Abstract
Tuberculosis (TB) control programmes of many low TB incidence countries of the European Union/European Economic Area (EU/EEA) perceive challenges in controlling TB due to high numbers of TB in migrants from high-incidence countries.
To assess the extent of TB transmission from the foreign-born to the native-born population, we quantitatively investigated the dynamics of TB transmission between these populations in the EU/EEA, using published molecular epidemiological studies. We searched PubMed and EMBASE databases from 1990 to August 2012.
We identified 15 studies performed during 1992–2007 covering 12,366 cases, of which median (range) 49.2% (17.7%–86.4%) were foreign-born. The proportion of clustered isolates ranged between 8.5% and 49.1% of the total number of TB cases genotyped and among these, foreign-born cases were equally or more likely to have unique isolates compared to native-born cases. One third of the clusters were “mixed”, i.e. composed of foreign- and native-born cases, involving 0–34.2% of all genotyped cases. Cross-transmission among foreign and native populations was bidirectional, with wide differences across studies.
This systematic review provides evidence that TB in a foreign-born population does not have a significant influence on TB in the native population in EU/EEA.
Short abstract
TB in foreign-born cases does not have a significant influence on TB in the native population in EU/EEA http://ow.ly/pTTXv
Introduction
Tuberculosis (TB) incidence is on a dwindling trajectory in the European Union (EU) and in the European Economic Area (EEA) member states. The overall notification rate in 2011 was 14.2 TB cases per 100 000 population, decreasing by 8% from 15.8 cases per 100 000 in 2009, and maintaining the downward trend observed in previous years [1]. An increasing number of EU/EEA member states reach notification rates below 10 TB cases per 100 000 population and progress towards TB elimination [2, 3]. Despite this favourable development, there are still member states with notification rates close to 100 TB cases per 100 000 [1].
As long as socio-economic determinants, together with political instability and internal conflicts and wars, drive international migration, the importation of latent TB infection and TB from other countries through human migration will contribute substantially to TB epidemiology in industrialised countries [4, 5]. An ecologic study on the distribution of notified TB cases in 2006 in EU countries showed that wealth inequality is directly correlated to TB notification, and in countries with lower TB notification rates, higher proportions of TB cases occur in foreign-born persons [6]. A descriptive analysis among 21 European countries using data from 1996–2005 found that TB notification rates increased in only three of them: the UK, Norway and Sweden, where approximately three quarters of cases were among the foreign-born [7]. Between 2001 and 2008 there has been a steady decline in the number of notified cases of national origin in most EU/EEA member states, while notifications of cases of foreign origin have generally increased [8]. Data from 1997–2009 provided by 41 GeoSentinel clinics in 19 countries on the spectrum of illness in international migrants, show that one-third of migrants were diagnosed with either latent TB infection or active TB. Active TB was reported in 11% of adults and 7% of children seen at GeoSentinel clinics, and TB was mostly (67%) detected more than 1 year after migration, and in 29% of the cases after more than 5 years [9].
Due to the increasing number of migrants arriving from high-incidence countries, national TB programmes in many low-incidence countries are challenged by the high number of TB cases in the foreign-born population (several of them harbouring resistant or multidrug-resistant strains of Mycobacterium tuberculosis [10]), with 25% of all notified TB cases reported as foreign-born in 2010 in the EU/EEA [1]. It has been reported that foreign-born individuals transmit Mycobacterium tuberculosis strains to the native-born population. The impact of TB transmission from migrants to the native-born population has been difficult to measure [11].
Numerous community-based studies have been conducted to estimate the incidence of recently transmitted disease and to identify risk factors for transmission [12–14]. These studies typically enrol cases with active TB and “fingerprint” their isolates based on distinct genomic polymorphisms. This is done through a number of molecular genotyping methods, including IS6110-based restriction fragment length polymorphism (RFLP), spoligotyping and mycobacterial interspersed repetitive units (MIRU)–variable-number tandem repeats (VNTR) analysis (12, 15 and 24 loci) [12, 13]. Based on fingerprinting results, TB cases are then classified as having shared (clustered) isolates or having unique (non-clustered) isolates. Cases with clustered isolates are assumed to belong to a transmission chain, and those with unique isolates are assumed to have reactivated disease. The identification of clusters of TB cases, and especially of those in which mixed native-born and foreign-born cases occur, enables to estimate the dynamics of transmission between the two populations, defined as “transmission permeability”, and the contribution, if any, of imported M. tuberculosis strains to the on-going transmission of TB in the native population [14].
Using published molecular epidemiological studies, we aimed to assess the extent of TB transmission from the foreign-born to the native-born population in the EU/EAA, by quantitatively investigating dynamics of TB transmission between these populations and to analyse the characteristics of reported mixed clusters, defined as clusters including both foreign-born and native-born cases.
Methods
Data sources
We searched the MEDLINE/PubMed and EMBASE databases (January 1990 to August 2012) for original, peer-reviewed molecular epidemiology studies that reported on the transmission of TB among foreign-born and native-born populations using strain clustering as the main outcome. We also searched bibliographies of identified articles and reviews for additional references. We considered articles in any language, but we extracted data only from those published in English, Spanish, French, Portuguese, Dutch, Italian, Swedish, Norwegian and Danish.
PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines were followed for reporting [15].
Search strategy
The search strategy included a combination of the following terms: “migration”, “tuberculosis” and “genotyping techniques”, and is fully reported in the online supplement.
Study selection
Original peer-reviewed articles were included if they were population- or hospital-based studies of TB on a local, regional or national level using molecular typing and if they reported on clustering results with the purpose of identifying and analysing transmission dynamics between foreign-born and native-born populations.
We only considered studies performed in countries of the EU/EEA where the study population was described in sufficient detail to be able to make valid estimates of TB clustering. The studies had to record the origin of the cases by country of birth or by nationality and had to provide the strain clustering results in disaggregated form based on case origin. To limit the bias, we excluded articles for the following reasons: 1) publications that analysed clustering rate only in selected TB risk groups such as studies on outbreaks, or of prisons and day-care centres; 2) publications that included only multidrug-resistant TB (MDR-TB) or HIV sero-positive cases, as this would not be a representative sample of the whole population; 3) publications in which the population overlapped with the study population described in another publication with a more representative sample; 4) publications which had a study observation period of less than 12 months; and 5) publications using only spoligotyping for determining clustering, as on its own, spoligotyping is less discriminatory than IS6110 RFLP analysis and MIRU–VNTR (12 and 15 loci) [12]. The selection of articles was done by two reviewers (A. Sandgren and M. Sañé Schepisi) working independently in a two-stage procedure: first selection based on titles and abstracts alone, and selection thereafter based on review of the full text to determine eligibility. Discrepancies were resolved by consensus.
After contacting most of the authors of eligible articles to verify published data or obtain additional information not provided in the published manuscript, we excluded studies when the data finally available did not provide sufficient data on the individuals in clusters for our analysis.
Data extraction
Data extraction was performed by two reviewers (A. Sandgren and M. Sañé Schepisi) working independently and discrepancies were resolved by consensus.
From each study, data were recorded on: study design, study site, study period, baseline TB incidence in the study area, total number of culture positive cases, number of foreign-born cases and country of origin of foreign-born cases, genotyping methods, cluster definition, total number of genotyped cases, total number of clusters, cluster sizes, total number and nationalities of clustered cases and nationality of the first diagnosed case in each cluster, number of clusters with both foreign-born and native-born populations (so-called mixed clusters), number of individuals in mixed clusters, number of foreign-born cases infected by natives and number of native-born cases infected by foreign-born subjects in mixed clusters defined as reported below, and presence of epidemiological links among cases in a given cluster.
Operational definitions
For the analysis the following classifications of clusters were used. “Native-born only clusters”: clusters constituted by natives of the study country only; “Foreign-born only clusters”: clusters constituted of foreign-born cases only; “Mixed clusters”: clusters including both foreign-born and native-born cases. Mixed clusters including at least two foreign-born cases and “foreign-born only clusters” were further classified, where enough information was available, as “uninational” or “multinational” if foreign-born cases in the cluster belonged to different nationalities.
For the purpose of this study the definition of foreign-born includes also foreign nationalities.
The sampling fraction was defined as the ratio of genotyped isolates to the study population, and it was estimated from the information on the study population given in the articles.
To analyse the direction of transmission in mixed clusters, we defined the index case as the first diagnosed case, and we assumed that this case was the source case in the cluster. Based on this assumption, we also calculated the following proportions: number of foreign-born cases possibly infected by a native-born index case (i.e. foreign-born cases included in a mixed cluster with a native-born index case) out of the total number of foreign-born cases in mixed clusters, and number of native-born possibly infected by a foreign-born index case (i.e. native-born cases included in a mixed cluster with a foreign-born index case) out of the total of native-born cases in mixed clusters.
Statistical analysis
The proportion of TB due to recent transmission was estimated using the n-1 method. This assumes that each cluster consists of one source case, due to reactivation disease, and that this “index” infectious case gave rise to the other cases in the cluster either by infecting them directly or by infecting a secondary case who then infected other members of the cluster, the rest thus being due to recent infection [16, 17]. We also estimated the crude extent of clustering by using the n method, without excluding the index case.
The main outcomes considered in the analysis were as follows:
1a) Proportion of all genotyped TB cases that are clustered, estimated by the n method: number of cases in clusters/total number of cases genotyped;
1b) Proportion of all genotyped TB cases that resulted from recent transmission, estimated by the n-1 method: (number of cases in clusters - number of clusters)/total number of cases genotyped;
2a) Proportion of all genotyped TB cases that are in mixed clusters, estimated by the n method: number of cases in mixed clusters/total number of cases genotyped;
2b) Proportion of all genotyped TB cases that resulted from recent transmission within mixed clusters, estimated by the n-1 method: (number of cases in mixed clusters - number of mixed clusters)/total number of cases genotyped;
2c) Proportion of clustered TB cases that resulted from recent transmission within mixed clusters, estimated by the n-1 method: (number of cases in mixed clusters - number of mixed clusters)/(number of cases in clusters - number of clusters).
Forest plots were generated using the metan command in Stata/IC 11.1 (Stata Corporation, College Station, TX, USA). The metan command was used for intervention effect estimates, for which the standard error of each outcome variable is required. We calculated the standard error by taking the square root of “p*(1-p)/N”, where p is the proportion clustered in the respective studies, and N the total number of study participants [18]. Heterogeneities for all main outcomes were calculated using the I2 index to determine whether pooled effect estimates are valid or not [19]. Subgroup analyses were performed to study the effect of the sampling fraction, as studies with less than 80% sampling fraction may introduce bias by underestimating the recent transmission. Subgroup analyses were also performed to study the effects of study design, study period, and the proportion foreign-born.
For the additional analysis included in the detailed description of studies and the characterisation of clusters, overall medians and ranges are reported for the included studies.
Quality assessment
Currently there is no validated tool for quality assessment of molecular epidemiological studies. We have therefore excluded studies that are more likely to be biased using strict inclusion and exclusion criteria. In addition, we present data relevant for determining the quality of studies in table 1.
Table 1– Summary of study design and epidemiological features of included studies.
| Author | Study design | Study site | Study period | Genotyping methods | Cluster definition | Sampling fraction % | Total genotyped N | Foreign-born % | |
| Primary | Secondary | ||||||||
| Allix-Béguec [20] | Prospective hospital-based | Brussels-Capital, Belgium | 1/1/2003 to 31/12/2004 | MIRU-VNTR-Spoligo. | RFLP | Identical spoligotypes and MIRU-VNTR type | 87.9 | 530 | 78.9 |
| Barniol [21] | Prospective population-based | Baden-Württemberg, Germany | 1/1/2003 to 31/12/2005 | RFLP | Spoligo. | Identical RFLP patterns | 41.1 | 308 | 56.2 |
| Bezanahary [22] | Retrospective hospital-based | Limousin region, France | 1/1/1998 to 31/12/2006 | Spoligo. | MIRU 15 | Identical spoligotypes and MIRU15 types | 63.7 | 165 | 32.7 |
| Borrell [23] | Population-based | Barcelona, Spain | 1/1/2003 to 31/12/2004 | RFLP | MIRU 12 | 1) Identical RFLP patterns with ≥6 fragments 2) <6 identical IS6110 bands and an identical MIRU 12 type | 67.4 | 463 | 32.0 |
| Brudey [24] | Population-based | Western Sweden | 1/6/1999 to 31/12/2002 | Spoligo. | VNTR 5 | Identical spoligotype and VNTR types | 50.9 | 112 | NA |
| Dahle [25] | Population-based | Norway | 1/1/1994 to 31/12/2001 | RFLP | Spoligo. | Identical RFLP patterns for strains with more than 4 copies of IS6110 | 100.0 | 538 | 70.8 |
| Diel [26] | Prospective population-based | Hamburg, Germany | 1/1/1997 to 30/6/2002 | RFLP | Identical RFLP patterns, isolates <5 bands were excluded | 93.3 | 796 | 42.0 | |
| Franzetti [27] | Prospective hospital-based | Milan and Brescia, Italy | 1/7/1993 to 30/6/2000 | RFLP | Spoligo. | Identical RFLP patterns containing ≥7 fragments or containing ≤6 band with identical spoligotypes | 90.1 | 1999 | 25.0 |
| Gil [28] | Prospective population-based | Castellon, Spain | 1/1/2004 to 31/12/2007 | RFLP | Identical RFLP patterns | 89.1 | 286 | 42.3 | |
| Gutierrez [29] | Prospective population-based | Paris, Ile de France, France | 1/1/1995 to 31/12/1995 | RFLP | DR | 1) Identical RFLP patterns with ≥6 fragments 2) RFLP patterns with ≥6 that differed by only one copy and indistinguishable DR-RFLP patterns, or 3) Identical RFLP patterns with two to five bands and identical DR-RFLP patterns | 83.2 | 272 | 59.2 |
| Inigo [30] | Prospective hospital-based | Madrid, Spain | 1/1/2002 to 31/12/2007 | RFLP | Spoligo. | 1) Identical RFLP patterns containing ≥6 fragments 2) <6 identical RFLP fragments and an identical spoligotypes | 72.2 | 1269 | 41.1 |
| Kamper-Jørgensen [31] | Prospective population-based | Denmark | 1/1/1992 to 31/12/2004 | RFLP | Spoligo. | 1) Identical RFLP pattern containing ≥5 bands in the RFLP pattern 2) <5 identical RFLP fragments and identical spoligotypes | 99.0 | 4631 | 61.6 |
| Kumar [32] | Prospective hospital-based | Northwest London, UK | 1/1/1993 to 31/12/1993 | RFLP | DR | RFLP differing by a single band If only one RFLP fragment: identical DR patterns | NA | 118 | 86.4 |
| López-Calleja [33] | Retrospective population-based | Zaragoza, Spain | 1/6/2001 to 31/5/2004 | RFLP | Spoligo. | 1) Identical RFLP pattern containing ≥5 fragments 2) <5 IS6110 bands that matched identically and identical spoligotyping pattern | 94.2 | 452 | 17.7 |
| Lucerna [34] | Prospective population-based | Almeria, Spain | 1/1/2003 to 31/12/2007 | RFLP | Spoligo. | 1) Identical RFLP pattern containing ≥6 bands 2) ≤6 identical RFLP fragments and identical spoligotypes | 76.9 | 427 | 56.2 |
MIRU: mycobacterial interspersed repetitive units; VNTR: variable-number tandem repeats; Spoligo.: spoligotyping; RFLP: IS6110 restriction fragment length polymorphism; NA: not available; DR: direct repeat restriction fragment length polymorphism.
Results
Our literature database search yielded 1644 records and, after selection, 15 articles were included for analysis. Articles were excluded for the reasons specified in Figure 1.
Figure 1–
Flowchart depicting methods for article inclusion and exclusion.
Description of studies
The included studies were performed in Spain (n=5) [23, 28, 30, 33, 34], Italy (n=1) [27], Germany (n=2) [21, 26], Belgium (n=1) [20], France (n=2) [22, 29], UK (n=1) [32], Sweden (n=1) [24], Denmark (n=1) [31], and Norway (n=1) [25] (table 1).
Five studies were hospital-based [20, 22, 27, 30, 32] and 10 were population-based [21, 23–26, 28, 29, 31, 33, 34], performed from 1992 [31] to 2007 [28, 30, 34], with either retrospective [22, 33] or prospective [20, 21, 26–32, 34] study designs. Two studies were nationwide (Denmark [31] and Norway [25]); the other 13 were regional or in an urban area. Two studies [21, 35] had rather low sampling fractions, i.e. only a small proportion of the population was genotyped and included in the study, thus there was a risk that the clustering and role of on-going transmission were underestimated in these studies [21, 24] Studies differed in how they categorised children born in the host country from at least one foreign-born parent: while Allix-Beguec et al. [20] and Barniol et al. [21] considered them as native-born, according to Dahle et al. [25], “non-Norwegian cases included first- and second-generation immigrants.”
In total, 12 366 culture-positive TB isolates (median 452, range 112–4631) were fingerprinted mainly by RFLP in combination with spoligotyping, over a median (range) study period of 48 (12–144) months. The median (range) percentage of fingerprinted foreign-born cases among all genotyped cases was 49.2% (17.7–86.4%).
Analyses of main outcomes
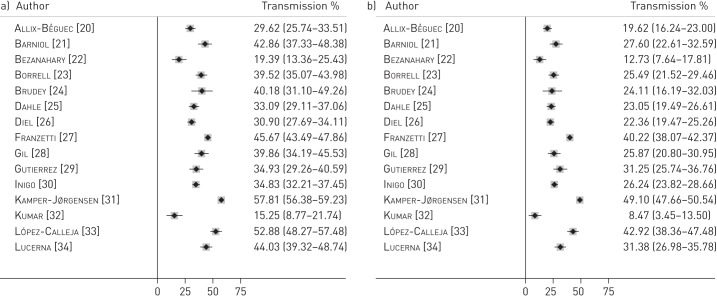
A total of 5659 cases were clustered and the proportion of clustered cases among the total number of genotyped cases (outcome 1a, estimated by the n method: number of cases in clusters/total number of cases genotyped) ranged between 15.3% and 57.8%; I2=98.1% (fig. 2a). The proportion of TB cases resulting from recent transmission, using the n-1 method (outcome 1b, estimated by the n-1 method: (number of cases in clusters - number of clusters)/total number of cases genotyped), ranged between 8.5% and 49.1%; I2=98.4% (fig. 2b).
Figure 2–
Forest plots for a) proportion of all genotyped tuberculosis (TB) cases that are clustered, estimated by the n method (outcome 1a), and b) proportion of all genotyped TB cases that resulted from recent transmission, estimated by the n-1 method (outcome 1b). Data are presented with 95% confidence intervals.
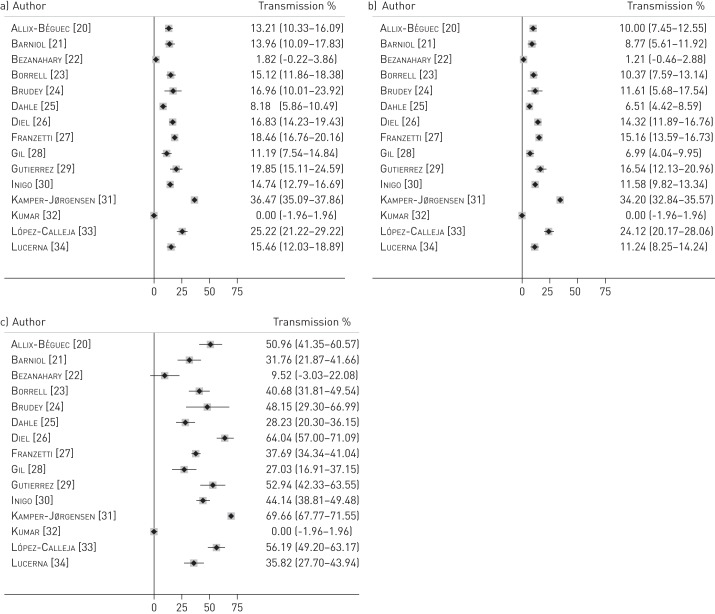
The proportion of genotyped TB cases in mixed clusters ranged between 0% and 36.5%; I2=99.7% (outcome 2a, estimated by the n method: number of cases in mixed clusters/total number of cases genotyped) (fig. 3a). The proportion of genotyped TB cases resulting from recent transmission within mixed clusters ranged between 0% and 34.2%; I2=99.6% (outcome 2b, estimated by the n-1 method: (number of cases in mixed clusters - number of mixed clusters)/total number of cases genotyped) (fig. 3b). This proportion was lower than 15% in 11 of the 15 studies. Finally, the proportion of clustered TB cases that resulted from recent transmission within mixed clusters (outcome 2c, estimated by the n-1 method: (number of cases in mixed clusters - number of mixed clusters)/(number of cases in clusters - number of clusters)) ranged from 0% to 69.7%; I2=99.8% (fig. 3c) and was below 50% in 10 out of 15 studies.
Figure 3–
Forest plots for a) proportion of all genotyped tuberculosis (TB) cases that are in mixed clusters, estimated by the n method (outcome 2a), b) proportion of all genotyped TB cases that resulted from a recent transmission within mixed clusters, estimated by the n-1 method (outcome 2b), and c) proportion of clustered TB cases that resulted from recent transmission within mixed clusters, estimated by the n-1 method (outcome 2c). Data are presented with 95% confidence intervals.
As substantial heterogeneity was found (I2>50%), pooled effect estimates were deemed not valid, and therefore only the range of the effect estimates, without the pooled values, are reported.
The subgroup analyses of the sampling fraction, study design, study period and the proportion foreign-born did not reveal any significant effects on the main outcomes (data not shown).
Detailed description of included studies
The median (range) proportion of foreign-born TB cases among all clustered cases was 48.9% (11.3–77.8%), with large differences between studies; in seven of the 15 studies the proportion of foreign-born clustered cases among the total clustered cases was over 50%. The median (range) proportion of clustered cases was 32.3% (13.7–51.6%) in foreign-born and 43.3% (18.7–68.7%) in native-born cases. The proportion of clustered cases among genotyped native-born cases was higher than in the foreign-born group in 12 of the 13 studies presenting this information. Of an overall 1094 clusters, a median (range) of 32.1% (0–90%) were mixed clusters, 27.3% (0–80%) were “native-born only clusters”, and 29.4% (8.9–75%) were “foreign-born only clusters”. Information on cluster composition was not available for a total of five of the 1094 clusters (range 0–36.4%). Among the 3140 native cases in a cluster, a median (range) of 35.0% (0–75%) were involved in mixed clusters and 65.0% (25–100%) in “native only clusters”. Similarly, 40.7% (0–70.4%) and 59.3% (29.6–100%) of the 2,522 foreign-born cases involved in clusters were part of mixed clusters and “foreign-born only clusters”, respectively.
Of the nine studies reporting detailed information on the nationality of clustered foreign-born cases, 0–80% (median 40%) of the “foreign-born only clusters”, and 40–100% (median 68.8%) of the mixed clusters, were multinational [20–27, 32].
table 2 summarises the information available on the nationality of the index case and the assumed direction of transmission in mixed clusters. Out of the 15 included studies, 10 reported, to a varying level of detail, the nationality of index cases of mixed clusters. The median (range) percentage of foreign-born index cases was 48.8% (33.3–90%). Based on data reported in four studies, the proportion of native-born individuals over the total of natives composing mixed clusters who could have acquired the infection from foreign-born cases ranged from 30.9 to 90.2%, while among foreign-born cases in mixed clusters, 10.7–43.8% could have acquired the infection from natives. When reporting the same cases to the total of native-born individuals in clusters, the proportion of native individuals who could have been infected by a foreign case ranged from 12 to 37.8% and the proportion of foreign-born cases (over the total of foreign-born individuals clustered) who could have been infected by a native case ranged from 2.7 to 22.2%.
Table 2– Summary of the information available on nationality of the index case in mixed clusters and on the likely direction of transmission between foreign-born and native-born cases.
| Author | Mixed clusters with identified index case/total number of mixed clusters | Foreign-born index case/total n identified index cases with known nationality | Native-born infected by foreign-born/native-born in mixed clusters | Foreign-born infected by native-born/foreign-born in mixed clusters |
| Allix-Béguec [20] | 10/17 (58.8) | 7/10 (70) | 17/24 (70.8) | 3/28 (10.7) |
| Barniol [21] | 16/16 (100) | 9/16 (56.3) | NA | NA |
| Borrell [23] | 22/22 (100) | 10/22 (45.5) | NA | NA |
| Dahle [25] | 9/9 (100) | 3/9 (33.3) | NA | NA |
| Diel [26] | 10/20 (50) | 9/10 (90) | 55/61 (90.2) | 3/24 (12.5) |
| Gil [28] | 12/12 (100) | 5/12 (41.7) | NA | NA |
| Inigo [30] | 40/40 (100) | 20/40 (50) | 38/123 (30.9) | 28/64 (43.8) |
| Kamper-Jørgensen [31] | 105/105 (100) | 50/105 (47.6) | NA | NA |
| López-Calleja [33] | 5/5 (100) | 2/5 (40) | NA | NA |
| Lucerna [34] | 18/18 (100) | 15/18 (83.3) | 22/32 (68.8) | 6/34 (17.6) |
Data are presented as n (%). NA: not available.
In two studies, in which estimates of direction of transmission were obtained through other formulae, the magnitude of transmission from foreign-born to natives was similar to that estimated for the opposite direction (foreign-born, infected by natives): 18.3% versus 16% and 5.8% versus 7.9%, for the respective studies [21, 31].
Discussion
This is the first systematic review on molecular epidemiological studies reporting transmission dynamics among foreign-born and native-born populations: it provides a snapshot of TB transmission in the EU/EEA and reveals some aspects with important implications for TB control and elimination. The analysis suggests that, although a large proportion of TB cases are found among foreign-born individuals in the studied low-incidence countries, transmission from the foreign-born population does not have the significant influence on the TB situation among native-born populations as has been hypothesised.
The proportion of TB cases due to transmission within mixed clusters using the n-1 method (outcome 2b) provides the most accurate estimate of the total attribution of mixed clusters to transmission, for the population with genotyping information. The proportion ranged between 0% and 34.2% (less than 15% in 11 of the included studies) and indicates that a maximum of 34.2% of the cases were involved in a transmission chain within mixed clusters. It should be noted that the estimates of transmission occurring in mixed clusters does not imply anything about the direction of transmission. Both native-born and foreign-born index cases seem to contribute to the same extent to TB transmission in mixed clusters (median percentage of foreign-born index cases was 48.8% in included studies providing this information). Thus, only a proportion of the estimated maximum of cases involved in a transmission chain within mixed clusters could be attributed to foreign-born to native-born direction of transmission. The best estimates available are from two of the included studies, which indicate that the magnitude of transmission from foreign-born cases to natives was 5.8–18.3% [21, 31].
It has been argued that as TB incidence in the host country decreases, the proportion of foreign-born cases will increase and the chance of transmission to the native population may increase. However, we have not found any evidence of an effect of the proportion of foreign-born cases on the proportion of cases due to transmission in mixed clusters (data not shown). It is possible that the number and structure of mixed clusters may change in the future. This will depend on contact rates and mixing patterns in the population, and it is difficult to predict how these will change.
Our analysis indicates that foreign-born TB cases are equally, or more, likely to have unique isolates than to have clustered isolates compared to native-born TB cases. Similar findings have been previously reported, among others, in a systematic review of molecular epidemiology studies from diverse industrialised settings [36]. Nevertheless, the proportion of foreign-born TB cases of all clustered cases is not negligible, especially within “foreign-born only clusters”, exceeding 50% in half of the included studies. Mixing may also occur between different foreign-born nationalities; in fact, up to 80% of the “foreign-born only clusters”, and the majority of mixed clusters, included foreign-born cases belonging to more than one nationality.
We observed a large heterogeneity across studies on almost all outcomes and therefore no pooled estimates were presented. The heterogeneity can be explained by the methodological diversity of the reviewed studies, including the design, study site, study period, cluster definition and fingerprinting techniques applied, which may have different discriminatory power [37]. The identified heterogeneity can also be explained by the different study populations: clustering may be related to social factors specific for nationalities, which may influence the interaction with the native-born population and with those of other nationalities [27]. Heterogeneity can also be introduced by differences in the level of social integration between native-born and foreign-born populations, as observed with Latin American populations in Spain [23, 38, 39], which share cultural and language similarities or Somali communities in Scandinavian countries [24, 25, 31], where limited transmission between migrant communities and the native-born resident population could be a reflection of poor integration. Indeed, one study quantified the degree of integration of foreign-born populations, through a questionnaire on socio-economic status and local language use for professional needs or social contacts. This study reported that the higher the degree of integration in the host country, the higher the likelihood to belong to a mixed cluster [21].
The introduction of imported strains into the autochthonous population may occur after a lag time (i.e. between the time of arrival in the host country and the moment imported strains appear among native-born contacts) [40]. Therefore, heterogeneity could also be attributed to the variability in duration of residence among recent or settled foreign-born cases [20, 41], and to differences in time of observation across included studies.
Limitations
The main limitation of the included studies and of this systematic review is that the general assumption that clustering is due to recent transmission has been only partly validated. The application of the n-1 method, when index cases were not in fact part of the cluster, and one member of the chain was instead identified as the index case, may have underestimated the total number of clustered cases. TB cases may occasionally be part of a cluster by chance, through a coincidental reactivation during the observation period of strains with stable fingerprint patterns [14, 25, 26, 40, 42, 43], especially if originating from relatively isolated populations with low variability in strain types [31]. Moreover, many factors, such as the choice of the fingerprinting method [16, 35, 42], adequate sampling of the population, length of the observation period [16, 42], population mobility and geographic coverage [44], may lead to either an over- or under-estimation of recent transmission. In addition, the discriminatory power of the fingerprinting methods employed will affect who is considered to be part of the same transmission chain. Therefore, more advanced methods using whole-genome sequencing with greater discriminatory power might be relevant to consider in order to get a more accurate reflection of transmission chains [45].
Another relevant weakness of the included studies is the extent of missing data on the direction of transmission among mixed clusters. When information on the index case was available, different definitions of place of origin for child cases were used [20, 21, 25, 31].
Furthermore, publication bias cannot be excluded, as positive results, considered as studies reporting on transmission dynamics from foreign-born to native-born populations in low-incidence countries, are more likely to be published. The opposite could also be true, as if the conventional wisdom is that foreign to local transmission is dominant, then studies that reach the opposite finding may be more likely to be published.
Conclusions
The molecular epidemiological evidence here presented in a systematic review for the first time shows that, although foreign-born cases are part of chains of on-going transmission involving both foreign-born and native-born cases, they are less likely to belong to a cluster than native-born cases are. Instead, when foreign-born cases are part of a cluster, they are more likely to belong to a cluster with foreign-born cases only. There is, however, a significant heterogeneity between studies. Our conclusions from this meta-analysis are that foreign-born cases can contribute to the TB burden in the native population in EU/EEA member states; however, the fear that TB control and elimination are greatly hampered by transmission from the foreign-born to the native-born population and that migration strongly influences the TB burden among existing residents of the countries is not supported by the available evidence.
Future research efforts need to be directed towards investigating the interaction between TB transmission and determinants of social mixing patterns among different nationalities in the EU/EEA. For this, there is a need to further integrate molecular typing in routine surveillance practices, to prospectively collect data on TB transmission. If Europe really wants to embark moving towards the elimination phase [2], all the recommended interventions need to be implemented [2, 3].
Acknowledgments
We thank the authors of the included articles who provided additional information or clarification: F. Franzetti, A.C. Carvalho, M. Gil, A. Gori, J. Inigo, Z. Kamper-Jørgensen, A.I. Lopez Calleja, M.A. Lucerna and A. Matteelli. We gratefully acknowledge the generous assistance and the valuable contributions in the development of the search strategy from R. Mancini, librarian at the National Institute for Infectious Diseases, IRCCS L. Spallanzani, Rome, Italy, and from I. Muñoz Guajardo, Medical Assistant Librarian at the European Centre for Disease Prevention and Control, Stockholm, Sweden.
Footnotes
This article has supplementary material available from www.erj.ersjournals.com
Support statement: A. Sandgren, E. Huitric and M.J. van der Werf are employees of the European Centre for Disease Prevention and Control. This study was supported in part by a grant from the Italian Ministry of Health, “Ricerca finalizzata” 2009 to E. Girardi.
Conflict of interest: Disclosures can be found alongside the online version of this article at www.erj.ersjournals.com
References
- 1.European Centre for Disease Prevention and Control/WHO Regional Office for Europe Tuberculosis Surveillance and Monitoring in Europe 2013. Stockholm, European Centre for Disease Prevention and Control, 2013 [Google Scholar]
- 2.Diel R, Loddenkemper R, Zellweger JP, et al. Old ideas to innovate tuberculosis control: preventive treatment to achieve elimination. Eur Respir J 2013; 42: 785–801 [DOI] [PubMed] [Google Scholar]
- 3.Veen J, Migliori GB, Raviglione M, et al. Harmonisation of TB control in the WHO European region: the history of the Wolfheze Workshops. Eur Respir J 2011; 37: 950–959 [DOI] [PubMed] [Google Scholar]
- 4.Broekmans JF, Migliori GB, Rieder HL, et al. European framework for tuberculosis control and elimination in countries with a low incidence. Recommendations of the World Health Organization (WHO), International Union Against Tuberculosis and Lung Disease (IUATLD) and Royal Netherlands Tuberculosis Association (KNCV) Working Group. Eur Respir J 2002; 19: 765–775 [DOI] [PubMed] [Google Scholar]
- 5.Dara M, de Colombani P, Petrova-Benedict R, et al. Minimum package for cross-border TB control and care in the WHO European region: a Wolfheze consensus statement. Eur Respir J 2012; 40: 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suk JE, Manissero D, Buscher G, et al. Wealth inequality and tuberculosis elimination in Europe. Emerg Infect Dis 2009; 15: 1812–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert RL, Antoine D, French CE, et al. The impact of immigration on tuberculosis rates in the United Kingdom compared with other European countries. Int J Tuberc Lung Dis. 2009; 13: 645–651 [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control Progressing Towards TB Elimination. Stockholm, European Centre for Disease Prevention and Control, 2010 [Google Scholar]
- 9.McCarthy AE, Weld LH, Barnett ED, et al. Spectrum of illness in international migrants seen at GeoSentinel clinics in 1997-2009, part 2: migrants resettled internationally and evaluated for specific health concerns. Clin Infect Dis 2013; 56: 925–933 [DOI] [PubMed] [Google Scholar]
- 10.Fattorini L, Mustazzolu A, Piccaro G, et al. Drug-resistant tuberculosis among foreign-born persons in Italy. Eur Respir J 2012; 40: 497–500 [DOI] [PubMed] [Google Scholar]
- 11.Dahle UR, Eldholm V, Winje BA, et al. Impact of immigration on the molecular epidemiology of Mycobacterium tuberculosis in a low-incidence country. Am J Respir Crit Care Med 2007; 176: 930–935 [DOI] [PubMed] [Google Scholar]
- 12.Mathema B, Kurepina NE, Bifani PJ, et al. Molecular epidemiology of tuberculosis: current insights. Clin Microbiol Rev 2006; 19: 658–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supply P, Allix C, Lesjean S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2006; 44: 4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzelli C, Rindi L. Molecular epidemiological approaches to study the epidemiology of tuberculosis in low-incidence settings receiving immigrants. Infect Genet Evol 2012; 12: 610–618 [DOI] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009; 6: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray M, Alland D. Methodological problems in the molecular epidemiology of tuberculosis. Am J Epidemiol 2002; 155: 565–571 [DOI] [PubMed] [Google Scholar]
- 17.Small PM, Hopewell PC, Singh SP, et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med 1994; 330: 1703–1709 [DOI] [PubMed] [Google Scholar]
- 18.Houben RM, Glynn JR. A systematic review and meta-analysis of molecular epidemiological studies of tuberculosis: development of a new tool to aid interpretation. Trop Med Int Health 2009; 14: 892–909 [DOI] [PubMed] [Google Scholar]
- 19.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21: 1539–1558 [DOI] [PubMed] [Google Scholar]
- 20.Allix-Béguec C, Supply P, Wanlin M, et al. Standardised PCR-based molecular epidemiology of tuberculosis. Eur Respir J 2008; 31: 1077–1084 [DOI] [PubMed] [Google Scholar]
- 21.Barniol J, Niemann S, Louis VR, et al. Transmission dynamics of pulmonary tuberculosis between autochthonous and immigrant sub-populations. BMC Infect Dis 2009; 9: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bezanahary H, Baclet MC, Sola C, et al. Interet du typage moleculaire pour l'epidemiologie de la tuberculose en Limousin (1998–2006) [Molecular strain typing contribution to epidemiology of tuberculosis in Limousin (1998 to 2006)]. Med Mal Infect 2008; 38: 309–317 [DOI] [PubMed] [Google Scholar]
- 23.Borrell S, Espanol M, Orcau A, et al. Tuberculosis transmission patterns among Spanish-born and foreign-born populations in the city of Barcelona. Clin Microbiol Infect 2010; 16: 568–574 [DOI] [PubMed] [Google Scholar]
- 24.Brudey K, Gordon M, Mostrom P, et al. Molecular epidemiology of Mycobacterium tuberculosis in western Sweden. J Clin Microbiol 2004; 42: 3046–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahle UR, Sandven P, Heldal E, et al. Continued low rates of transmission of Mycobacterium tuberculosis in Norway. J Clin Microbiol 2003; 41: 2968–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diel R, Rusch-Gerdes S, Niemann S. Molecular epidemiology of tuberculosis among immigrants in Hamburg, Germany. J Clin Microbiol 2004; 42: 2952–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franzetti F, Codecasa L, Matteelli A, et al. Genotyping analyses of tuberculosis transmission among immigrant residents in Italy. Clin Microbiol Infect 2010; 16: 1149–1154 [DOI] [PubMed] [Google Scholar]
- 28.Gil M, Moreno R, Marin M, et al. Influencia de la inmigracion en los patrones de transmision de la tuberculosis en Castellon (2004–2007) [Influence of immigration on tuberculosis transmission patterns in Castellon, Spain (2004–2007)]. Gac Sanit 2011; 25: 122–126 [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez MC, Vincent V, Aubert D, et al. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J Clin Microbiol 1998; 36: 486–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inigo J, Garcia de Viedma D, Arce A, et al. Differential findings regarding molecular epidemiology of tuberculosis between two consecutive periods in the context of steady increase of immigration. Clin Microbiol Infect 2013; 19: 292–297 [DOI] [PubMed] [Google Scholar]
- 31.Kamper-Jørgensen Z, Andersen AB, Kok-Jensen A, et al. Migrant tuberculosis: the extent of transmission in a low burden country. BMC Infect Dis 2012; 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar D, Saunders NA, Watson JM, et al. Clusters of new tuberculosis cases in North-west London: a survey from three hospitals based on IS6110 RFLP typing. J Infect 2000; 40: 132–137 [DOI] [PubMed] [Google Scholar]
- 33.López-Calleja AI, Lezcano MA, Vitoria MA, et al. Genotyping of Mycobacterium tuberculosis over two periods: a changing scenario for tuberculosis transmission. Int J Tuberc Lung Dis. 2007; 11: 1080–1086 [PubMed] [Google Scholar]
- 34.Lucerna MA, Rodriguez-Contreras R, Barroso P, et al. Epidemiologia molecular de la tuberculosis en Almeria. Factores asociados a transmision reciente [Molecular epidemiology of tuberculosis in Almeria (Spain). Factors associated with recent transmission]. Enferm Infecc Microbiol Clin 2011; 29: 174–178 [DOI] [PubMed] [Google Scholar]
- 35.Brudey K, Driscoll JR, Rigouts L, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 2006; 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fok A, Numata Y, Schulzer M, et al. Risk factors for clustering of tuberculosis cases: a systematic review of population-based molecular epidemiology studies. Int J Tuberc Lung Dis 2008; 12: 480–492 [PubMed] [Google Scholar]
- 37.Kato-Maeda M, Metcalfe JZ, Flores L. Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future Microbiol. 2011; 6: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inigo J, Garcia de Viedma D, Arce A, et al. Analysis of changes in recent tuberculosis transmission patterns after a sharp increase in immigration. J Clin Microbiol 2007; 45: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Lirola M, Alonso-Rodriguez N, Sanchez ML, et al. Advanced survey of tuberculosis transmission in a complex socioepidemiologic scenario with a high proportion of cases in immigrants. Clin Infect Dis 2008; 47: 8–14 [DOI] [PubMed] [Google Scholar]
- 40.Ellis BA, Crawford JT, Braden CR, et al. Molecular epidemiology of tuberculosis in a sentinel surveillance population. Emerg Infect Dis 2002; 8: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng E, Kreiswirth B, Driver C, et al. Changes in the transmission of tuberculosis in New York City from 1990 to 1999. N Engl J Med 2002; 346: 1453–1458 [DOI] [PubMed] [Google Scholar]
- 42.Glynn JR, Vynnycky E, Fine PE. Influence of sampling on estimates of clustering and recent transmission of Mycobacterium tuberculosis derived from DNA fingerprinting techniques. Am J Epidemiol 1999; 149: 366–371 [DOI] [PubMed] [Google Scholar]
- 43.Dale JW, Nor RM, Ramayah S, et al. Molecular epidemiology of tuberculosis in Malaysia. J Clin Microbiol 1999; 37: 1265–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braden CR, Templeton GL, Cave MD, et al. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis 1997; 175: 1446–1452 [DOI] [PubMed] [Google Scholar]
- 45.Walker TM, Ip CL, Harrell RH, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 2013; 13: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]