoxic induction of the human erythropoietin gene: cooperation between the promoter and enhancer, each of which contains steroid receptor response elements. Mol Cell Biol. 1992;12(12):5373–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki N, Obara N, Pan X, Watanabe M, Jishage K-I, Minegishi N, et al. Specific contribution of the erythropoietin gene 3′ enhancer to hepatic erythropoiesis after late embryonic stages. Mol Cell Biol. 2011;31(18):3896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, et al. Differentiating the functional role of hypoxia-inducible factor (HIF)-1α and HIF-2α (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2α target gene in Hep3B and Kelly cells. FASEB J. 2004;18(12):1415–7 [DOI] [PubMed] [Google Scholar]
- 13.Wollenick K, Hu J, Kristiansen G, Schraml P, Rehrauer H, Berchner-Pfannschmidt U, et al. Synthetic transactivation screening reveals ETV4 as broad coactivator of hypoxia-inducible factor signaling. Nucleic Acids Res. 2012;40(5):1928–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279(37):38458–65 [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki S, Souma T, Hirano I, Pan X, Minegishi N, Suzuki N, et al. A mouse model of adult-onset anaemia due to erythropoietin deficiency. Nat Commun. 2013;4:1950. [DOI] [PubMed] [Google Scholar]


