Abstract
Autophagy, a vital catabolic process that degrades cytoplasmic components within the lysosome, serves as an essential cytoprotective response to pathologic stresses that occur during diseases such as cancer, ischemia, and infection. In addition to its role as a stress response pathway, autophagy plays an essential quality control function in the cell by promoting basal turnover of long-lived proteins and organelles as well as selectively degrading damaged cellular components. This homeostatic function protects against a wide variety of diseases including neurodegeneration, myopathy, liver disease, and diabetes. This review discusses our current understanding of these two principal functions for autophagy as a physiologic stress response and quality control mechanism within mammalian cells and details how alterations in autophagy promote human disease.
Keywords: Macroautophagy, lysosome, mitophagy, inflammation, neurodegeneration, myopathy
INTRODUCTION
Autophagy (literally “self-eating”) is an essential catabolic pathway that degrades cellular components within the lysosome. There are three principal routes of autophagic degradation, mainly differing in the means of cargo delivery to the lysosome: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (1). During macroautophagy, bulk cytoplasmic components are sequestered in a double membrane structure called the autophagosome (AP); the AP is subsequently trafficked to the lysosome where its outer membrane fuses to the lysosome, leading to degradation of its contents. In microautophagy and CMA, cargo is directly taken up by the lysosome, either through invagination of the lysosomal membrane (in microautophagy) or by unfolding and translocation of proteins with a specific signal sequence that is recognized by the LAMP2A receptor on the lysosome (in CMA) (1). This review focuses on macroautophagy, hereafter called “autophagy”.
Autophagy was first identified in mammalian cells by electron microscopy studies in the 1960s, but an understanding of the molecular pathways involved was not achieved until almost thirty years later, following the discovery of the first autophagy genes (ATGs) through landmark genetic screens in yeast. These initial experiments identified a set of highly conserved ATGs that control autophagy (2), many of which have mammalian orthologues. Further studies have elucidated the molecular pathways that control autophagy in mammalian systems (Figure 1) and have revealed the importance of autophagy in both normal physiology and disease (1). The identification of yeast ATGs and their mammalian homologues has also enabled experimental manipulation of the autophagy pathway and the development of assays for monitoring autophagic activity and flux (Figure 2) (3).
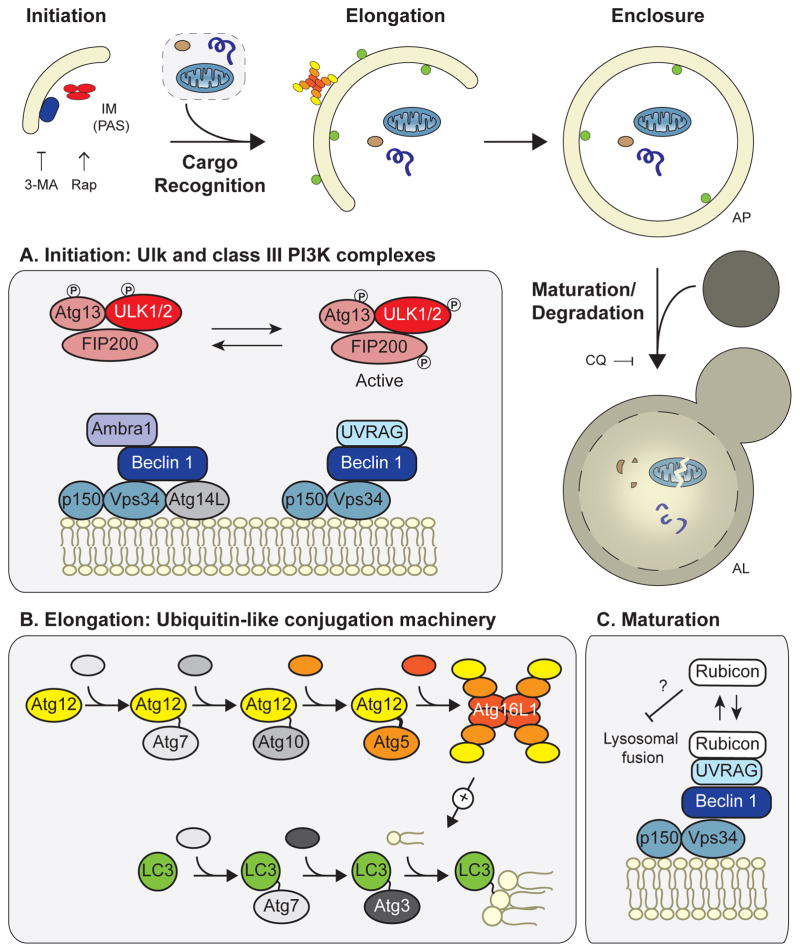
Figure 1. Overview of the mammalian autophagy machinery.
Autophagy occurs in a series of distinct stages: 1) initiation of the isolation membrane, also known as the phagopore assembly site; 2) cargo recognition; 3) elongation; 4) enclosure of the double-membrane structure to form the autophagosome; and 5) maturation/degradation, in which the AP fuses to the lysosome to form an autolysosome, upon which its contents are degraded. The effects of autophagy inhibitors 3-MA and chloroquine and the autophagy inducer rapamycin are shown. (A) The Ulk and class III PI3K complexes are necessary for autophagy initiation. Beclin 1 forms two distinct autophagy-promoting complexes with the PI3K, Vps34. (B) The ubiquitin-like conjugation machinery attaches Atg12 to Atg5 and LC3 to the lipid phosphotidylethanolamine; this machinery mediates autophagosome elongation. Loss of any of these core components (e.g. Beclin 1, Atg5, etc.) leads to autophagy deficiency. (C) In addition to its role in autophagy induction, Beclin 1 promotes autophagic maturation via interactions with Rubicon. Abbreviations: IM, isolation membrane; PAS, phagopore assembly site; AP, autophagosome; AL, autolysosome; 3-MA, 3-methyladenine; CQ, chloroquine; Rap, rapamycin; PI3K, phosphatidylinositol 3-kinase.
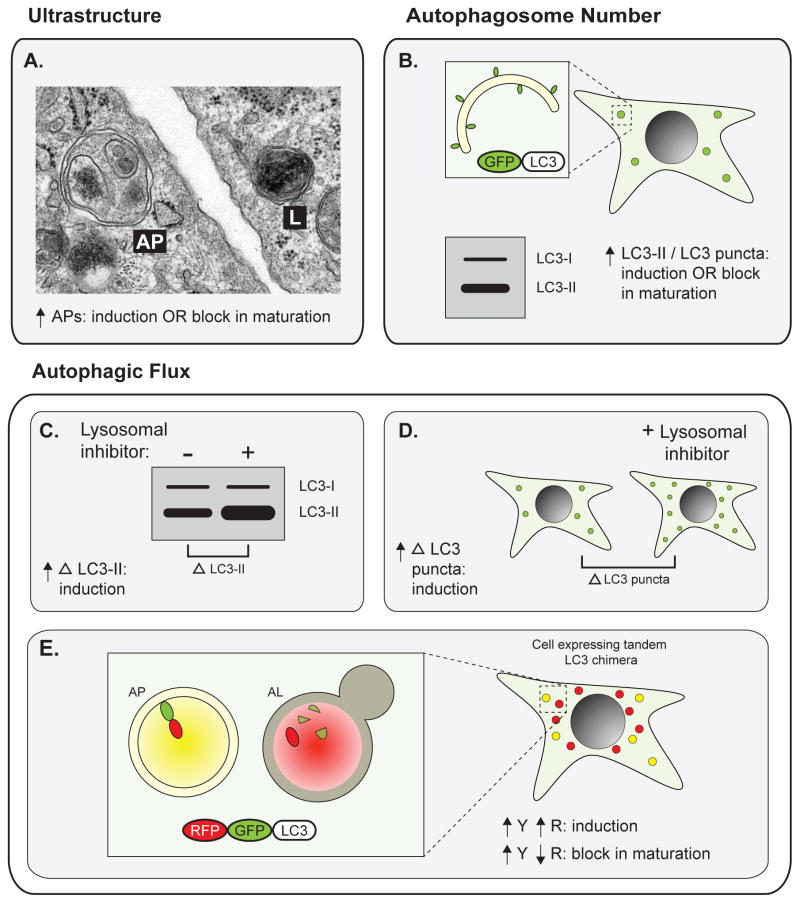
Figure 2. Methods for monitoring autophagy.
Early studies implicating autophagy in disease primarily measured total autophagosome number, either (A) by identification of APs and ALs by electron microscopy or (B) by measuring total levels of lipidated LC3 (LC3-II) or number of LC3 puncta (which mark APs) in cells expressing GFP-tagged LC3 (GFP-LC3). Importantly, in both of these cases, an increase in APs can indicate either autophagy induction or a block in AP maturation. Autophagic flux assays more directly measure total autophagic activity. In the presence of lysosomal inhibitors, (C) increased accumulation of lipidated LC3 or (D) increased accumulation of GFP-LC3 puncta indicates autophagic induction. (E) In cells expressing tandem RFP-GFP-tagged LC3, APs are identified as yellow puncta and ALs are detected as red puncta following quenching of GFP fluorescence in the lysosome. An increase in both signals indicates autophagic induction, whereas an increase in yellow with a decrease in red indicates a block in maturation. Abbreviations: AP, autophagosome; AL, autolysosome; LC3-I, non-lipidated LC3; LC3-II, lipidated LC3; Δ LC3-II, accumulation of lipidated LC3; Δ LC3 puncta, accumulation of GFP-LC3 puncta; Y, yellow; R, red.
Autophagy machinery components are mutated in a variety of human diseases, highlighting the importance of autophagy in human health (Table 1). Tissue-specific deletion of ATGs and autophagy-related genes in mice has also provided further insight into the role of autophagy in maintaining normal tissue homeostasis and in disease pathogenesis (Table 2). In this review, we will discuss the role of autophagy in the cellular stress response and in cellular homeostasis, and how alterations in autophagy promote disease pathogenesis.
Table 1.
Human disease mutations that affect autophagy
| Gene | Function in autophagy | Disease | Likely disease defect | Reference |
|---|---|---|---|---|
| Core autophagy component | ||||
|
| ||||
| Atg16L1 | Core autophagy component; essential for autophagosome elongation | Crohn’s disease | Defect in bacterial clearance, increased proinflammatory cytokine secretion | Hampe, et al. 2006. Nature genetics 39: 207–11 |
|
| ||||
| Autophagy induction/initiation | ||||
|
| ||||
| IRGM | Autophagy induction in response to IFNγ | Crohn’s disease | Defect in bacterial clearance, abnormal lymphocytes and macrophages | Parkes, et al. 2007. Nature genetics 39: 830–32 |
| Jumpy (MTMR14) | PI3P phosphatase, negative regulator of autophagy induction | Centronuclear myopathy | Overactive autophagy | Tosch, et al. 2006. Human molecular genetics 15: 3098 |
|
| ||||
| Cargo Recognition/ Autophagosome (AP) targeting | ||||
|
| ||||
| NOD2 | Autophagy induction in response to MDP; targeting of Atg16L1 to bacteria | Crohn’s disease | Defect in bacterial clearance, increased proinflammatory cytokine secretion | Ogura, et al. 2001. Nature 411: 603–06 |
| PINK1 | Mitophagy | PD | Defect in mitochondrial clearance, accumulation of damaged mitochondria | Valente, et al. 2004. Science 304: 1158 |
| Parkin | Mitophagy | PD | Defect in mitochondrial clearance, accumulation of damaged mitochondria | Kitada, et al. 1998. Nature 392: 605–08 |
|
| ||||
| Lysosome function | ||||
|
| ||||
| presenilin 1 | Targeting of v-ATPase V0a1 to the lysosome | AD | Defect in lysosomal acidification, accumulation of protein aggregates | Sherrington, et al. 1995. Nature 375: 754–60 |
| lysosomal α-glucosidase | Glycogen breakdown in the lysosome | Pompe diseasea | Lysosomal glycogen accumulation, defect in AP-lysosome fusion | Shen, et al. 1996. Journal of Clinical Investigation 98: 352 |
| LAMP2B | Maintenance of lysosome function | Danon diseaseb | Defect in AP-lysosome fusion | Nishino, et al. 2000. Nature 406: 906–10 |
glycogen storage disorder, characterized by muscle atrophy and cardiomyopathy
X-linked hereditary disorder, characterized by cardiomyopathy and myopathy
Table 2.
Disruption of autophagy in mouse models of disease
| Genotype | Tissue | Phenotype | References |
|---|---|---|---|
| Beclin1+/− | All | Spontaneous malignancies; Resistance to pressure-induced heart failure; Decreased infarct size during cardiac I/R | Qu, et al. 2003. JCI 112: 1809–20; Zhu, et al. 2007. JCI 117: 1782; Matsui, et al. 2007. Circ Res 100: 914–22 |
| Atg5f/f: MerCreMera | Cardiomyocytes | In adult mice: hypertrophy, contractile dysfunction, and accumulation of ubiquitinated proteins | Nakai, et al. 2007. Nat Med 13: 619–24 |
| Atg5−/− | Thymus (transplant) | Altered T-cell selection and multiorgan inflammation | Nedjic, et al. 2008. Nature 455: 396–400 |
| Atg5f/f: villin-Cre | Intestinal epithelium | Paneth cell and granule secretion defects | Cadwell, et al. 2008. Nature 456: 259–63 |
| Atg7f/f: villin-Cre | Intestinal epithelium | Paneth cell and granule secretion defects | Cadwell, et al. 2009. Autophagy 5: 250 |
| Atg16L1HM | Intestinal epithelium | Paneth cell and granule secretion defects, increased susceptibility to DSS-induced colitis | Cadwell, et al. 2008. Nature 456: 259–63; Cadwell, et al. 2010. Cell 141: 1135–45 |
| Atg5f/f: nestin-Cre | Neurons | Neurodegeneration and motor defects, accumulation of protein aggregates and inclusions | Hara, et al. 2006. Nature 441: 885–89 |
| Atg7f/f: nestin-Cre | Neurons | Neurodegeneration and motor defects, accumulation of protein aggregates and inclusions | Komatsu, et al. 2006. Nature 441: 880–84 |
| Becn1+/− APPb | All | Increased APP/Aβ aggregation, severe neurodegeneration | Pickford, et al. 2008. JCI 118: 2190; Jaeger, et al. 2010. PloS One 5: e11102 |
| Parkin−/− | All | Mitochondrial defects; no neurodegeneration | Palacino, 2004. JBC 279: 18614 |
| Pink1−/− | All | Age-dependent mitochondrial defects and loss of spontaneous motor activity; no neurodegeneration | Gispert, et al. 2009. PloS One 4: e5777 |
| Slc6a3- Parkin- Q311Xc | Dopaminergic neurons | Progressive motor defects, substantia nigra degeneration, α-synuclein accumulation | Lu, et al. 2009. J Neurosci 29: 1962–76 |
| Atg5f/f: HSA-Cre | Muscle | Accumulation of abnormal mitochondria and protein aggregates and inclusions | Raben, et al. 2008. Hum Mol Gen 17: 3897 |
| Atg5f/f: HSA-Cre GAA KOd | Muscle | Severe muscle atrophy, accumulation of abnormal mitochondria and protein aggregates and inclusions | Raben, et al. 2008. Hum Mol Gen 17: 3897 |
| Atg7f/f: MLC1f-Cre | Muscle | Muscle atrophy, accumulation of abnormal mitochondria and protein aggregates and inclusions | Masiero, et al. 2009. Cell Metabolism 10: 507–15 |
| Atg7f/f: MX1-Cre | Liver | Hepatomegaly; accumulation of abnormal membranous structures, mitochondria, and peroxisomes, and protein inclusions | Komatsu, et al. 2005. JCB 169: 425 |
| Atg7f/f: Alb-Cre | Liver | Increased TG and cholesterol levels, increased TG accumulation following starvation | Singh, et al. 2009. Nature 458: 1131–35 |
| Atg7f/f: RIP-Cre | β cells | β cell degeneration, loss of insulin production, accumulation of damaged mitochondria and protein aggregates | Ebato, et al. 2008. Cell Metabolism 8: 325–32; Jung, et al. 2008. Cell Metabolism 8: 318–24 |
tamoxifen-inducible, cardiomyocyte specific
Transgenic overexpression of human APP
Transgenic overexpression of C-terminal truncated human Parkin
Lysosomal alpha-glucosidase knockout, a mouse model of Pompe disease
Abbreviations: I/R, ischemia-reperfusion; TG, triglyceride
AUTOPHAGY IN STRESS RESPONSE
Autophagy is an essential pro-survival pathway induced by a wide variety of stresses including nutrient deprivation, growth factor withdrawal, oxidative stress, infection, and hypoxia (4). During periods of stress, autophagy maintains cellular biosynthetic capacity and ATP levels by supplying amino acids for de novo protein synthesis and providing substrates for the tricarboxylic acid (TCA) cycle, such as amino acids and free fatty acids (5). In the mouse, autophagy is upregulated in almost all tissues except the brain following starvation (6). Mice lacking the essential autophagy genes Atg5 or Atg7 (Figure 1B) have decreased plasma and tissue amino acid concentrations and die within one day after birth, likely due to nutrient depletion during the neonatal starvation period (7, 8). Figure 1 details the molecular functions of ATGs discussed throughout this review. The importance of autophagy as a pro-survival catabolic pathway has also been demonstrated in cell culture systems. In Bax/Bak double knockout cells, which are unable to undergo apoptosis, intact autophagy protects cells from death following prolonged growth factor withdrawal. In growth factor deprived cells, RNAi-mediated Atg7 depletion or treatment with the autophagy inhibitors 3-methyladenine (3-MA) or chloroquine (CQ) (Figure 1) leads to cell death. Supplying autophagy-deficient cells with the TCA cycle substrate methylpyruvate rescues ATP production and cell viability (9), demonstrating that the catabolic function of autophagy likely mediates cytoprotection. Importantly, self-eating does not protect a cell indefinitely; rather, autophagy is proposed to function as a “battery” that buys cells time, allowing them to survive if the stress is resolved in a timely manner. In this context, autophagy is viewed as a salvage mechanism that provides basic components to sustain core metabolic functions during starvation or stress (10).
Signaling pathways that control autophagy
Stress-induced autophagy is primarily controlled at two critical nodes: mammalian target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK). TOR was initially identified as a negative regulator of autophagy in yeast and has been corroborated as a major regulator of mammalian autophagy (4). In mammalian cells, under normal nutrient conditions active mTOR phosphorylates Ulk1 (the mammalian Atg1 orthologue) and sequesters it in a complex with Atg13 and FIP200, inhibiting autophagy (11–13). Starvation, amino acid deprivation, or growth factor withdrawal inhibit mTOR activity, leading to autophagy induction (Figure 3A) (14). AMPK is a major positive regulator of autophagy that is activated by a high ratio of AMP to ATP (4). Under conditions of low intracellular energy, activated AMPK induces autophagy both by phosphorylating Ulk1, activating it, and by inhibiting mTORC1 via phosphorylation of Raptor (Figure 3A) (11, 15). Both AMPK and mTOR also control cell growth and metabolism, coupling autophagy to these processes.
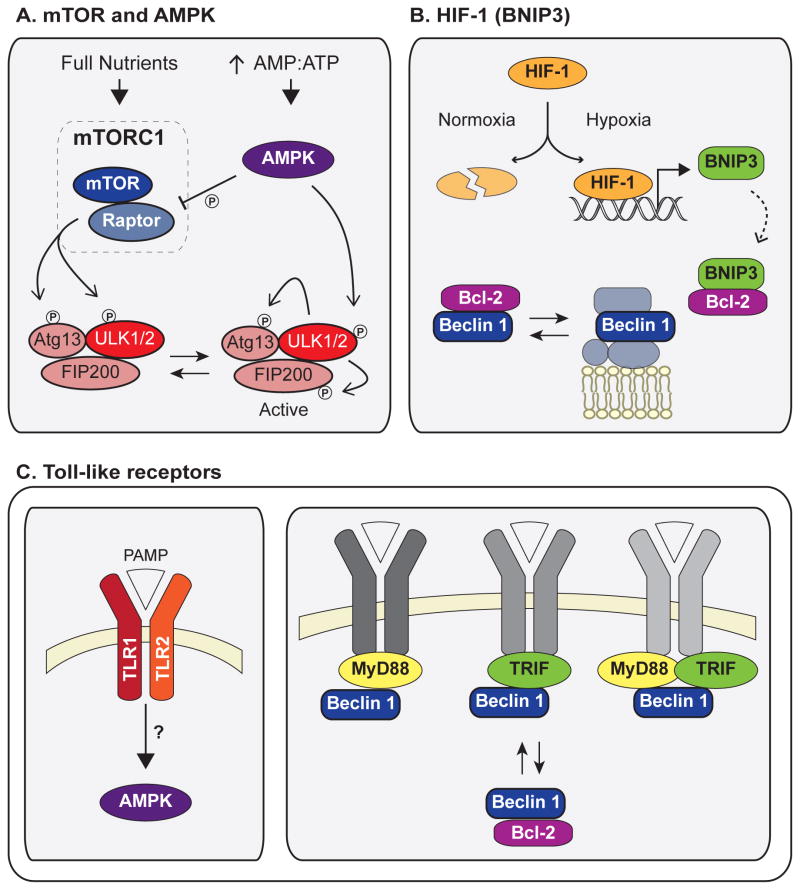
Figure 3. Major stress pathways that induce autophagy.
(A) As part of the mTORC1 complex, active mTOR phosphorylates and inactivates Ulk1. Starvation inactivates mTOR, leading to formation of an active Ulk1 complex in which Ulk1 phosphorylates Atg13 and FIP200. During conditions of low cellular energy, AMPK is activated by high AMP:ATP and induces autophagy both by phosphorylation and activation of Ulk1 and by inhibition of mTORC1 via phosphorylation of Raptor. (B) HIF-1 and BNIP3 induce autophagy following hypoxia. HIF-1 is stabilized under hypoxic conditions, leading to increased BNIP3 transcription. BNIP3 binds Bcl-2 and disrupts its inhibitory interaction with Beclin 1, leading to autophagy induction. (C) Following infection, activation of toll-like receptors (TLRs) by pathogen-associated molecular patterns (PAMPs) leads to autophagy induction. Induction downstream of TLR1/2 stimulation depends on AMPK signaling. The downstream TLR signaling molecules MyD88 and TRIF interact with Beclin 1 following TLR activation and disrupt the inhibitory Bcl-2/Beclin 1 complex. Abbreviations: PAMP, pathogen-associated molecular pattern.
Other important stress response pathways that induce autophagy include HIF-1 signaling in response to hypoxia, p53 signaling downstream of DNA damage, and pattern recognition receptor (PRR) signaling following infection (4). The transcription factor HIF-1 is stabilized under hypoxic conditions, leading to induction of a hypoxia-associated gene expression pattern. The BH3-only protein BNIP3 is one major HIF-1 target, and it is essential for autophagy induction following hypoxia. BNIP3 induces autophagy by binding to Bcl-2 and consequently disrupting the inhibitory interaction between Bcl-2 and Beclin 1 (the mammalian orthologue of yeast Atg6) (Figure 3B) (16). The tumor suppressor p53 is induced by a wide variety of cellular stresses including DNA damage and plays dual roles in autophagy induction. Multiple transcriptional targets of p53 activate autophagy, including BAX and PUMA. On the other hand, cytosolic p53 also has transcription-independent functions, and cytosolic p53 inhibits autophagy (4). The consequences of this balance between the pro- and anti-autophagic roles of p53 are not well understood. Finally, PRRs recognize molecular patterns associated with different pathogens and induce autophagy following infection (Figure 3C) (17). The signaling pathways leading from PRR activation to autophagy induction remain unclear, but there is evidence for AMPK and Beclin 1 as downstream effectors in some contexts (18, 19). The induction of autophagy via PRRs is discussed in greater detail below.
AUTOPHAGY ACTIVATION IN RESPONSE TO DISEASE-RELATED CELL STRESS
Given the central role that autophagy plays in responding to cellular stresses, it is not surprising that autophagy is an essential response to disease-induced stress. Autophagy generally plays a pro-survival role during disease, although there are also contexts in which either overactive autophagy or autophagy induction coupled to lysosomal or maturation defects may be cytotoxic, as discussed below. Stresses such as starvation, hypoxia, and oxidative or genotoxic stress contribute to the etiology of multiple diseases including cancer, stroke, heart disease, and infection. Notably, growing evidence supports key roles for autophagy during cancer progression and metastasis. As a result, there is immense interest in discerning how to most effectively modulate autophagy to treat cancer. Accordingly, the multifaceted roles of autophagy in cancer progression and response to therapy have been recently covered in multiple excellent reviews (10, 20). Below, we focus on three specific examples that poignantly illustrate the diverse roles for autophagy as a response to disease-induced cell stress: 1) cardiac ischemia-reperfusion injury, 2) infection, and 3) chronic inflammation during Crohn’s disease.
Role of autophagy during cardiac ischemia and reperfusion injury
Cardiac ischemia-reperfusion (I/R) injury results from a blockage in blood supply to the heart. It is characterized by two phases: ischemia, during which impaired coronary circulation leads to a state of nutrient and oxygen deprivation, and reperfusion, during which circulation is restored. The primary tissue damage occurs during the reperfusion phase (21).
Autophagy induction during ischemia is likely a response to nutrient withdrawal and subsequent energy depletion caused by impaired blood supply. Since starvation is a classical inducer of autophagy, it is not surprising that autophagy plays an important role in responding to the starved state during I/R. Moreover, HIF-1 signaling and BNIP3 upregulation following hypoxia also may also induce autophagy following ischemia (Figure 3B), although the relative role of this pathway in cardiac ischemia is unknown (16). Autophagy during the ischemic phase is thought to be cytoprotective, because it is able to supply amino acids for use in the TCA cycle to maintain cellular ATP levels. Autophagy may also protect against oxidative damage during ischemia by supplying amino acids for glutathione synthesis (21). Using glucose deprivation in primary cultured myocytes as an in vitro model of ischemia, Matsui et al. demonstrated that autophagy induction following ischemia is AMPK-dependent (Figure 3A). Inhibiting AMPK or inhibiting autophagosome formation with 3-MA during glucose deprivation severely depletes cellular ATP and reduces cell viability (22). Further confirming this result in vivo, transgenic mice with cardiac-specific expression of a dominant-negative AMPK have reduced autophagy induction during ischemia and increased infarct size (23). Although these data suggest that autophagy is upregulated following ischemia and plays a cytoprotective role, there may be certain pathological contexts in which an ischemic cell is unable to induce autophagy. In the cardiomyocyte tumor line HL-1, although autophagy is induced by glucose deprivation alone, autophagosome formation is severely impaired upon combining glucose deprivation with hypoxia (24). Autophagy induction may depend on the degree of energy depletion following prolonged or severe ischemia, since autophagy is an ATP-dependent process (21).
Autophagosome accumulation is further increased during the reperfusion phase (22). However, this accumulation is likely due to decreased AP turnover rather than increased induction, since I/R leads to a profound impairment in autophagic flux as measured by buildup of GFP-LC3 puncta following treatment with lysosomal inhibitors (Figure 2D) (24). The precise role of autophagy during reperfusion remains somewhat controversial. Beclin 1 is upregulated during reperfusion, and overexpression of Beclin 1 enhances autophagic flux in an in vitro model of I/R, which is cytoprotective (22, 24). However, inhibiting autophagy by treatment with 3-MA or Beclin 1 knockdown also increases cell viability in an in vitro model of I/R (22, 25), and Beclin1 (Beclin 1) heterozygous mice have decreased infarct size following I/R (Table 2) (22). These conflicting data may be attributed to the dual roles of Beclin 1 in regulating both autophagosome formation and autophagic flux. Beclin 1 is an essential component of the phosphatidylinositol 3-kinase (PI3K) complex that mediates autophagosome initiation (Figure 1A); at the same time, Beclin 1 also promotes autophagic flux via interactions with Rubicon, although the precise mechanism for this process is unclear (Figure 1C) (26, 27). This suggests a model in which autophagy is cytoprotective during I/R, but the severe block in autophagic flux following reperfusion is cytotoxic; this block in autophagosome maturation leads to accumulation of APs, expansion of the lysosomal compartment, and eventual leakage of lysosomal proteases and cell death (21). Inhibiting autophagy at an early step by 3-MA treatment or RNAi-mediated depletion of Beclin 1 decreases autophagosome accumulation, increasing cell viability. Similarly, enhancing AP turnover via overexpression of Beclin 1 reduces AP accumulation and is also cytoprotective. Additional studies that more carefully monitor changes in autophagic flux and dissect the effects of inhibiting the early versus late steps in autophagy during I/R remain to be done.
Further evidence for the cytoprotective role of autophagy during I/R comes from studies of cardiac preconditioning. Cardiac preconditioning consists of repeated brief periods of ischemia induced before I/R and is one of the most reproducible methods of preventing tissue damage following I/R (21). Autophagy is upregulated during preconditioning, and perfusion of the dominant negative TAT (HIV-1 transactivator of transcription) fusion protein Atg5-K130R inhibits autophagy and blocks the protective effect of preconditioning (28). In vitro, the cardioprotective drugs uridine-5′-triphosphate, diazoxide, and ranolazine also induce autophagy, and Atg5-K130R expression similarly blocks their cytoprotective effects (28).
Overall, the current data suggest that autophagy plays a cytoprotective role during cardiac I/R, but that induction of autophagy coupled to a block in flux during reperfusion leads to a cytotoxic buildup of the lysosomal compartment. Because of this, therapies that induce AP formation without increasing autophagic flux may cause more harm than good in I/R; in such contexts, inhibitors that block early steps of autophagy may be cytoprotective, at least during reperfusion. Further research is necessary to determine whether autophagy modulation is a viable therapy in cardiac I/R.
Autophagy and the host response to infection
Autophagy represents one of the most ancient innate immune responses and serves as the first line of defense against bacterial, protozoan, and viral pathogens (17). Xenophagy, or degradation of foreign pathogens by autophagy, is the most direct innate immune response in the cell, and multiple studies have confirmed this important role for autophagy following infection. For example, a subset of Salmonella typhimurium are targeted to APs following infection, and autophagy-deficient Atg5 −/− cells are more permissive for cytosolic growth (29). Similarly, APs are targeted to Sindbis virus in infected neurons, and Atg5 knockdown leads to delayed viral clearance and increased cell death (30).
Intriguingly, numerous pathogens have evolved mechanisms to evade, suppress, or subvert the host autophagic machinery. Cytosolic Listeria monocytogenes evades autophagic recognition by recruitment of host cytoskeletal proteins; the bacterial surface protein ActA interacts with the Arp2/3 complex and actin, masking the bacteria behind host proteins. Unlike wild type L. monocytogenes, ActA mutant bacteria are efficiently targeted to APs (31). Likewise, Shigella flexineri evade autophagy by secreting IcsB, which competitively binds the bacterial surface protein VirG and prevents its interaction with the autophagy machinery component Atg5 (32). Alternatively, Mycobacterium tuberculosis blocks autophagosome maturation, suppressing autophagic flux altogether. Rapamycin treatment or starvation overcomes this block and leads to decreased intracellular survival of bacteria (33). Other pathogens such as Coxiella burnetii subvert the autophagic machinery and use autophagic vacuoles as sites of replication (34).
Through its role in xenophagic clearance of pathogens, autophagy acts as a direct and critical effector of pattern recognition receptor (PRR) signaling following infection. PRRs are activated by pathogen-associated molecular patterns (PAMPs) and cellular stress signals called danger-associated molecular patterns (DAMPs), allowing recognition of specific pathogens or stress conditions associated with infection (17). The four main classes of PRRs are toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD) leucine-rich repeat containing receptors (NLRs), retinoic acid-inducible gene 1 protein (RIG-I)-like helicase receptors (RLRs), and C-type lectin receptors (CLRs). Autophagy induction downstream of TLR activation has been best characterized. TLR1, TLR2, TLR4, TLR5, and TLR6 mainly recognize bacterial PAMPs, whereas TLR3, TLR7, TLR8, and TLR9 primarily recognize viral molecules. Autophagy is induced downstream of both bacterial and viral TLRs. The mycobacterial lipoprotein LpqH activates autophagy downstream of TLR1 and TLR2 (19). The TLR4 ligand lipopolysaccharide (LPS), another bacterial molecule, induces autophagy and enhances autophagic degradation of M. tuberculosis (35). Activation of TLR3 with the synthetic double-stranded RNA analogue polyinosinic-polycytidylic acid or activation of TLR7 with single-stranded RNA stimulate formation of GFP-LC3 puncta and formation of LC3-II (Figure 2B) (36).
Our understanding of the signaling pathways connecting TLR activation to autophagy induction remains incomplete. Autophagy induction following TLR1/2 stimulation with LpqH is mediated by AMPK signaling (Figure 3C), and depletion or inhibition of AMPK decreases autophagosome formation following stimulation (19). Furthermore, Shi and Kehrl demonstrated an interaction between the TLR downstream signaling molecules MyD88 and TRIF and the autophagy protein Beclin 1, suggesting that Beclin 1 recruitment to TLR signaling components mediates autophagy induction. The interaction between Beclin 1 and MyD88 or TRIF is enhanced by TLR signaling and leads to disruption of the inhibitory Bcl-2/Beclin 1 complex, similarly to BNIP3-dependent disruption of Bcl-2/Beclin 1 following hypoxia (Figures 3B, C) (18).
Autophagy machinery components also directly interact with PRRs and DAMPs. The NLRs NOD1 and NOD2 recruit the essential autophagy protein Atg16L1 to sites of bacterial entry at the plasma membrane, leading to AP formation at these sites (37). High mobility group box 1 (HMGB1) is a nuclear chromatin-associated protein that is released into the cytosol and extracellular space following oxidative stress or necrotic cell death, acting as a DAMP. Interestingly, HMGB1 was recently identified as a novel binding partner of Beclin 1. Following release into the cytosol, HMGB1 competitively binds Beclin 1, disrupting the inhibitory Bcl-2/Beclin 1 complex (38). Finally, the Atg12-Atg5 conjugate directly interacts with the RLRs RIG-I, MDA5, and MAVS. This result points to a potential non-canonical role for Atg12-Atg5 in infection, because in this case autophagy components negatively regulate RLR signaling and subsequent type I interferon (IFN) production, hence suppressing the antiviral immune response; in contrast, autophagy itself generally plays a positive role in antiviral immunity (39). Additional non-canonical roles of ATGs have emerged in recent years and are discussed in the sidebar.
SIDEBAR: Non-canonical functions of ATGs.
Although ATGs are traditionally thought to function solely as autophagy mediators, several recent studies have identified autophagy-independent roles for ATGs. As described in the main text, the Atg12-Atg5 conjugate directly interacts with RLRs, suppressing the antiviral immune response (39). Interestingly, non-canonical ATG functions may be a common feature of the immune response. Following infection, the protozoa Toxoplasma gondii induces formation of a parasitophorous vacuole that acts as a replication site. Atg5 mediates recruitment of the IFNγ-inducible GTPase IIGP1 to the vacuole, leading to membrane disruption. Surprisingly, this does not involve AP formation, suggesting that this anti-parasitic role of Atg5 is autophagy-independent (147). Similarly, coronaviruses induce formation of double membrane vesicles (DMVs) that act as sites of replication. Non-lipidated LC3 decorates the DMV surface and is necessary for coronavirus infection; this process occurs independently of Atg7 (148).
ATGs may also have autophagy-independent roles outside the context of infection. Recently, a novel conjugation of Atg12 to Atg3 was uncovered. The Atg12-Atg3 complex does not affect classic macroautophagy but instead modulates mitochondrial homeostasis and cell death (149). Rubinstein et al. recently identified a novel proapoptotic role for free Atg12. Atg12 binds and negatively regulates antiapoptotic Bcl-2 family members via a BH3-like motif (150).
(Note to editor: Please insert sidebar near “Autophagy and the host response to infection” section discussing Atg12-Atg5 interaction with RIG-I, MDA5, and MAVS)
Abundant evidence points to a crucial role for autophagy adaptors, also known as cargo receptors, in targeting pathogens to autophagosomes. These adaptors include p62/sequestosome 1 (SQSTM1), nuclear dot protein 52 kDa (NDP52), neighbor of BRCA1 gene 1 (NBR1), and Optineurin (OPTN), all of which bind both ubiquitin and LC3 family members such as LC3 and GABARAP-1, mediating recruitment of LC3-positive isolation membranes to ubiquitinated targets (Figure 4A) (40–44). Accordingly, ubiquitinated proteins decorate the surface of cytosolic bacteria and are one of the main signals recognized by autophagy adaptor proteins (Figure 4B) (45). Autophagic degradation of several pathogens including S. typhimurium, L. monocytogenes, S. flexineri, and the Sindbis virus is mediated by p62 (30, 31, 46, 47); NDP52 and OPTN have both been shown to promote autophagic degradation of S. typhimurium (43, 44).
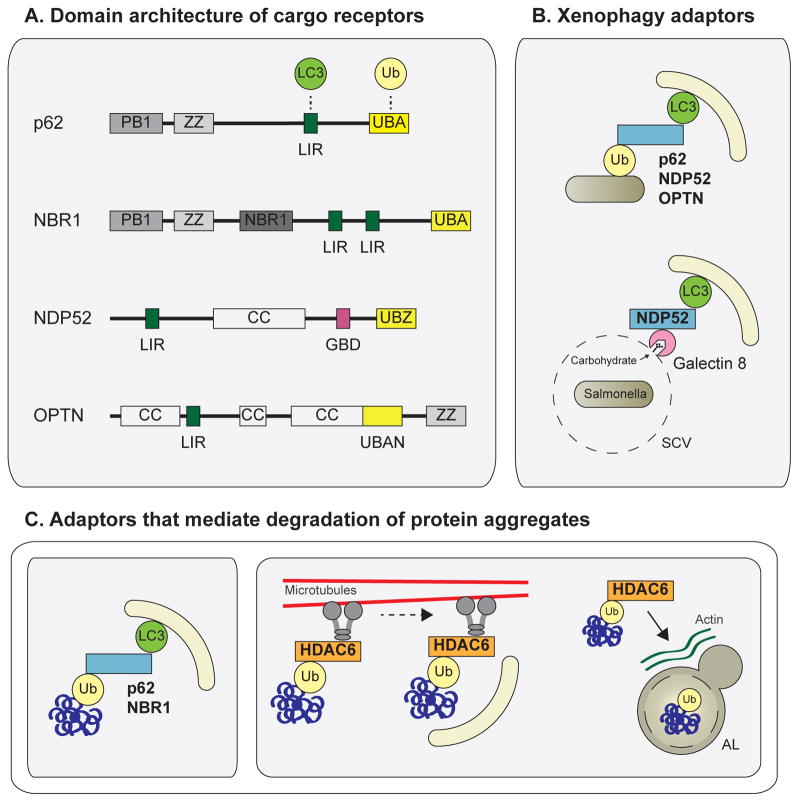
Figure 4. Degradation of ubiquitinated autophagy substrates.
(A) Domain architecture of the autophagy cargo receptors p62, NBR1, NDP52, and Optineurin. These autophagy adaptors contain an ubiquitin-binding domain (UBA, UBZ, UBAN) and an LC3-interacting region (LIR), mediating recruitment of LC3-containing APs to ubiquitinated cargo (B) The adaptors p62, NDP52, and OPTN mediate degradation of ubiquitinated pathogens via ubiquitin-binding domain and LIR interactions. During Salmonella infection, Galectin 8 binds host carbohydrates exposed on ruptured Salmonella-containing vesicles and recruits NDP52 to these sites, mediating AP recruitment (C) p62 and NBR1 also mediate degradation of ubiquitinated protein aggregates via UBA and LIR interactions. HDAC6 is important for clearance of ubiquitinated aggregates and mitochondria. HDAC6 interacts with ubiquitin, dynein motors, and the actin remodeling machinery to promote dynein-mediated transport of ubiquitinated substrates to APs and enhanced AP-lysosome fusion at these sites. Abbreviations: OPTN, optineurin; Ub, ubiquitin; UBA/UBZ/UBAN, ubiquitin-binding domain; LIR, LC3-interacting region; SCV, Salmonella-containing vesicle
In addition to these cargo receptors, other regulators of clearance have been identified that do not depend upon an ubiquitin signal. As described above, the essential autophagy protein Atg5 directly binds to VirG on the surface of S. flexineri (32). The adaptor Tecpr1 interacts with both Atg5 and WIPI-2 (the mammalian orthologue of Atg18), mediating targeting of early isolation membranes to S. flexineri (48). Recently, galectin 8 was identified as a novel adaptor that mediates autophagic degradation of S. typhimurium. Galectin 8 binds host glycans exposed on ruptured Salmonella-containing vesicles and recruits NDP52 to these sites; galectin-mediated NDP52 recruitment precedes and is independent of ubiquitin-mediated recruitment (Figure 4B) (49). Interestingly, galectin 8—as well as galectins 3 and 9—also bind host glycans on damaged endosomes and lysosomes in the absence of infection, suggesting that they act as general sensors of vesicle damage (49).
Autophagy plays other less direct roles in responding to infection in addition to its primary role in pathogen clearance. Autophagy acts as a topological inversion mechanism that engulfs and delivers cytosolic PAMPs to endosomal PRRs and to major histocompatibility complex class II (MHC II) compartments for antigen loading. The topological inversion function of autophagy is important for multiple components of the immune response including PRR activation in the innate immune response, activation of helper T cells in the adaptive immune response, and preventing autoimmunity (17). Evidence of the latter was demonstrated in thymus-specific Atg5-deficient mice. Presentation of self-derived antigens by MHC II complexes on thymic epithelial cells is essential for generation of self-tolerant T cells, and deletion of Atg5 leads to altered T-cell selection and multiorgan inflammation (Table 2) (50). Finally, autophagy modulates the proinflammatory response. Secretion of the proinflammatory cytokines IL-1β and IL-18 is enhanced in Atg16L1 and Atg7 null macrophages, suggesting that basal autophagy suppresses the proinflammatory response (51). This is mediated at least in part by accumulation of damaged mitochondria in autophagy-deficient cells (discussed in detail below) and release of mitochondrial DNA into the cytosol, leading to increased signaling through the NALP3 (a NOD-like receptor family member) inflammasome (52). Interestingly, although basal autophagy suppresses IL-1β production, the opposite is true of autophagy induction. Dupont et al. recently identified a novel autophagy-dependent export pathway for extracellular secretion of IL-1β that requires Atg5, the mammalian Golgi reassembly stacking protein paralogue GRASP55 (GORASP2), and Rab8a, a regulator of exocytosis. Autophagy induction by starvation increases IL-1β secretion in response to multiple proinflammatory agonists, and inhibiting autophagic flux reduces IL-1β secretion (53). These data suggest that autophagy is uniquely able to both suppress proinflammatory signaling during basal conditions and upregulate proinflammatory signaling when induced during times of stress.
IL-1β lacks the signal sequence directing it to the conventional secretory pathway via the Golgi and ER. Interestingly, recent studies suggest that autophagy may play a general role in unconventional protein secretion. Autophagy is involved in unconventional secretion of Acyl-CoA binding protein (Acb1/AcbA) in yeast and Dictyostelium; this process is mediated by delivery of APs containing Acb1/AcbA to the plasma membrane rather than the lysosome (54). In mammalian cells, unconventional secretion of the DAMP HMGB1 and the cytokine IL-18 are increased following autophagy induction, similarly to IL-1β (53). Together, these data suggest that autophagy may play a more general role in secretion during the immune response.
Autophagy in Crohn’s disease
Crohn’s disease is a chronic form of inflammatory bowel disease that affects the intestines. Mechanisms driving Crohn’s pathogenesis include an abnormal inflammatory response, abnormal Paneth cell granule secretion, and impaired intracellular bacterial clearance (55, 56). Autophagy has been implicated in the etiology of this disease by genome-wide association studies identifying disease-related polymorphisms in Atg16L1, IRGM (immunity-related GTPase family M protein), and NOD2 (Table 1) (57). Atg16L1 forms a complex with the Atg12-Atg5 conjugate and is necessary for proper autophagosome elongation (Figure 1B). IRGM and NOD2 are both involved in the autophagic response to infection. IRGM induces autophagy in response to interferon γ (IFNγ), leading to increased clearance of bacterial pathogens (58). As described above, the PRR NOD2 recruits Atg16L1 to bacterial entry sites, targeting bacteria for autophagic degradation (37).
Studies of Atg16l1, Irgm1, and Nod2 knockout mice have provided critical insight into the mechanisms of Crohn’s disease progression. Atg16l1 hypomorphic mice have Paneth cell abnormalities and defects in granule secretion similar to that of Crohn’s patients homozygous for the ATG16L1 disease allele (Table 2) (55). Interestingly, Paneth cell abnormalities are not seen when Atg16l1 hypomorphic mice are raised in gnotobiotic conditions. Administration of a specific strain of the enteric murine norovirus (MNV) capable of establishing persistent infection recapitulates Paneth cell abnormalities. Combined MNV infection and treatment with the inflammatory agent dextran sodium sulfate (DSS) induces intestinal injury and leads to further abnormalities characteristic of Crohn’s disease including blunted intestinal villi and increased intestinal inflammation (59). Surprisingly, Atg16l1 hypomorphic mice have equivalent rates of MNV replication as wild type mice, suggesting that defects in viral clearance do not underlie disease pathogenesis. Instead, intestinal damage is dependent upon TNFα, IFNγ, and the presence of commensal bacterial flora, suggesting that an abnormal proinflammatory response of Atg16l1 hypomorphic mice—in the context of viral infection in combination with commensal bacteria—plays a major role in development of Crohn’s-like abnormalities (59). Another study has demonstrated that macrophages from chimeric mice lacking Atg16L1 in hematopoietic cells have increased IL-1β production in response to LPS stimulation or infection with non-invasive enteric bacteria. These mice are highly sensitive to DSS-induced colitis, suggesting that increased proinflammatory cytokine secretion from hematopoietic cells may also promote intestinal damage in Atg16L1-dependent Crohn’s (51).
Irgm1 null mice have impaired clearance of intracellular bacteria, lymphopenia, and defects in macrophage function (Table 2) (60, 61). Together with data demonstrating that IRGM is important for induction of autophagy and autophagic degradation of bacteria following infection (58), this suggests that autophagy defects underlie the impaired bacterial clearance seen in Crohn’s patients. NOD2 disease variants are also associated with reduced autophagic clearance of bacteria (Table 2) (62). Nod2 null mice have defects in responding to the bacterial PAMP muramyl dipeptide (MDP) and increased susceptibility to bacterial infection (63). Mutant mice expressing the disease-associated allele of NOD2 have increased proinflammatory cytokine secretion in response to NOD2 stimulation and increased sensitivity to DSS-induced intestinal inflammation (64). Interestingly, Atg16L1 directly interacts with NOD2, supporting a common role for the two proteins in Crohn’s pathogenesis. As described above, NOD2 recruits Atg16L1 to sites of bacterial entry (37). Both NOD2 and Atg16L1 are necessary for autophagy induction in intestinal epithelial cells following NOD2 stimulation with MDP, and the disease-variant of Atg16L1 blocks MDP-mediated bacterial killing (65). Together, these data suggest that the defects in Nod2 deficient or mutant mice are at least partly due to impaired autophagic clearance of bacteria.
Further direct support for the role of autophagy in Crohn’s disease pathogenesis comes from Atg5 and Atg7 conditional knockout mice (Table 2). Mice lacking Atg5 or Atg7 in intestinal epithelial cells develop Paneth cell and granule secretion defects that phenocopy Atg16L1 hypomorphic mice and resemble abnormalities seen in Crohn’s patients homozygous for the ATG16L1 disease allele (55, 66). Overall, given the important role of autophagy in xenophagic bacterial clearance, modulation of proinflammatory cytokine secretion, and contribution to extracellular secretion pathways, defects in autophagy may underlie multiple mechanisms that promote Crohn’s pathogenesis.
AUTOPHAGY IN CELLULAR QUALITY CONTROL AND HOMEOSTASIS
Although autophagy was initially identified as a cellular response to stress, it has long been recognized that cells exhibit a basal level of autophagy independent of nutrient and stress status. The two principal degradation pathways in the cell are the ubiquitin-proteasome system (UPS) and autophagy. Whereas the UPS mainly degrades short-lived proteins, autophagy specializes in the removal of long-lived proteins, and, unlike the UPS, it is uniquely able to degrade whole organelles such as mitochondria, peroxisomes, and endoplasmic reticulum (ER) (67). The homeostatic role of autophagy involves both non-selective degradation that supports basal turnover of cytoplasmic components and selective degradation that specifically targets damaged or aggregated organelles and proteins. Selective autophagy serves an essential cellular quality control function in cells (67).
The generation of tissue-specific conditional knockout Atg5 and Atg7 mice first revealed the essential role for basal autophagy in the degradation of protein aggregates and damaged organelles (Table 2). Atg7-deficient hepatocytes and β cells, Atg5-deficient cardiomyocytes, and Atg5- or Atg7-deficient muscle cells accumulate abnormal mitochondria and ubiquitin-positive protein aggregates and inclusion bodies (8, 68–72). Ubiquitinated protein aggregates also accumulate in the brains of neuron-specific Atg5 and Atg7 knockout mice (73, 74). In addition to mitochondrial abnormalities, Atg7 null hepatocytes accumulate peroxisomes, and Atg7 null β cells have abnormally distended ER and Golgi (8, 69). Importantly, in Atg5-deficient neurons, diffuse ubiquitin positive proteins accumulate before protein aggregates are observed, supporting the idea that autophagy plays a crucial role in basal protein turnover in addition to degrading damaged or aggregated proteins (73).
As in xenophagy, autophagy adaptors target the autophagy machinery to specific substrates such as ubiquitinated proteins or depolarized and/or damaged mitochondria; p62 and NBR1, which bind both LC3 family members and ubiquitin and promote autophagic degradation of ubiquitinated substrates, also mediate the removal of protein aggregates (Figure 4A, C). Hence, autophagy-deficient cells accumulate ubiquitin-positive aggregates containing p62 and NBR1 (41, 42). Another ubiquitin-binding protein, HDAC6, is important for autophagic clearance of both protein aggregates and mitochondria. In contrast with other adaptors that interact with components of the autophagy machinery, HDAC6 recruits dynein microtubule motors and actin remodeling machinery to ubiquitinated substrates, promoting transport of ubiquitinated substrates to autophagosomes and leading to enhanced fusion of autophagosomes to lysosomes at these sites (Figure 4C) (75, 76).
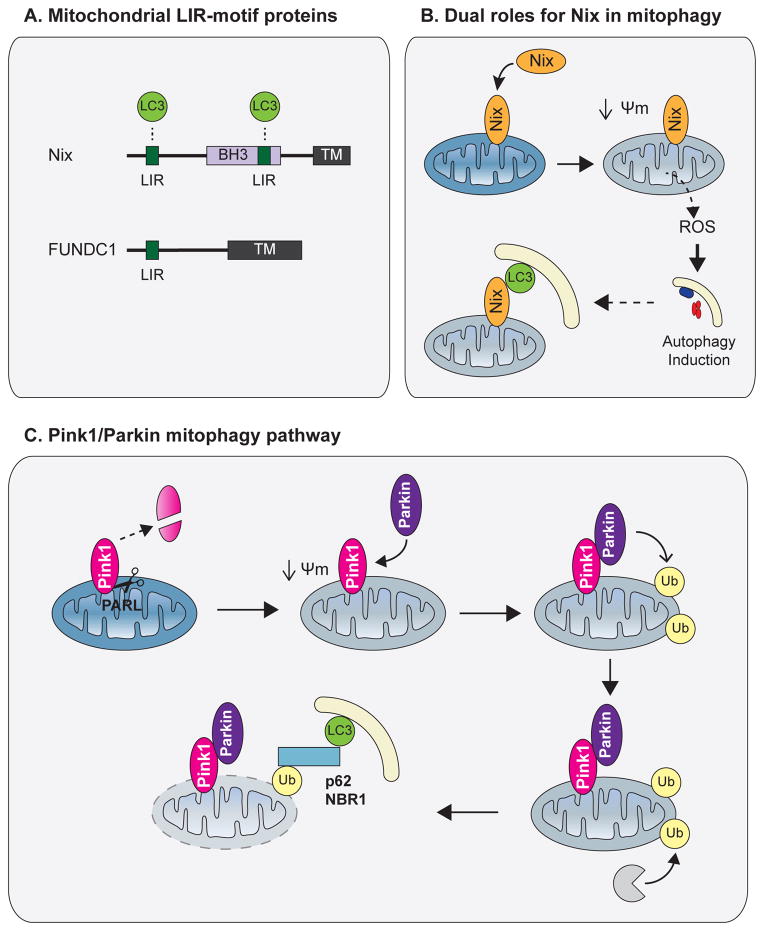
Multiple adaptors have been identified that mediate specific degradation of mitochondria by autophagy, termed mitophagy (77). Nix, also known as BNIP3L1, is a mitochondria-localized BH3-only protein that is upregulated during hypoxia. In response to mitochondrial stressors, Nix promotes mitochondrial depolarization and subsequent formation of reactive oxygen species (ROS), leading to autophagy induction via stress response pathways. Moreover, Nix binds LC3 family members, particularly GABARAP-L1, which mediates targeting of autophagosomes to depolarized mitochondria (Figure 5A, B) (78, 79). Another recently described mitochondrial protein, FUNDC1 (Figure 5A), interacts with LC3 following its hypoxia-induced dephosphorylation, acting as a mitophagy receptor following oxygen deprivation (80).
Figure 5. Mitophagy.
(A) Domain architecture of the mitophagy adaptors Nix and FUNDC1. Both proteins contain a transmembrane domain embedded in the outer mitochondrial membrane (OMM) as well as a LIR, which mediates recruitment of APs to mitochondria. (B). Following mitochondrial stress, Nix promotes mitochondrial depolarization and ROS formation, leading to autophagy induction. Nix also mediates targeting of APs to mitochondria via its LIR. (C) In healthy polarized mitochondria, PINK1 is constitutively degraded by the inner mitochondrial membrane protease PARL. Following depolarization, full length PINK1 accumulates on the OMM and recruits the E3 ligase Parkin. Parkin ubiquitinates multiple OMM proteins, leading to proteasomal degradation of OMM components and p62/NBR1-mediated AP recruitment to mitochondria. Abbreviations: TM, transmembrane domain; Ψm, mitochondrial membrane potential; ROS, reactive oxygen species.
The most well described mitophagy pathway involves the proteins PINK1 (PTEN-induced putative kinase 1) and Parkin (Figure 5C). The mitochondrial resident protein PINK1 is constitutively degraded in healthy mitochondria; this is mediated by membrane potential-dependent cleavage of PINK1 by the inner mitochondrial membrane protease presenilin-associated rhomboid-like protein (PARL) (81). Following mitochondrial depolarization, PINK1 accumulates on the outer mitochondrial membrane (OMM) and recruits the E3 ubiquitin ligase Parkin. Parkin ubiquitinates multiple proteins on the OMM, leading to proteasomal degradation of OMM components followed by autophagosome recruitment and autophagic degradation of damaged mitochondria (82–87). Notably, other mitophagy adaptors also play a role in Parkin-mediated mitophagy. Because Nix promotes mitochondrial depolarization, it also promotes Parkin translocation (78). NBR1 and p62 colocalize with Parkin-positive mitochondria and mediate autophagosome targeting to damaged, ubiquitin positive mitochondria (82). Interestingly, on depolarized mitochondria, stabilized PINK1 also phosphorylates Miro, an adaptor protein that connects the motor protein kinesin to the OMM surface. Phosphorylation of Miro leads to its Parkin-dependent degradation and subsequently to mitochondrial arrest, suggesting that the PINK1-Parkin pathway serves as a mechanism to isolate damaged mitochondria in addition to promoting mitophagic degradation (88).
THE HOMEOSTATIC AND QUALITY CONTROL ROLES OF AUTOPHAGY PREVENT CELL INJURY AND DISEASE
Basal autophagy may be particularly important in post-mitotic cells such as neurons and myocytes, because, unlike dividing cells, they are unable to dilute out damaged proteins and organelles into daughter cells (1). Indeed, loss of autophagy in neurons or muscle generally leads to atrophy and cell death, as detailed below (70, 72–74). Highly secretory cells such as hepatocytes and β cells may also be more dependent upon intact autophagy since they produce large quantities of protein in the ER, leading to higher levels of misfolded proteins and aggregates (89). Below, we discuss the role of autophagy in these tissues and describe how impaired basal autophagy may contribute to diseases such as neurodegeneration, myopathy, liver disease, and diabetes.
Autophagy in neurodegenerative disease
Basal autophagy is essential for proper neuron function and generally plays a cytoprotective role, preventing buildup of protein aggregates and damaged mitochondria. Neuron-specific Atg5 and Atg7 knockout mice develop progressive motor defects accompanied by neurodegeneration and accumulation of polyubiquitinated proteins and aggregates (Table 2) (73, 74). Interestingly, Beclin 1 expression decreases with age in the human brain, suggesting that decreased autophagy may underlie the observed association between advanced age and increased incidence of neurodegenerative disease (90). Autophagy has been implicated in numerous neurodegenerative diseases; here, we focus on Parkinson’s, Alzheimer’s, and Huntington’s disease.
Autophagy in Parkinson’s disease
Parkinson’s disease (PD) is caused by the degeneration of dopaminergic neurons in the substantia nigra. Characteristic features of PD include accumulation of autophagic vacuoles, abnormal mitochondrial function, and the presence of Lewy bodies—intracellular inclusions containing the protein α-synuclein and ubiquitin (77). Although most Parkinson’s cases are sporadic, about 10% of cases are hereditary. Studies of familial inherited forms of PD have identified multiple disease-associated mutations, providing insight into the etiology of both hereditary and sporadic PD and implicating alterations in autophagy in disease pathogenesis (Table 1).
The first identified PD-associated polymorphism was in α-synuclein, the major component of Lewy bodies. PD-associated mutations in α-synuclein cause it to form amyloid like fibrils associated with disease progression. Transgenic mice overexpressing a disease-associated mutant A53T α-synuclein develop protein inclusions accompanied by neurodegeneration and motor defects, mimicking PD (91). Wild type and disease-associated mutant α-synuclein are degraded by macroautophagy, chaperone-mediated autophagy (CMA), and the proteasome (92–94). Mutant α-synuclein inhibits CMA by blocking the LAMP2A receptor, preventing protein translocation into the lysosome and subsequent degradation; the cell compensates for this defect in CMA by upregulating macroautophagy (92). Autophagy upregulation in A53T-expressing neurons in vitro leads to profound loss of mitochondrial mass accompanied by depletion of cellular ATP and cell death (95), and treatment with 3-MA or knockdown of Parkin or Beclin 1 partially protects against A53T-mediated cytotoxicity (95, 96). Overall, these data suggest that, in contrast to its usual cytoprotective role, the compensatory upregulation of macroautophagy—and increased mitophagy—in response to pathogenic α-synuclein is cytotoxic and may worsen progression of familial or sporadic forms Parkinson’s disease caused by mutations in α-synuclein.
Interestingly, gene duplication of wild type α-synuclein has also been implicated in familial Parkinson’s disease (97). Whereas mutant α-synuclein induces macroautophagy, increased expression of the wild type protein has the opposite effect. Overexpression of wild type α-synuclein in a human neuroblastoma cell line causes mislocalization of Atg9 and suppresses autophagosome biogenesis via inhibition of Rab1a, a GTPase involved in ER-Golgi trafficking (98). Transgenic mice overexpressing wild type α-synuclein accumulate α-synuclein aggregates and abnormal APs and lysosomes in neurons leading to neuron death and motor defects (99, 100). Lentiviral delivery of Beclin 1 to the brains of these mice reduces α-synuclein accumulation and disease pathology (100). Overall, these data suggest that imbalanced autophagy—either overactive or suppressed—can be detrimental to neuron survival in the context of α-synuclein pathology. Further research into α-synuclein pathology and alterations in autophagic flux in sporadic PD will be necessary to determine whether autophagy modulation might be beneficial or harmful in these contexts.
Mutations in PINK1 and Parkin cause an autosomal recessive form of early-onset PD (Table 1) (101, 102). As described above, PINK1 recruits Parkin to depolarized mitochondria, leading to ubiquitination of OMM proteins, autophagosome recruitment, and mitophagic degradation (Figure 5C) (82–87). In cell culture systems, PD-associated mutations in PINK1 or Parkin impair mitophagy via several mechanisms, including: 1) defects in targeting of Parkin to depolarized mitochondria, 2) disruption of Parkin’s E3 ligase activity, or 3) formation of non-functional proteins that aggregate in intracellular inclusions (86, 103, 104). Fibroblasts isolated from patients with Parkin mutations have decreased mitochondrial complex I activity suggestive of impaired mitochondrial function; complex I activity defects are also seen in the substantia nigra of patients with sporadic PD (105, 106). Together, these data suggest that mitophagy defects may underlie PD pathogenesis.
Surprisingly, Parkin and PINK1 knockout mice do not recapitulate PD (Table 2). Mice lacking Parkin or PINK1 have mitochondrial defects including decreased complex I activity, reduced respiration, and increased sensitivity to oxidative stress, but these defects occur in the absence of substantia nigra degeneration (107, 108). Aged PINK1−/− mice have progressive mitochondrial dysfunction, weight loss, and reduction in spontaneous locomotive activity similar to PD patients. However, they lack Lewy body accumulation, neurodegeneration, and decreased lifespan that are seen in PD patients (109). These data suggest that patients with Parkin or PINK1 mutations have other genetic variations that modify neuronal sensitivity to decreased mitophagy. Alternatively, polymorphisms in Parkin and PINK1 in patients may not strictly act as loss-of-function mutations; rather, they may also exert dominant negative or dominant toxic effects. Supporting the latter hypothesis, transgenic mice overexpressing a truncated mutant human Parkin in dopaminergic neurons develop progressive motor defects accompanied by substantia nigra degeneration and α-synuclein accumulation, more closely mimicking PD progression (110). Overall, the alterations in Parkin and PINK1 in patients with hereditary PD suggest that a mitophagy deficit contributes to PD pathology, although it may not be the sole cause.
One of the most well-characterized mouse models of PD uses MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) treatment. Administration of MPTP leads to Parkinson-like disease in both primates and mice. The toxic metabolite of MPTP, MPP+, accumulates in dopaminergic neurons, leading to neurodegeneration and motor defects similar to those seen in PD including rigidity, tremor, and akinesia (77). In MPP+ treated neurons, an increase in autophagosomes precedes cell death; both Atg5 or Atg7 knockdown as well as pretreatment with the early autophagy inhibitor 3-MA abrogate MPP+ cytotoxicity (96, 111), suggesting that overactive autophagy enhances MPP+-mediated cell death. However, because these studies have not measured autophagic flux, they are unable to distinguish between autophagy induction and a late block in autophagy in response to MPP+ (Figure 2). Additional studies indicate that MPTP treatment in mice or MPP+ treatment in cell culture leads to lysosomal membrane permeabilization and lysosomal depletion in neurons, presumably causing a block in autophagic flux and AP accumulation. Treatment with rapamycin restores lysosomal levels, enhances AP clearance, and blocks MPP+-mediated cell death in vitro and neurodegeneration in vivo (112). The mechanism by which rapamycin enhances lysosomal activity is unclear. Overall, similar to cardiac I/R, the results above suggest that autophagy protects against MPTP-induced cell death but that a block in autophagic flux and resulting lysosomal permeabilization is cytotoxic. As a result, inhibiting the initial steps of autophagy via 3-MA treatment or ATG knockdown reduces AP accumulation and is cytoprotective. Similarly, promoting autophagic flux via rapamycin treatment (by an unknown mechanism) reduces AP accumulation and also increases cell viability. Overall, given the conflicting results regarding the cytotoxic versus cytoprotective effects of autophagy in different models of PD (e.g. α-synuclein, MPTP, etc.), further research clarifying autophagic activity and flux in PD patients and model systems will be required to determine whether autophagy modulation is a viable therapy for this disease.
Autophagy in Alzheimer’s disease
Alzheimer’s disease (AD) is a form of dementia characterized by neuronal death in the cerebral cortex. The hallmarks of AD are the presence of intracellular neurofibrillary tangles containing hyperphosphorylated tau and presence of extracellular β-amyloid (Aβ) plaques generated by cleavage of amyloid precursor protein (APP) by γ-secretase (77). Accumulation of autophagosomes and abnormal mitochondria are seen in postmortem brains of AD patients, suggesting that autophagy plays a role in disease progression (113). Several studies suggest that defects in autophagic maturation may be a general feature of Alzheimer’s pathology. AD-associated mutations in the transmembrane protein presenilin 1, that cause a form of familial early-onset AD, lead to defects in autophagic flux due to faulty targeting of the v-ATPase V0a1 subunit to the lysosome and resulting defects in lysosomal acidification (Table 1) (114). More broadly, immature forms of autophagic vacuoles such as isolation membranes (IMs) and autophagosomes (APs) rather than autolysosomes (ALs) (Figure 1) accumulate in AD brains. The accumulated autophagic vacuoles resemble those seen in primary cortical neurons following inhibition of lysosomal proteases or vinblastine treatment—both of which block autophagosome maturation—suggesting that autophagosome accumulation in AD brains is at least partly caused by a block in autophagic flux (113, 115, 116).
Other studies have demonstrated that autophagy plays a role in degradation of pathogenic APP and tau. In cells stably expressing wild type APP or an AD-associated mutant APP, Beclin 1 knockdown leads to accumulation of APP and Aβ, whereas autophagy induction via Beclin 1 overexpression or rapamycin treatment decreases APP and Aβ levels (117, 118). Supporting this in vitro data, reduced Beclin 1 levels accompanied by increased APP and Aβ levels are seen in AD patients. Additionally, Beclin1 heterozygous mice expressing AD-associated mutant APP have increased APP and Aβ aggregation and more severe neurodegeneration compared to wild type mice (118, 119). In keeping with the putative autophagic flux defect seen in brains of AD patients, APP overexpression itself leads to autophagosome accumulation in cell culture models; this is likely due to a maturation defect, although further studies more directly measuring autophagic flux (Figure 2C) remain to be done. Interestingly, both APP and γ-secretase accumulate in autophagosomes, suggesting that the autophagic compartment can also act as a source of pathogenic Aβ when autophagosome maturation is impaired (116). Pathogenic tau is also degraded by autophagy. Autophagy inhibition by 3-MA increases tau aggregation and toxicity, and autophagy induction with rapamycin decreases toxicity in cells overexpressing mutant tau (120, 121).
There is conflicting data about whether autophagy is cytoprotective or cytotoxic in AD. Autophagy inhibition has generally been found to increase tau toxicity, whereas rapamycin decreases toxicity (120, 121). Similarly, Atg7 knockdown increases Aβ toxicity in a neuroblastoma cell line, and Beclin1 heterozygous mice have increased susceptibility to APP-induced neurodegeneration (118, 119, 122). However, another study indicates that inhibition of autophagy by 3-MA treatment or Beclin 1 knockdown decreases Aβ toxicity in human neuroblastoma and glioma cell lines (123). Once again, an intriguing possibility is that the cytoprotective versus cytotoxic effect of autophagy inhibition in AD models depends upon the level of autophagic flux and degree of lysosomal impairment in each system. In this case, autophagy inhibition will be cytoprotective in cells with profound impairment in autophagic maturation—in which there is expansion of the autophagosomal/lysosomal compartment and increased Aβ production—but cytotoxic in cells with less severe impairment in flux.
Although there is conflicting data about autophagy inhibition in AD models, autophagy induction using rapamycin has generally been found to play a cytoprotective role. In a mouse model of AD using transgenic mice expressing both pathogenic APP and tau, autophagy induction by rapamycin rescues cognitive deficits and decreases accumulation of Aβ and tau aggregates (117). One might speculate that rapamycin promotes both autophagy and lysosomal function in AD models, as is the case in MPTP models of PD, and is thus able to rescue autophagic flux defects. Overall, further research is required to determine the feasibility of autophagy modulation as a treatment for AD, including interrogating the changes in autophagic flux that occur during AD pathogenesis and dissecting the effects of specifically manipulating discrete stages of autophagy in AD patients.
Autophagy in Huntington’s disease
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease caused by a trinucleotide CAG (polyglutamine) expansion within the protein huntingtin (htt). HD causes death of striatal and cortical neurons accompanied by mitochondrial dysfunction and accumulation of mutant htt within inclusion bodies (77). Polyglutamine-expanded (PolyQ) htt is degraded by autophagy, suggesting that increased autophagy may provide a therapeutic benefit in HD. Mutant htt accumulates in autophagosomes (124, 125), and autophagy inhibition by Beclin 1 knockdown increases levels of polyQ-htt (90). In contrast, autophagy induction by rapamycin enhances clearance of a polyQ-htt fragment and reduces htt-dependent cell death (125).
Interestingly, in mouse and human cellular models of HD and cells from HD patients, autophagosome formation and clearance are normal but autophagic vacuoles contain little cargo, suggesting that impaired cargo recognition and the resulting defect in engulfment of cytoplasmic components underlies the accumulation of dysfunctional mitochondria and protein aggregates seen in HD (126). The cargo recognition defect may be in part caused by sequestration of p62 by mutant htt (126). Given that the inefficient recognition of autophagic substrates may cause certain defects in Huntington’s disease, either increasing the efficiency of autophagic cargo degradation or simply increasing overall levels of autophagy may be therapeutically useful in these patients. Indeed, treatment with rapamycin or a rapamycin analogue reduces htt aggregates and slows neurodegeneration in fly and mouse models of HD (127).
Autophagy in myopathy
Myopathy is defined as muscle weakness resulting from muscle fiber dysfunction. There are many forms of myopathy with distinct pathologic features including muscle cell degeneration, cellular changes leading to reduced contractile function, mitochondrial alterations leading to energy deficits, metabolic defects, and abnormal inflammation. Basal autophagy is essential for maintenance of proper muscle mass and function, and defects in autophagy are sufficient to cause myopathy in mice. Muscle-specific Atg5 and Atg7 knockout mice develop severe muscle atrophy and progressive muscle weakness accompanied by accumulation of abnormal mitochondria, accumulation of ubiquitinated proteins, and abnormal lysosome distribution (Table 2) (70, 72). Autophagy also protects against stress-induced myopathy; it is induced in muscle cells in response to muscle atrophy induced by starvation or denervation (128, 129), and mice lacking Atg7 in myocytes have increased muscle loss in response to either of these stresses (70). The autophagic response to muscle atrophy is in part mediated by BNIP3 (Figure 3B) (130).
Direct evidence for the importance of autophagic flux for muscle maintenance in humans can be found in a group of myopathies characterized by lysosomal defects (Table 1). Pompe disease is caused by a defect in lysosomal α-glucosidase, leading to a defect in lysosomal glycogen degradation and severe myofibril loss. In a mouse model of Pompe disease, muscle-specific loss of α-glucosidase leads to muscle atrophy along with autophagy induction coupled to a defect in autophagosome-lysosome fusion. However, the loss of Atg5 in addition to α-glucosidase further increases the degree of muscle loss, suggesting that the block in AP-lysosome fusion is partial and that autophagy induction may still provide therapeutic benefit to these patients (72). Danon disease is caused by defects in the lysosomal protein LAMP2B, leading to cardiomyopathy and myopathy. The loss of LAMP2B function in human patients, as well as mice, results in the accumulation of autophagosomes due to impaired autophagosome-lysosome fusion (131). Other lysosomal myopathies include X-linked myopathy with excessive autophagy. In each of these diseases, defects in autophagic flux are associated with severe forms of myopathy.
Although autophagy defects can lead to myopathy, excessive autophagy may also promote muscle loss under certain conditions. High levels of oxidative stress contribute to muscle disease, and transgenic mice with muscle-specific expression of mutant G93A superoxide dismutase 1 (SOD1), one of the major antioxidant enzymes, develop progressive muscle atrophy and mitochondrial dysfunction associated with autophagy induction (132). In this context, muscle cell degeneration may be in part due to excessive autophagy; in vivo RNAi-mediated knockdown of LC3 via electrotransfer partially rescues muscle atrophy (132). Jumpy (myotubularin related protein 14, MTMR14) is a PI3P phosphatase that negatively regulates autophagy by reducing PI3K-dependent recruitment of WIPI-1 to autophagosomes (133). A R336Q mutation in Jumpy that renders it catalytically inactive was identified in a patient with centronuclear myopathy, a congenital myopathy in which nuclei are abnormally localized in skeletal muscle cells, leading to decreased contraction (Table 1). Mutant R336Q Jumpy is unable to negatively regulate autophagy (133), suggesting that overactive autophagy may contribute to muscle dysfunction in this type of myopathy. Overall, these data suggest that homeostatic levels of autophagy in muscle are cytoprotective, but that either too much or too little can lead to muscle dysfunction.
Autophagy in liver disease
As in muscle, proper liver function requires basal autophagy. Hepatocytes may be particularly dependent upon basal autophagy due to their high biosynthetic activity and role in protein and carbohydrate storage. Mice lacking Atg7 in the liver develop hepatomegaly and accumulate abnormal concentric membranous structures, abnormal organelles such as mitochondria and peroxisomes, and ubiquitin-positive protein inclusions (Table 2) (8). Liver-specific Atg7 knockout mice also have altered lipid profiles including higher total triglyceride (TG) levels, increased TG accumulation following nutrient deprivation, and increased total cholesterol levels, suggesting that basal autophagy controls lipid metabolism (134).
One of the main causes of liver disease is chronic alcohol use. Interestingly, chronic alcohol consumption inhibits liver cell autophagy, suggesting that decreased autophagy may underlie the pathogenesis of alcoholic liver disease. Rats chronically fed alcohol exhibit decreased numbers of hepatic autophagic vacuoles and the accumulation of abnormal lysosomes in parallel with decreased degradation of long-lived proteins and accumulation of protein and lipid (135, 136). Consistent with an autophagy defect, patients with a form of alcoholic liver disease called alcoholic steatohepatitis accumulate protein aggregates called Mallory-Denk bodies (MDBs) primarily composed of keratin 8 and keratin 18 that are positive for ubiquitin and p62 (137). In a mouse model of MDB formation, autophagic vacuoles contain keratins 8 and 18 as well as ubiquitin, and rapamycin treatment decreases the number of MDBs (138), suggesting that autophagy is involved in clearance of these aggregates. Together, these data suggest that alcohol-mediated autophagy inhibition may promote the development of alcoholic liver disease. One potential mechanism by which alcohol inhibits autophagy is through the reduction of AMPK activity. As discussed above, AMPK induces autophagy in response to low cellular ATP (Figure 3A). Ethanol administration reduces hepatic AMPK activity in both mice and cell culture, and the AMPK activator AICAR protects against alcohol-induced fatty liver in rats, suggesting that autophagy induction via AMPK activation may be a viable therapy in alcoholic liver disease (139, 140).
Fatty liver disease, characterized by triglyceride accumulation in liver cells, can also occur in the absence of chronic alcohol consumption. The main risk factors for development of non-alcoholic fatty liver disease (NAFLD) are obesity and insulin resistance. As described above, basal autophagy regulates intracellular lipid content in hepatocytes. Following starvation, lipid droplets stored by hepatocytes are degraded by autophagy to supply free fatty acids for energy production; loss of Atg5 or 3-MA treatment leads to increased TG storage similar to that seen in fatty liver disease (134). Autophagy levels also modulate insulin signaling. Atg5 and Atg7 null MEFs have decreased insulin receptor signaling following insulin stimulation, and in vivo knockdown of Atg7 in the liver of lean mice leads to severe insulin resistance (89). Together, these data suggest that decreases in hepatic autophagy can cause TG accumulation and insulin resistance and may contribute to fatty liver disease.
What mechanisms might underlie reduced autophagy in patients with fatty liver disease? Insulin is a major hormonal regulator of autophagy that suppresses autophagy induction, and obesity itself may elicit reductions in autophagy. In both genetic (ob/ob) and high fat diet (HFD)-induced models of obesity and insulin resistance, hepatic autophagy is decreased (89, 141). This may be partly due to hyperinsulinemia, although insulin depletion by ablation of pancreatic β cells does not restore autophagy levels (89). Instead, autophagy inhibition is partly mediated by an increase in calpain 2, which degrades Atg7, and overexpression of Atg7 in the liver of ob/ob mice partially rescues glucose tolerance and insulin sensitivity (89). Taken as a whole, the data suggest that alcohol- or obesity-mediated defects in autophagy contribute to progression of some forms of liver disease.
Autophagy in type 2 diabetes
As previously discussed, autophagy plays a key role in removing damaged mitochondria and ubiquitinated proteins and aggregates. Both of these processes—mitophagy and autophagic removal of damaged proteins—are implicated in β cell homeostasis and survival (69, 142). The removal of damaged or aggregated proteins via autophagy is especially important following endoplasmic reticulum (ER) dysfunction, known as ER stress, and β cells are particularly sensitive to ER stress because of their role in insulin secretion (69).
Direct evidence for the importance of autophagy in β cell function comes from β cell-specific Atg7 knockout mice (ATG7:RIP-Cre) (Table 2). ATG7:RIP-Cre mice fed a high fat diet (HFD) phenocopy type 2 diabetes with progressive β cell degeneration, hyperglycemia, loss of insulin production, and cellular hypertrophy. The β cells of ATG7:RIP-Cre mice have characteristics associated with both autophagy defects and diabetes pathology, including accumulation of damaged mitochondria, reduced respiration, increased oxidative stress, and accumulation of ubiquitinated p62-positive aggregates (68, 69, 142). Ebato et al. demonstrated that autophagy maintains both β cell number and function (i.e. insulin secretion) in mice fed a high fat diet. In mice with intact autophagy, a compensatory increase in β cell mass is seen following HFD; this is not seen in mice lacking pancreatic Atg7 (68). Autophagy-deficient islet cells also have decreased insulin secretion. This defect can be partially rescued by antioxidant treatment, suggesting that increased ROS production by damaged mitochondria may play a role in suppressing secretion (142).
Data suggests that patients with type 2 diabetes have autophagy defects; patient samples have greatly increased numbers of autophagic vacuoles along with reduced LAMP2, suggesting that these patients have impaired lysosome activity and autophagosome maturation (143). The same holds true in multiple rodent models of diabetes. In a Rab3A −/− mouse model of diabetes, autophagosomes accumulate in islet β cells in conjunction with decreased LAMP2 (144). Increased autophagosomes are also seen by electron microscopy in db/db mice and ZDF rats, along with p62 accumulation in db/db mice and age-dependent accumulation of polyubiquitinated GFP-LC3-positive protein aggregates ZDF rats, again suggesting that autophagosomes accumulate due to a decrease in autophagic flux (68, 145, 146). Together, these data suggest that autophagy is essential for proper β cell function, particularly in response to a high fat diet. Autophagic flux is suppressed in diabetic patients and in multiple models of diabetes, suggesting that decreased autophagy contributes to the development of this disease.
CONCLUSION
Autophagy plays an essential role in the adaptive response to cell stress and in the maintenance of cellular homeostasis and quality control. Both functions of autophagy are important in disease: in responding to disease-induced cell stress and in preventing homeostatic imbalances that lead to cell injury and disease. On the whole, most evidence supports that autophagy has a beneficial role in human health and disease prevention. Defects in discrete steps in autophagy—either in induction, cargo recognition, or flux—are a common feature of multiple diseases including I/R, Crohn’s disease, neurodegeneration, myopathy and diabetes; such deficiencies may drive or exacerbate disease progression (Figure 6). However, as is the case in some rare forms of myopathy and PD, too much autophagy can also be pathogenic, leading to excessive self-digestion of cellular components. A more thorough understanding of the perturbations that occur in autophagy induction and autophagic flux in these diseases will be important for development of therapeutics that target autophagy. Autophagy inhibitors may be useful in treating diseases in which there is either overactive autophagy or a cytotoxic late block in autophagic flux. Similarly, treatments that induce autophagy may be beneficial in diseases where there is a block in autophagy initiation but will be harmful in diseases that have a severe block in autophagic maturation (Figure 6). Further research into the basic autophagic and lysosomal machinery and upstream signaling pathways will be necessary to identify novel targets for disease therapies that induce or inhibit autophagy at its different steps.
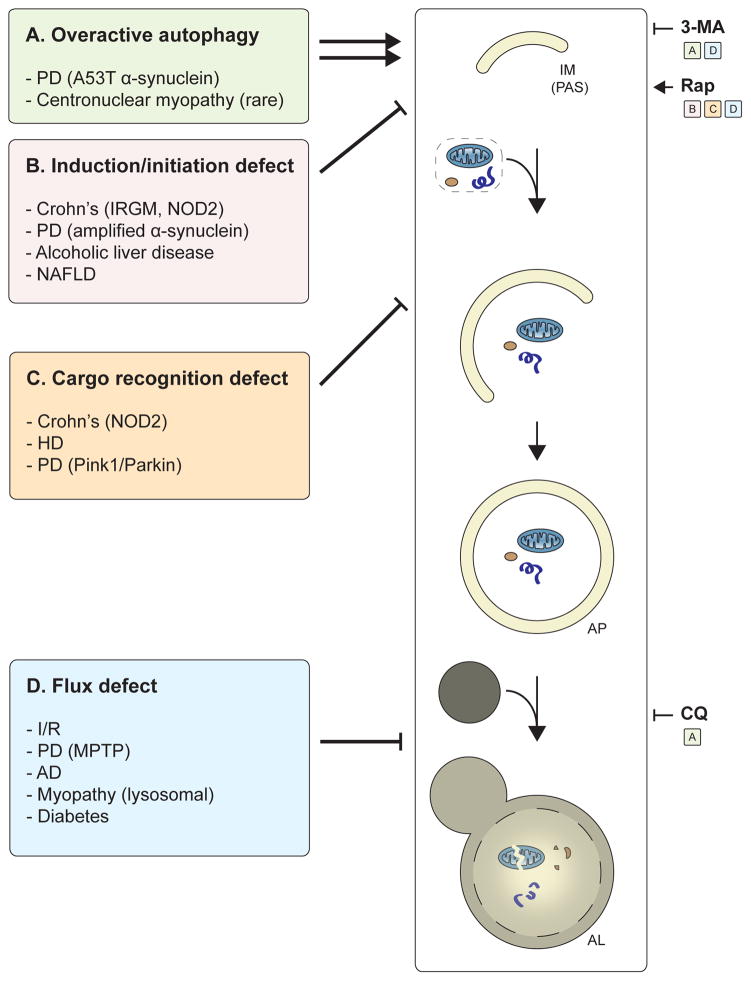
Figure 6. Summary of autophagy alterations in disease and potential treatments.
The major autophagy alterations in human disease and examples of diseases for each class are listed. (A) In diseases with “overactive” autophagy, inhibition of autophagy (via 3-MA or CQ) may provide therapeutic benefit. In diseases that lead to a defect in (B) autophagy induction/initiation or (C) cargo recognition, patients may benefit from drugs such as rapamycin (Rap) that induce autophagy. (D) In diseases that lead to a block in autophagic maturation, Rap treatment may partially restore flux and provide a cytoprotective effect. Alternatively, in diseases with a severe block in autophagic maturation, blocking autophagy at an early step (e.g., via 3-MA) may be therapeutically useful because they mitigate the cytotoxic accumulation of autophagosomes and lysosomes.
SUMMARY POINTS.
Autophagy is an essential pro-survival pathway in cells induced following diverse stresses such as starvation, hypoxia, infection, and oxidative stress.
Autophagy also maintains cellular homeostasis in the absence of stress by promoting basal (non-selective) turnover of cytoplasmic components including whole organelles.
Selective autophagy, including mitophagy and degradation of ubiquitinated substrates, is an essential cellular quality control mechanism that degrades damaged cellular components.
Specific autophagy adaptors, also known as cargo receptors, are important mediators or selective autophagy.
Alterations in autophagy in human disease include: 1) defects in autophagy induction, 2) defects in cargo recognition, 3) defects in autophagic maturation, and 4) overactive autophagy.
Autophagy induction (e.g. by rapamycin) may provide a therapeutic benefit in diseases with defects in autophagy induction, cargo recognition, or autophagic maturation.
Inhibiting the early steps in the autophagy pathway may be therapeutically useful in diseases with a severe block in autophagic maturation; in contrast, inhibiting either early or late steps in the autophagy trafficking process may be useful in diseases notable for “overactive” autophagy.
A more complete understanding of the autophagy alterations that occur in different diseases will be important for development of therapeutics that target autophagy.
Acknowledgments
Grant support to JD includes the NIH (CA126792 and ARRA Supplement CA126792-S1), an Era of Hope Scholar Award (W81XWH-11-1-0130) from the DOD Breast Cancer Research Program, an HHMI Physician-Scientist Early Career Award, and a New Frontiers Award from the UCSF Program for Breakthrough Biomedical Research. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1144247.
GLOSSARY
- Macroautophagy
Catabolic pathway involving sequestration of cytoplasmic components into a double membrane structure called the autophagosome, and subsequent lysosomal degradation
- Basal autophagy
Constitutive autophagic degradation that proceeds in the absence of any overt stress or stimulus and serves an important housekeeping mechanism
- Xenophagy
Selective autophagic degradation of intracellular pathogens in a host cell
- Mitophagy
Selective autophagic degradation of damaged or superfluous mitochondria
- Pattern recognition receptor
Receptor activated by pathogen-specific molecules or damage signals following infection
- Autophagy adaptor (cargo receptor)
Adaptor protein that mediates autophagosome targeting to cargo (e.g. mitochondria, aggregates), often via ubiquitin- and LC3-binding domains
LIST OF ACRONYMS USED
- AP
autophagosome
- ATG
autophagy gene
- mTOR
mammalian target of rapamycin
- 3-MA
3-methyladenine
- PRR
pattern recognition receptor
- PAMP
pathogen-associate molecular pattern
- DAMP
danger-associated molecular pattern
- I/R
ischemia-reperfusion
- PD
Parkinson’s disease
- AD
Alzheimer’s disease
- HD
Huntington’s disease
- NAFLD
non-alcoholic fatty liver disease
BIBLIOGRAPHY
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M. A Unified Nomenclature for Yeast Autophagy-Related Genes. Developmental cell. 2003;5:539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Molecular cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular biology of the cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–36. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology. 2005;169:425. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Debnath J. Autophagy and Tumorigenesis. Seminars in Immunopathology. 2010;32:383–96. doi: 10.1007/s00281-010-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Molecular biology of the cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Molecular biology of the cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–23. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Molecular and cellular biology. 2009;29:2570. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, Ornatowski W, Vergne I, Deretic V. Autophagy and pattern recognition receptors in innate immunity. Immunological reviews. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. Journal of Biological Chemistry. 2008;283:33175–82. doi: 10.1074/jbc.M804478200. Identifies a direct mechanism for TLR-mediated autophagy induction via interaction between the TLR downstream effectors MyD88/Trif and Beclin 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin DM, Yuk JM, Lee HM, Lee SH, Son JW, Harding CV, Kim JM, Modlin RL, Jo EK. Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cellular microbiology. 2010;12:1648–65. doi: 10.1111/j.1462-5822.2010.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb RA, Mentzer RM., Jr Autophagy during cardiac stress: joys and frustrations of autophagy. Annual review of physiology. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion. Circulation research. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 23.Takagi H, Matsui Y, Hirotani S, Sakoda H, Asano T, Sadoshima J. AMPK mediates autophagy during myocardial ischemia in vivo. Autophagy. 2007;3:405. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 24.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. Journal of Biological Chemistry. 2006;281:29776. doi: 10.1074/jbc.M603783200. Along with (22), demonstrates that ischemia induces autophagy, but that reperfusion leads to a severe block in autophagic flux. [DOI] [PubMed] [Google Scholar]
- 25.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. Journal of molecular and cellular cardiology. 2006;40:846–52. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 26.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature cell biology. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 27.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nature cell biology. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM, Gottlieb RA. Autophagy induced by ischemic preconditioning is essential for cardioprotection. Journal of cardiovascular translational research. 2010;3:365–73. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. Journal of Biological Chemistry. 2006;281:11374. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 30.Orvedahl A, MacPherson S, Sumpter R, Jr, Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host & Microbe. 2010;7:115–27. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature cell biology. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 34.Gutierrez MG, Vázquez CL, Munafó DB, Zoppino F, Berón W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cellular microbiology. 2005;7:981–93. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. The EMBO journal. 2008;27:1110–21. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travassos LH, Carneiro LAM, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature immunology. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 38.Kang R, Livesey KM, Zeh H, Loze MT, Tang D. HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6:1209. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- 39.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K. The Atg5–Atg12 conjugate associates with innate antiviral immune responses. Proceedings of the National Academy of Sciences. 2007;104:14050. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–33. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 41.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Molecular cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. Journal of Biological Chemistry. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 43.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature immunology. 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 44.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrin AJ, Jiang X, Birmingham CL, So NSY, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Current biology. 2004;14:806–11. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 46.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host & Microbe. 2009;6:137–49. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. The Journal of Immunology. 2009;183:5909. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host & Microbe. 2011;9:376–89. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–18. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 51.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta; production. Nature. 2008;456:264–68. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 52.Nakahira K, Haspel JA, Rathinam VAK, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2010;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. The EMBO journal. 2011;30:4701–11. doi: 10.1038/emboj.2011.398. Identifies a role for autophagy in unconventional secretion in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manjithaya R, Subramani S. Autophagy: a broad role in unconventional protein secretion? Trends in cell biology. 2011;21:67–73. doi: 10.1016/j.tcb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. With (59) and (66), confirms a role for autophagy in Crohn’s pathogenesis and identifies a novel environment-plus-susceptibility gene interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sewell GW, Marks DJB, Segal AW. The immunopathogenesis of Crohn’s disease: a three-stage model. Current opinion in immunology. 2009;21:506–13. doi: 10.1016/j.coi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011;60:1580–88. doi: 10.1136/gut.2009.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 59.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng CG, Collazo-Custodio CM, Eckhaus M, Hieny S, Belkaid Y, Elkins K, Jankovic D, Taylor GA, Sher A. Mice deficient in LRG-47 display increased susceptibility to mycobacterial infection associated with the induction of lymphopenia. The Journal of Immunology. 2004;172:1163. doi: 10.4049/jimmunol.172.2.1163. [DOI] [PubMed] [Google Scholar]
- 61.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, Oliver T, Taylor GA. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) The Journal of Immunology. 2007;179:6963. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 62.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJP, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature medicine. 2009;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 63.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 64.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn’s disease potentiates NF-κB activity and IL-1β processing. Science. 2005;307:734. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 65.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–41. e2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cadwell K, Patel KK, Komatsu M, Virgin HW, IV, Stappenbeck TS. A common role for Atg16L1, Atg5, and Atg7 in small intestinal Paneth cells and Crohn’s disease. Autophagy. 2009;5:250. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nature cell biology. 2010;12:836–41. doi: 10.1038/ncb0910-836. Reviews the molecular mechanisms involved in selective autophagy, particularly degradation of ubiquitinated substrates. [DOI] [PubMed] [Google Scholar]
- 68.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell metabolism. 2008;8:325–32. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW. Loss of Autophagy Diminishes Pancreatic [beta] Cell Mass and Function with Resultant Hyperglycemia. Cell metabolism. 2008;8:318–24. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 70.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell metabolism. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 71.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nature medicine. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 72.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Human molecular genetics. 2008;17:3897. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–89. doi: 10.1038/nature04724. Together, (73) and (74) strikingly demonstrate the importance of basal autophagy in normal tissue homeostasis—in this case, in neurons. [DOI] [PubMed] [Google Scholar]
- 74.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–84. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 75.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–38. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 76.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. The EMBO journal. 2010;29:969–80. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheung ZH, Ip NY. Autophagy deregulation in neurodegenerative diseases – recent advances and future perspectives. Journal of neurochemistry. 2011;118:317–25. doi: 10.1111/j.1471-4159.2011.07314.x. [DOI] [PubMed] [Google Scholar]
- 78.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. Journal of Biological Chemistry. 2010;285:27879–90. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 2009;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 81.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. The Journal of cell biology. 2010;191:933. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RLJ, Hess S, Chan DC. Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Human molecular genetics. 2011;20:1726. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou Y, Saiki S, Kawajiri S, Sato F. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of cell biology. 2010;189:211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences. 2010;107:378. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. Journal of Biological Chemistry. 2011;286:19630. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell metabolism. 2010;11:467–78. doi: 10.1016/j.cmet.2010.04.005. Provides evidence linking autophagy defects to ER stress and insulin resistance in obesity and diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. Journal of Biological Chemistry. 2006;281:14474. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 91.Giasson BI, Duda JE, Quinn SM, Zhang B, Trojanowski JQ, Lee VMY. Neuronal [alpha]-synucleinopathy with severe movement disorder in mice expressing A53T human [alpha]-synuclein. Neuron. 2002;34:521–33. doi: 10.1016/s0896-6273(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 92.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 93.Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type α-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. Journal of Biological Chemistry. 2008;283:23542. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. α-Synuclein is degraded by both autophagy and the proteasome. Journal of Biological Chemistry. 2003;278:25009. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 95.Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, Zharkovsky A, Kaasik A. Mutant A53T α-synuclein induces neuronal death by increasing mitochondrial autophagy. Journal of Biological Chemistry. 2011;286:10814. doi: 10.1074/jbc.M110.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wong ASL, Lee RHK, Cheung AY, Yeung PK, Chung SK, Cheung ZH, Ip NY. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson’s disease. Nature cell biology. 2011;13:568–79. doi: 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 97.Ross OA, Braithwaite AT, Skipper LM, Kachergus J, Hulihan MM, Middleton FA, Nishioka K, Fuchs J, Gasser T, Maraganore DM. Genomic investigation of α-synuclein multiplication and parkinsonism. Annals of neurology. 2008;63:743–50. doi: 10.1002/ana.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S. α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. The Journal of cell biology. 2010;190:1023–37. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, Mucke L. Dopaminergic loss and inclusion body formation in α-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 100.Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in α-synuclein models of Parkinson’s and Lewy body diseases. The Journal of Neuroscience. 2009;29:13578–88. doi: 10.1523/JNEUROSCI.4390-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–08. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 102.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 103.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature cell biology. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 104.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. The Journal of cell biology. 2010;189:671–79. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mortiboys H, Thomas KJ, Koopman WJH, Klaffke S, Abou-Sleiman P, Olpin S, Wood NW, Willems PHGM, Smeitink JAM, Cookson MR. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Annals of neurology. 2008;64:555–65. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schapira A, Cooper J, Dexter D, Clark J, Jenner P, Marsden C. Mitochondrial complex I deficiency in Parkinson’s disease. Journal of neurochemistry. 1990;54:823–27. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 107.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proceedings of the National Academy of Sciences. 2008;105:11364. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. Journal of Biological Chemistry. 2004;279:18614. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 109.Gispert S, Ricciardi F, Kurz A, Azizov M, Hoepken HH, Becker D, Voos W, Leuner K, Müller WE, Kudin AP. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PloS one. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu XH, Fleming SM, Meurers B, Ackerson LC, Mortazavi F, Lo V, Hernandez D, Sulzer D, Jackson GR, Maidment NT. Bacterial artificial chromosome transgenic mice expressing a truncated mutant parkin exhibit age-dependent hypokinetic motor deficits, dopaminergic neuron degeneration, and accumulation of proteinase k-resistant α-synuclein. The Journal of Neuroscience. 2009;29:1962–76. doi: 10.1523/JNEUROSCI.5351-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu J, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. The American journal of pathology. 2007;170:75. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dehay B, Bové J, Rodríguez-Muela N, Perier C, Recasens A, Boya P, Vila M. Pathogenic lysosomal depletion in Parkinson’s disease. The Journal of Neuroscience. 2010;30:12535–44. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. Journal of Neuropathology & Experimental Neurology. 2005;64:113. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 114.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. The Journal of Neuroscience. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, Mohan PS, Mercken M, Farmery MR, Tjernberg LO. Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. The Journal of cell biology. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caccamo A, Majumder S, Richardson A, Strong R, Oddo S. Molecular Interplay between Mammalian Target of Rapamycin (mTOR), Amyloid-β, and Tau. Journal of Biological Chemistry. 2010;285:13107. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jaeger PA, Pickford F, Sun CH, Lucin KM, Masliah E, Wyss-Coray T. Regulation of amyloid precursor protein processing by the Beclin 1 complex. PloS one. 2010;5:e11102. doi: 10.1371/journal.pone.0011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. The Journal of clinical investigation. 2008;118:2190. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ. Rapamycin alleviates toxicity of different aggregate-prone proteins. Human molecular genetics. 2006;15:433. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Martinez-Vicente M, Krüger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Human molecular genetics. 2009;18:4153. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Abeta-induced cytotoxicity. Autophagy. 2009;5:502–10. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 123.Wang H, Ma J, Tan Y, Wang Z, Sheng C, Chen S, Ding J. Amyloid-β 1–42 Induces Reactive Oxygen Species-Mediated Autophagic Cell Death in U87 and SH-SY5Y Cells. Journal of Alzheimer’s Disease. 2010;21:597–610. doi: 10.3233/JAD-2010-091207. [DOI] [PubMed] [Google Scholar]
- 124.Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. The Journal of Neuroscience. 2000;20:7268–78. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Human molecular genetics. 2002;11:1107. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 126.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, De Vries R, Arias E, Harris S, Sulzer D. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nature neuroscience. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature genetics. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 128.Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–44. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 129.Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes & development. 2005;19:1715–22. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J. FoxO3 controls autophagy in skeletal muscle in vivo. Cell metabolism. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 131.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Path Mech Dis. 2008;3:427–55. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 132.Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Bonconpagni S, Belia S, Wannenes F, Nicoletti C, Del Prete Z. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell metabolism. 2008;8:425–36. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 133.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. The EMBO journal. 2009;28:2244–58. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–35. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Donohue TM, Jr, McVicker DL, Kharbanda KK, Chaisson ML, Zetterman RK. Ethanol administration alters the proteolytic activity of hepatic lysosomes. Alcoholism: Clinical and Experimental Research. 1994;18:536–41. doi: 10.1111/j.1530-0277.1994.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 136.Pösö A, Hirsimäki P. Inhibition of proteolysis in the liver by chronic ethanol feeding. Biochemical journal. 1991;273:149. doi: 10.1042/bj2730149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. The American journal of pathology. 2002;160:255. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47:2026–35. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 139.Tomita K, Tamiya G, Ando S, Kitamura N, Koizumi H, Kato S, Horie Y, Kaneko T, Azuma T, Nagata H. AICAR, an AMPK Activator, Has Protective Effects on Alcohol-Induced Fatty Liver in Rats. Alcoholism: Clinical and Experimental Research. 2005;29:240S–45S. doi: 10.1097/01.alc.0000191126.11479.69. [DOI] [PubMed] [Google Scholar]
- 140.You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798–808. doi: 10.1053/j.gastro.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 141.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia. Journal of Biological Chemistry. 2009;284:31484. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1:425. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Masini M, Bugliani M, Lupi R, Del Guerra S, Boggi U, Filipponi F, Marselli L, Masiello P, Marchetti P. Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia. 2009;52:1083–86. doi: 10.1007/s00125-009-1347-2. [DOI] [PubMed] [Google Scholar]
- 144.Marsh BJ, Soden C, Alarcón C, Wicksteed BL, Yaekura K, Costin AJ, Morgan GP, Rhodes CJ. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine β-cells. Molecular Endocrinology. 2007;21:2255–69. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- 145.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic β-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes. 2007;56:930. doi: 10.2337/db06-1160. [DOI] [PubMed] [Google Scholar]
- 146.Li X, Zhang L, Meshinchi S, Dias-Leme C, Raffin D, Johnson JD, Treutelaar MK, Burant CF. Islet microvasculature in islet hyperplasia and failure in a model of type 2 diabetes. Diabetes. 2006;55:2965. doi: 10.2337/db06-0733. [DOI] [PubMed] [Google Scholar]
- 147.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host & Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reggiori F, Monastyrska I, Verheije MH, Calě T, Ulasli M, Bianchi S, Bernasconi R, de Haan CAM, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host & Microbe. 2010;7:500–08. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The Autophagy Protein Atg12 Associates with Antiapoptotic Bcl-2 Family Members to Promote Mitochondrial Apoptosis. Molecular cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]