Abstract
Regeneration of active glucocorticoids within liver and adipose tissue by the enzyme 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) may be of pathophysiological importance in obesity and Metabolic Syndrome and is a therapeutic target in type 2 diabetes. Polymorphisms in HSD11B1, the gene encoding 11β-HSD1, have been associated with metabolic phenotype in humans, including type 2 diabetes and hypertension. Here we have tested the functional consequences of 2 single nucleotide polymorphisms located in contexts that potentially affect tissue levels of 11β-HSD1. We report no effect of allelic variation at rs846910, a polymorphism within the 5′-flanking region of the gene on HSD11B1 promoter activity in vitro. However, compared to the common G allele, the A allele of rs13306421, a polymorphism located 2 nucleotides 5′ to the translation initiation site, gave higher 11β-HSD1 expression and activity in vitro and was translated at higher levels in in vitro translation reactions, possibly associated with a lower frequency of “leaky scanning”. These data suggest that this polymorphism may have direct functional consequences on levels of 11β-HSD1 enzyme activity in vivo. However, the rs13306421 A sequence variant originally reported in other ethnic groups may be of low prevalence as it was not detected in a population of 600 European caucasian women.
Keywords: steroid metabolism, glucocorticoid, obesity, SNP, translation, regulation
INTRODUCTION
The microsomal enzyme 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) generates active glucocorticoids (cortisol, corticosterone) from intrinsically inert 11keto substrates (cortisone, 11-dehydrocorticosterone) thus amplifying glucocorticoid action in cells and tissues in which it is expressed (1, 2). Recent evidence has indicated a pathogenic role for 11β-HSD1 in metabolic disease. A number of studies have demonstrated a strong association in humans between the level of 11β-HSD1 expression in adipose tissue and body mass index (BMI) (reviewed (2, 3)). Moreover, 11β-HSD1 expression in omental adipose tissue correlates with fat cell size independently of obesity (4). A causative role is suggested by the phenotype of transgenic mice which overexpress 11β-HSD1 in adipose tissue (5). These mice develop all the major features of metabolic syndrome including central obesity, insulin resistance, dyslipidaemia and hypertension (5, 6). Mice over-expressing 11β-HSD1 in liver also show insulin resistance and hypertension but remain lean (7). Conversely, 11β-HSD1 inhibition increases hepatic insulin sensitivity in humans (8) and its deficiency or inhibition ameliorates the metabolic consequences of obesity, increasing insulin sensitivity and reducing blood glucose levels in obese or diabetic mice (9-13).
Sequence variation in HSD11B1, the human gene encoding 11β-HSD1, has been linked with cardiovascular risk factors associated with obesity in adults, although not with obesity per se (14). HSD11B1 is transcribed from 2 promoters (Figure 1), with the P2 promoter predominating in metabolically active tissues, where it is potently regulated by the transcription factor C/EBPα (15). Polymorphisms in the P2 promoter region (rs846910) and an intronic enhancer (rs12086634) are associated with type 2 diabetes and/or hypertension in 3 different populations (16-18), and the G allele of rs12086634, associated with lower 11β-HSD1 transcriptional activity in vitro (19), may be protective against obesity amongst patients with polycystic ovary syndrome (PCOS) (20). Moreover, the combination of less common allelic variants at rs846910 and rs12086634 is associated with higher levels of 11β-HSD1 mRNA and activity in adipose tissue in southern European caucasian women with and without PCOS (Gambineri, A., Tomassoni, F., Munarini, A., Stimson, R.H., Pagotto, U., Mioni, R., Chapman, K.E., Andrew, R., Pasquali, R. and Walker, B.R., manuscript submitted). However, with the exception of rs12086634, the functional relevance of these and other non-exonic polymorphisms has not been reported.
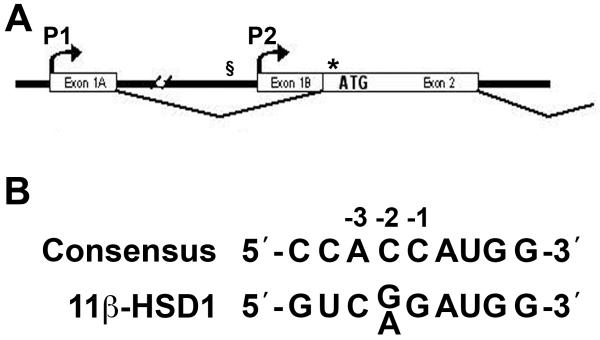
Figure 1. Locations of relevant SNPs: rs846910 at −2937 with respect to the P2 promoter and rs13306421, close to the translation start of the human HSD11B1 gene.
(A) Schematic representation of the 5′ end of the HSD11B1 gene. Exons 1A and 1B (and the associated P1 and P2 promoters, respectively) (15) are indicated, as is exon 2, containing the translation start of 11β-HSD1. The position of SNP rs846910 is indicated by § and SNP rs13306421 is indicated by *.
(B) SNP rs13306421 is located at −2 (numbering with respect to the AUG translation start codon at +1), within the ribosome binding site of 11β-HSD1 mRNA. The ribosome binding site of HSD11B1 is a poor match to the consensus sequence (21). When a pyrimidine replaces the preferred purine at position −3, translation becomes more sensitive to changes at other positions, including −2 (21).
Here we have investigated the effect of 2 polymorphisms upon 11β-HSD1 transcription and translation; rs846910 located 2937 nucleotides 5′ to the transcription start of the HSD11B1 P2 promoter, and rs13306421, a polymorphism situated at −2 with respect to the translation start site (Figure 1). The translation start of HSD11B1 lies in a sub-optimal context, with deviation from the consensus ribosome binding site (21) (Figure 1B) and with 2 additional AUG codons located close downstream. When a pyrimidine occupies position −3 (as it does in HSD11B1), translation becomes sensitive to changes at other positions, including −2 (21). Moreover, even small departures from the consensus ribosome binding site allow nearby AUG codons to be reached by leaky scanning (22). Accordingly, we have tested the effect of the rs13306421 polymorphism on activity of 11β-HSD1 and its translation in vitro.
MATERIALS AND METHODS
Plasmids
To test the effect of rs846910, the 5′-flanking region (−4643 to +88) of HSD11B1 was amplified from BAC DNA (RP1-28O10, encompassing the HSD11B1 gene, obtained from the Sanger Institute, Hinxton, UK) using primers 5′-TCCCTAGCAGAGGTTCTCCATGAGG-3′ and 5′-AGCTGGCCTGAAGACTCCTGTAGG-3′ and Accuprime Pfx (Invitrogen, Paisley, UK), a high fidelity thermostable polymerase. Sequencing confirmed the presence of the more common G allele in the cloned product. The PCR product was subcloned into pSV0L (23) and the A allele of rs846910 introduced by site-directed mutagenesis using a QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, California, USA) according to the Manufacturer’s instructions, creating plasmids pHSD11B1(−2937A)-Luc and pHSD11B1(−2937G)-Luc, differing only at the SNP at −2397. Plasmids to test the effect of rs13306421, the translation start site SNP were constructed from pBS-SK+-h11β-HSD1, encoding human 11β-HSD1 (24), a gift from Dr Perrin White. pBS-SK+-h11β-HSD1 was first digested with Eco0109I to remove a fragment of lac DNA originating from the lambda vector used to clone the cDNA. Sequencing confirmed the presence of the G allele at −2 with respect to the translation start site; this plasmid was named pBS-h11β-HSD1(G). Site-directed mutagenesis to change the G allele to A was carried out (as above), to create pBS-h11β-HSD1(A). The 2 alleles of 11β-HSD1 were subcloned into pcDNA3.1(−) (Invitrogen, Paisley, UK) using standard techniques to generate pCMV-h11β-HSD1(G) and pCMV-h11β-HSD1(A). To test whether the translation start polymorphism could affect translation of a heterologous protein, we replaced the coding sequence of 11β-HSD1 with luciferase, creating pCMV-(G)Luc and pCMV-(A)Luc, as follows: the 11β-HSD1 leader sequence with either the G or the A allele of SNP rs13306421 was amplified by PCR from pCMV-h11β-HSD1(G) and pCMV-h11β-HSD1(A) using a T7 (forward) primer and a reverse primer; either 5′-ATGGCGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCCATCCGACAGGGAG-3′ (G allele; the translation start and −2 nucleotide are underlined) or 5′-ATGGCGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCCATCTGACAGGGAG-3′ (A allele). The product included an NheI restriction enzyme site from the polylinker in pCMV-h11β-HSD1(G)/(A) and a KasI site in the reverse primer. pCMV-(G)Luc and pCMV-(A)Luc were then assembled in pcDNA3.1(−) from an NheI-KasI fragment from the PCR product and a KasI-SpeI fragment encoding luciferase from pGL3-basic (Promega, Southampton, UK). To create the N162Q glycosylation mutant, site-directed mutagenesis was used to change asparagine 162 of 11β-HSD1 to glutamine, as described above. All constructs were verified by sequencing.
Cell culture and transfection
The effect of rs846910 on HSD11B1 promoter activity was tested in A549 cells which express the endogenous HSD11B1 gene (25). A549 cells were maintained and transfected as described (25). Briefly, 1.5 × 105 cells per well of a 6 well plate were transfected using lipofectamine 2000 (Invitrogen, Paisley, UK) with 250ng pHSD11B1(−2937A)-Luc or pHSD11B1(−2937G)-Luc and 250ng pRSV-LacZ (encoding β-galactosidase, as internal control). Where appropriate, 50ng pMSV-C/EBPα (gift from S. McKnight) was added. 48h after transfection, cells were lysed and luciferase and β-galactosidase activity measured as previously described (15). Transfections were carried out in triplicate with at least 2 independent preparations of each plasmid and the experiment was repeated 7 times.
The effect of rs13306421 on enzyme activity of 11β-HSD1 in mammalian cells was tested in chinese hamster ovary (CHO) cells, which do not express endogenous 11β-HSD1 activity (26, 27). CHO cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100IU/mL penicillin and 100 g/mL streptomycin. Twenty-four hours prior to transfection, cells were seeded at 1.5x105 cells/well in a 6-well plate and transfected with 500ng DNA using lipofectamine 2000 (Invitrogen, Paisley, UK), according to the manufacturer’s instructions. For assay of 11β-HSD1 activity, cells were transfected with 250ng H6PD expression plasmid (27) and 250ng pCMV-h11β-HSD1(G) or pCMV-h11β-HSD1(A). Controls contained pcDNA3.1(−) (“empty vector”) instead of the H6PD plasmid, the pCMV-h11β-HSD1 plasmid, or both. 11β-HSD1 reductase activity was measured by incubating intact cells with 200nM cortisone containing 5nM [3H]-cortisone tracer for 16–24h (preliminary experiments established that conversion was linear up to at least 24h and the time point used for each assay gave 20–40% conversion of added substrate). After incubation, steroids were extracted with ethyl acetate, separated by thin layer chromatography using a mobile phase of chloroform and ethanol (92:8) and quantitated using a phosphorimager (Fuji FLA-2000, Raytek Scientific Ltd, Sheffield UK). For assay of luciferase activity, cells were transfected with 250ng pRSV-LacZ (to monitor transfection efficiency) together with 250ng either pCMV-(G)Luc or pCMV-(A)Luc. 48h after transfection, cells were lysed and luciferase and β-galactosidase activity measured as above. All transfections were carried out in triplicate and for each plasmid (A or G allele), at least 3 independent plasmid preparations were also used. Transfections were repeated 4–6 times. Data were normalised between experiments by arbitrarily setting the mean value of the G allele to 100%.
Immunoblotting was carried out on CHO cells transfected as above, but with 5x105 cells/well seeded in a 6-well plate and 1μg pCMV-h11β-HSD1(G) or pCMV-h11β-HSD1(A) DNA (3 independent preparations of each plasmid). Cells were harvested 48h after transfection, lysed in Laemmli buffer and proteins (50μg) separated by electrophoresis on a discontinuous 15% SDS-polyacrylamide resolving gel. Proteins were transferred to Hybond nitrocellulose membrane (GE Healthcare Life Sciences, Little Chalfont, UK) at 200mA for 2h. Membranes were blocked with 5% milk block (Biorad, Hemel Hempstead, UK) in TBST (0.015M Tris, 0.15M NaCl, 0.1% Tween 20, pH7.4) for 1h at room temperature followed by overnight incubation at 4°C with 1:1000 dilution of sheep anti-human 11β-HSD1 antibody (The Binding Site Ltd, Birmingham, UK). Membranes were washed in TBST then incubated with Alexa Fluor 680-labelled donkey anti-sheep antibody (Invitrogen) used at 1:10,000 dilution. Immunolabeling was detected and quantified using an Odyssey infrared imager and Odyssey software (LI-COR Biosciences UK, Cambridge, UK).
In vitro translation
The mRNA encoding either the A or the G allele of rs13306421 was transcribed and translated in vitro from pBS-h11β-HSD1(G)/(A) or pCMV-h11β-HSD1(G)/(A) using the TNT T7 Quick Coupled Transcription/Translation System (Promega), according to the manufacturer’s instructions. Three independent plasmid preps for each allele were tested. Reactions (50μl) contained 1μg template DNA, 20μCi 35S-methionine (37.0TBq/mmol; Perkin Elmer, Beaconsfield, UK), 1μl T7 TNT PCR enhancer (supplied with the kit), 40μl TNT Quick Master Mix (supplied with the kit). Positive controls contained 1μg of a plasmid encoding luciferase (supplied with the kit); negative controls omitted DNA template. Reactions to investigate glycosylated 11β-HSD1 isoforms were carried out using pCMV-h11β-HSD1(A) or the N162Q mutant of 11β-HSD1 as template, with or without inclusion of 4μl canine pancreatic microsomes (Promega). Proteinase K sensitivity was tested by incubating aliquots of the in vitro translation reactions with 0.2mg/ml proteinase K on ice for 30min, in the presence or absence of 0.5% Triton X-100. To inactivate proteinase K, 1mM PMSF was added, followed by heating at 100 °C, 5 min in SDS loading buffer (50mM TrisHCl pH 6.8, 100mM DTT, 2% SDS, 0.1% Bromophenol blue, 10% glycerol). Proteins were separated by SDS-PAGE in either 15% or 10% discontinuous polyacrylamide gels. Gels were fixed for 30 min in 50% methanol, 10% glacial acetic acid followed by 5 min in 10% glycerol, then dried and exposed to autoradiographic film or to a phosphorimager screen. Quantification was carried out using a phosphoimager (Fuji BAS FLA-2000, Raytek, Sheffield, UK) and Aida 3.44 software (Raytek, Sheffield, UK).
Genotyping of SNP rs13306421 within a population of Southern European women with and without PCOS
Three hundred unmedicated caucasian women with PCOS, aged 18–45 yr, and 300 caucasian controls recruited from the general population in Nothern Italy and comparable for age and body weight were genotyped for rs13306421. PCOS was diagnosed according to the Rotterdam consensus conference criteria (28). Controls had no signs of hyperandrogenism and regular ovulatory menstrual cycles (29).
Blood samples for DNA extraction were collected in EDTA and stored at 4°C. DNA was extracted using a QIAamp DNA Blood Kit (Qiagen Inc., Valencia, CA). Genotyping of rs13306421 was undertaken by single nucleotide primer extension (SNuPE) and DHPLC, adapted from (30). HSD11B1 gene fragments were amplified by PCR for 35 cycles, each consisting of 30s at 95°C, 30s at 65°C and 20s at 72°C. Primers for rs13306421 were 5′-GCTGCCTGCTTAGGAGGTTGTAG-3′ (forward) and 5′-AACACATCTTGGTCCTCAGGAACAC-3′ (reverse). Reactions (10βl) contained 25pmol each primer, 40ng genomic DNA, 200βM dNTPs, 2mM MgCl2 and 0.08U AmpliTaq Gold (Applied Biosystems, Warrington, UK) in the buffer provided by the manufacturer. PCR products were incubated for 60 min at 37°C with 2βl of Exo-SAP-IT (GE Healthcare Europe GmbH, Milan, Italy) to hydrolyze unincorporated nucleotides and degrade excess primers; the reaction was terminated by incubation at 80°C for 15 min. Primer extension reactions were carried out in a 25βl reaction containing the purified PCR products, 50βM of the appropriate ddNTPs (ddATP, ddGTP, ddCTP), 200pmol primer (5′-TTTTTTTTTTTTTTTTGGAGTCTTCAGGCCAGCTCCCTGTC-3′) and 1.25U Thermo Sequenase (GE Healthcare Europe GmbH, Milan, Italy) in the buffer provided by the manufacturer. SNuPE reactions were performed in a thermal cycler with 35 cycles each consisting of 30s at 94°C, 15s at 55°C, 60s at 60°C, followed by 2s at 15°C. Primer extension products (10βl) were loaded on SaraSep DNASep column (Transgenomic, Glasgow, UK) at 80°C and separated by DHPLC on a Wave system (Transgenomic, Glasgow, UK) using a linear acetonitrile gradient (over 7 min from 20–37% acetonitrile in 0.1M triethylamine acetate buffer, pH 7) at a constant flow rate of 0.9ml/min. Data were acquired using a UV-detector at 260nm.
RESULTS
SNP rs846910, located 2937 nucleotides 5′ to the translation start of HSD11B1, does not influence HSD11B1 promoter activity
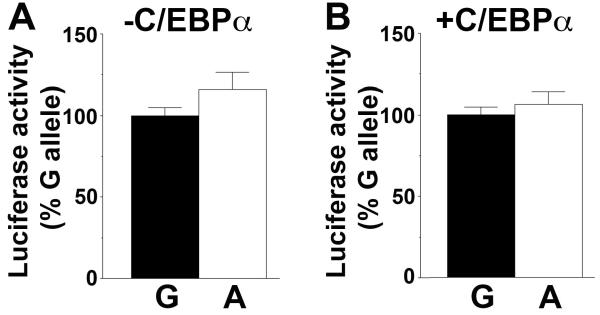
A549 (human lung epithelial) cells in which the endogenous HSD11B1 promoter is active (15) were used to test the effect of allelic variation at −2937. Luciferase reporter plasmids encoding −4643 to +88 of HSD11B1, differing only by A/G at −2937, showed similar promoter activity in A549 cells (Figure 2A). Moreover, co-transfection of C/EBPα, a transcription factor important for expression of 11β-HSD1 in some tissues (15) but absent from A549 cells (25), did not differentially regulate the A and G alleles of rs846910 (Figure 2B) although it did markedly increase HSD11B1 promoter activity (fold induction with C/EBPα, mean ± sem: A allele, 8.1±1.0 vs G allele, 8.9±1.8; p=0.7, n=7 experiments, each with ≥6 replicates), consistent with previous data (15).
Figure 2. Allelic variation at rs846910 does not affect HSD11B1 P2 promoter activity.
(A and B) Site-directed mutagenesis was used to change G at −2937 (with respect to the transcription start site of the P2 promoter, +1) to A, generating reporter plasmids pHSD11B1(−2937G)-Luc and pHSD11B1(−2937A)-Luc. Following transfection into A549 cells there was no difference in luciferase activity directed by pHSD11B1(−2937G)-Luc encoding the G allele (black bars) and pHSD11B1(−2937A)-Luc, encoding the A allele (white bars), either in the absence (A) or in the presence (B) of C/EBPα. Data are expressed relative to the more common G allele, arbitrarily set to 100% for each experiment and are means ± SEM from 7 independent transfections, each carried out in triplicate and with ≥2 independent plasmid preps of either the A allele or the G allele. Significance was tested with unpaired Students t-test.
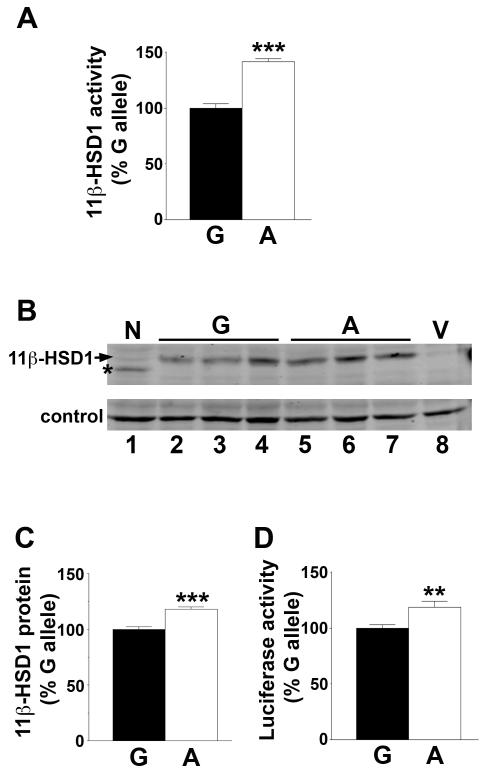
The A allele of SNP rs13306421 is associated with higher 11β-HSD1 activity and confers higher expression upon a heterologous protein
Expression plasmids encoding 11β-HSD1 cDNA, differing only by A/G at −2 with respect to the translation start site, were each co-transfected into CHO cells (which lack endogenous 11β-HSD1 activity (26, 27)) together with an expression plasmid encoding H6PD, required for reductase activity of 11β-HSD1 (27). “Empty vector” or H6PD alone gave no 11β-HSD1 reductase activity in transfected cells (data not shown). 11β-HSD1 reductase activity was higher when encoded by the A allele than the G allele (Figure 3A), suggesting that the leader sequence of HSD11B1, including the Kozak sequence, determines the translation efficiency of 11β-HSD1. Consistent with this, immunoblotting of transfected cells showed more 11β-HSD1 protein expressed from the A allele compared to the G allele (Figure 3B, C). To test whether translation of a heterologous protein was affected by the polymorphism, the leader sequence of 11β-HSD1, including the translation start, was used to drive luciferase expression. Higher luciferase activity was produced from the A allele than the G allele (Figure 3D).
Figure 3. The A allele of SNP rs13306421 is associated with higher enzyme activity.
(A) Site-directed mutagenesis was used to change G at −2 of 11β-HSD1 (with respect to the ATG translation start) to A. 11β-Reductase activity was measured by conversion of 200nM [3H]-cortisone to cortisol following co-transfection of CHO cells with expression plasmids encoding H6PD and either the G (black bar) or A allele (white bar) of 11β-HSD1. 11β-HSD1 activity was normalised between experiments by arbitrarily setting the mean conversion of the G allele plasmids to 100% for each experiment.
(B) Representative western blot showing levels of 11β-HSD1 protein encoded by 3 independent preparations of pCMV-h11β-HSD1(G) (lanes 2–4), pCMV-h11β-HSD1(A) (lanes 5–7), 11β-HSD1 N162Q (lane 1; a mutation at one of the glycosylation sites of human 11β-HSD1) or pcDNA3.1 vector (V, lane 8). 50μg protein was loaded per lane. The membrane was incubated with sheep anti-human 11β-HSD1 antibody. A cross-reacting band of ~58kDa, present in untransfected cells, was used as a convenient loading control.
(C) Quantification of immunoblot shown in (B). Levels of 11β-HSD1 are expressed relative to the loading control, with the G allele arbitrarily set to 100%. Data are means ± SEM from 3 independent transfections, with 3 independent plasmid preps of either the A allele or the G allele. Significance was tested with unpaired Student’s t-test; ***, p<0.001.
(D) The A allele directs higher expression of a heterologous protein, luciferase. The leader sequence of luciferase (to the ATG translation start site) was replaced with the 11β-HSD1 leader sequence; either the A or G allele. Luciferase activity was measured in transfected CHO cells. To normalise between experiments the mean luciferase activity of the G allele plasmid was arbitrarily set to 100% for each experiment.
Data are means ± SEM from 4–6 independent transfections, each carried out in triplicate and with 3 independent plasmid preps of either the A allele or the G allele. Significance was tested with unpaired Student’s t-test; **, p<0.01; ***, p<0.001.
The A allele of SNP rs13306421 directs more efficient translation of full-length 11β-HSD1 in vitro
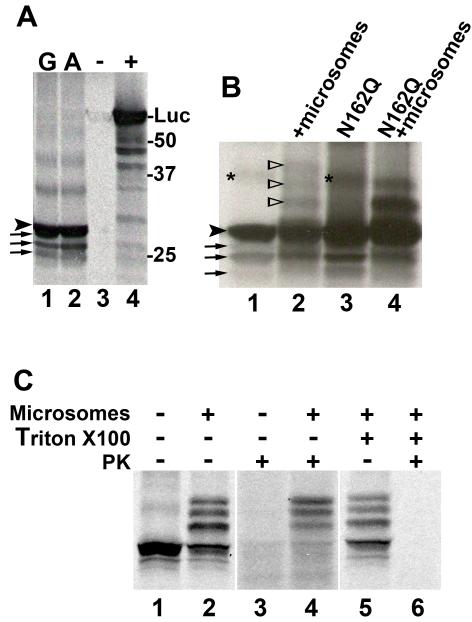
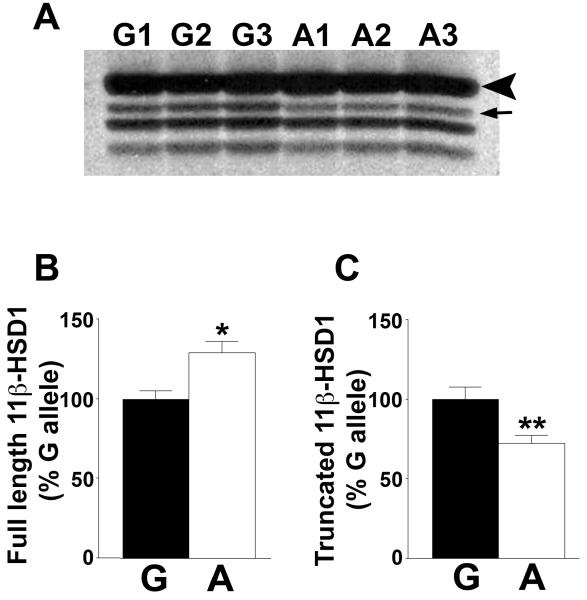
To determine whether the A allele confers higher enzyme activity through increased translation, mRNA encoding either the A allele or the G allele of rs13306421 was transcribed and translated in vitro. Four products were obtained with both the A and the G allele, with the largest and most abundant migrating with a relative molecular size of ~29kDa (Figure 4, and see also Figure 5), smaller than the predicted Mr of 32.4kDa, but corresponding to the size previously observed for in vitro translated human 11β-HSD1 (31). This suggested that truncated 11β-HSD1 was synthesised in addition to the full length, giving rise to the smaller products. Addition of unlabelled methionine together with 35S-labelled methionine had no effect on the pattern of labelled products (data not shown) indicating that they were not due to C-terminal truncation because of limiting amounts of methionine in the reactions. The N-terminus of 11β-HSD1 determines the location and orientation of 11β-HSD1 in the endoplasmic reticulum (31), and N-terminally truncated proteins would not be predicted to translocate into microsomes. To test this, in vitro transcription-translation was carried out in the presence of microsomes, which results in glycosylation of 29kDa human 11β-HSD1 at 3 sites, N123, N162 and N207 (31). Addition of microsomes resulted in the appearance of 3 products of ~31, 33 and 35kDa (Figure 4B; Lanes 1 and 2), consistent with previous reports of glycosylated products of 31, 33 and 35kDa (31, 32). Moreover, when a mutant 11β-HSD1 in which one of the 3 glycosylation sites was mutated to glutamine (N162Q) was translated in vitro, only 2 additional products were produced in the presence of microsomes (Figure 4B), consistent with the effect of the mutation in transfected cells (lane 1, Figure 3B). The mutation had no effect on the products in the absence of microsomes. Whereas the 31, 33 and 35kDa proteins were protected from proteinase K digestion, the 29kDa and smaller products were fully digested by proteinase K (Figure 4C; Lanes 3 and 4), supporting their exclusion from microsomes. Addition of Triton X100 to solubilise membranes rendered all the 11β-HSD1 products susceptible to proteinase K digestion (Figure 4C; Lanes 5 and 6). Quantification of in vitro transcription-translation products from 3 independent plasmid preps of each of the G allele and A allele of rs13306421 showed a significant increase in the amount of full length 11β-HSD1 produced from the A allele, with a concomitant decrease in the amount of the largest truncated product (Figure 5). Overall, the ratio of full length 11β-HSD1 to total truncated 11β-HSD1 (amount of full lenth/the sum of the 3 truncated products) was significantly lower for the G allele than for the A allele of 11β-HSD1 (A allele, 3.29 ± 0.09 vs G allele, 2.23 ± 0.13; p<0.05).
Figure 4. The A allele of rs13306421 produces more full-length 11β-HSD1 in coupled in vitro transcription-translation reactions.
(A) Representative autoradiograph showing 35S-labelled products of in vitro transcription-translation of 11β-HSD1. Reactions contained 1 μg of plasmid encoding the G allele (lane 1) or the A allele (lane 2) of rs13306421, vector (negative control, indicated by - above the lane) or luciferase (positive control, indicated by + above the lane). An arrowhead indicates full-length 11β-HSD1 migrating at ~29kDa (relative to molecular weight markers, indicated at the right of the gel) and arrows indicate the 3 truncated products obtained from the 11β-HSD1 plasmids. Luciferase (Mr, 61kDa) is indicated (Luc).
(B) Representative autoradiograph showing 35S-labelled products of in vitro translation reactions carried out with plasmids encoding 11β-HSD1 (lanes 1 and 2; A allele of rs13306421) or 11β-HSD1 with a mutation in one of the glyosylation sites, N162Q (lanes 3 and 4) in the absence (lanes 1 and 3) or the presence (lanes 2 and 4) of microsomes. An arrowhead indicates full-length 11β-HSD1 and arrows indicate the 3 truncated products obtained from the 11β-HSD1 plasmids. Open arrowheads indicate the positions of glycosylated 11β-HSD1. An asterisk indicates a non-specific band.
(C) Representative autoradiograph showing 35S-labelled products of in vitro transcription-translation of 11β-HSD1 carried out in the presence or absence of microsomes, Triton X100 and proteinase K (PK) (indicated above the lanes).
Unglycosylated 11β-HSD1 was susceptible to proteinase K digestion, whereas the glycosylated products were protected (lanes 3, 4). Addition of Triton X100 to solubilise microsomal membranes rendered all products susceptible to PK digestion (lanes 5, 6).
Figure 5. The A allele of rs13306421 generates a greater proportion of full length 11β-HSD1 in vitro.
(A) Representative autoradiograph showing 35S-labelled products of in vitro transcription-translation of 11β-HSD1 carried out with 3 independent preparations of each of the A and G allele of rs13306421 (indicated above the lanes; G1–G3 and A1–A3). An arrowhead indicates full length 11β-HSD1 and the arrow indicates the largest of the truncated products.
(B and C) Quantification by phosphorimager showing more full-length non-glycosylated 11β-HSD1 produced from the A allele (white bars) than the G allele (black bars) of rs13306421 (B), but less of the largest truncated product from the A allele (C). Levels of the 2 smaller truncated products did not differ significantly between the A and G alleles. Data are means ± sem (n=3) and are expressed relative to the normal G allele, arbitrarily set to 100%. Significance was tested with unpaired Student’s t-test; *, p<0.05, ** p<0.01.
The A allele of SNP rs13306421 is present at <1% in a Southern European population of women with and without PCOS
The 600 women genotyped (300 PCOS and 300 controls comparable for age and body weight) were found to be homozygous for the G allele at rs13306421. Accordingly, the frequency of the A allele is considerably less than 1% in this population.
DISCUSSION
We find no functional consequences of allelic variation at rs846910, located in the 5′ flanking region of the P2 promoter of HSD11B1 in a context where it could alter promoter activity. Thus, despite its association with metabolic phenotype (16-18), this polymorphism does not appear to directly influence 11β-HSD1 expression, although it remains possible that it modifies response to stimuli not tested here, for example TNFα. The lack of effect on promoter activity is consistent with our recent data showing no independent association of rs846910 with metabolic phenotype in the same Southern European population of women as examined here (Gambineri, A., Tomassoni, F., Munarini, A., Stimson, R.H., Pagotto, U., Mioni, R., Chapman, K.E., Andrew, R., Pasquali, R. and Walker, B.R., manuscript submitted). SNP rs846910 is unlikely to affect the potential binding site of a transcription factor as no sites for mammalian transcription factors were predicted in this region (using the AliBaba2 programme, which uses binding sites in TRANSFAC; www.gene-regulation.com). Moreover, rs846910 lies outside of the region that is highly conserved between human and mouse, so is unlikely to be in an important conserved regulatory region, although it may exert human-specific regulation. However, it may be in linkage disequilibrium with another, functional, polymorphism that accounts for the association with metabolic phenotype. Alternatively, the effect of the polymorphism on promoter activity may have been too small to detect in the current study, but might become apparent in combination with other functional polymorphisms. Indeed, in our recent study, the combination of less common alleles at both rs846910 and at rs12086634, a polymorphism previously shown to influence transcription (19), was associated with more prevalent Metabolic Syndrome and higher adipose 11β-HSD1 activity (Gambineri, A., Tomassoni, F., Munarini, A., Stimson, R.H., Pagotto, U., Mioni, R., Chapman, K.E., Andrew, R., Pasquali, R. and Walker, B.R., manuscript submitted).
The reported polymorphism rs13306421, close to the translation start of 11β-HSD1, modulates translation of the enzyme, with the A allele resulting in greater enzyme expression in transfected cells and in in vitro translation reactions, with a higher ratio of full-length enzyme to truncated products synthesised in vitro. The G allele results in less full length 11β-HSD1 translated in vitro, but more of the truncated products. The latter finding suggests that the A allele increases translation initiation at the ATG initiator codon, concomitantly decreasing “leaky scanning” where translation initiates at nearby internal AUG codons rather than the AUG translation start codon (22). The N-terminus of 11β-HSD1 is required for its insertion into the endoplasmic reticulum membrane (31) and N-terminally truncated 11β-HSD1 is enzymatically inactive (33, 34). Accordingly, the truncated products of “leaky scanning” are predicted to be enzymatically inactive. The context of the initiator AUG dictates the extent to which leaky scanning occurs (22). The translation start of 11β-HSD1 is both in a sub-optimal context (deviating from the consensus sequence at all 3 of the critical nucleotides immediately preceding the start codon) and has several nearby AUG codons (Met4, Met16 and Met31, being the closest). According to the Kozak rules (21), translation initiation is sensitive to the base pair at −2 when the −3 nucleotide is a pyrimidine (as in HSD11B1) rather than the optimal purine. Thus, the A allele of rs13306421 may favour translation initiation at Met1, concomitantly decreasing initiation from downstream AUGs. Only the fully glycosylated full-length protein was detected in transfected cells. It is likely that any truncated proteins would be subject to rapid degradation.
Whether the polymorphism rs13306421 is truly a polymorphism or may more properly be called a mutation is an open question. We failed to find a single case of the A allele in our screen of 600 subjects. It is therefore highly unlikely that this allele contributes to the alterations in 11β-HSD1 activity associated with obesity and diabetes. Furthermore, although the A allele was reported at 0.5–1% in the original African American and Japanese populations (http://www.jmdbase.jp/snp_info.asp?targetkey=imcj-snp&keyword=JMDBase_000702), in a further study, genotyping of a total of 210 individuals in four different populations (Nigeria, Japan, China, Europe) detected only the G allele (HapMap database). Thus, further studies in other populations are required to confirm whether the A allele is indeed a polymorphism. Nevertheless, our studies show the clear potential to influence levels of active 11β-HSD1.
ACKNOWLEDGEMENTS
We thank Perrin White for providing a plasmid encoding human 11β-HSD1 cDNA and Federica Tomassoni for genotyping.
GRANT SUPPORT: This work has been supported by a Wellcome Trust Programme grant, a BHF Programme grant, and the Sixth EC Program grant (LSHM-CT-2003-503041).
ABBREVIATIONS
- 11β-HSD1
11β-hydroxysteroid dehydrogenase type 1
- PAGE
polyacrylamide gel electrophoresis
- SNP
single nucleotide polymorphism
Footnotes
DISCLOSURES: ELVM, VK, NN, AG, RD, UP, RP and KEC have nothing to declare. BRW is an inventor on patents concerning 11β-HSD1 inhibitors owned by the University of Edinburgh.
STATEMENT: This is an un-copyedited author manuscript copyrighted by The Endocrine Society. This may not be duplicated or reproduced, other that for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright owner, The Endocrine Society. From the time of acceptance following peer review, the full text of this manuscript is made freely available by The Endocrine Society at http://www.endojournals.org/. The final copy edited article can be found at http://www.endojournals.org/. The Endocrine Society disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The citation of this article must include the following information: author(s), article title, journal title, year of publication, and DOI.
REFERENCES
- 1.Tomlinson JW, Walker EA, Bujalska IJ, Draper N, Lavery GG, Cooper MS, Hewison M, Stewart PM. 11β-hydroxysteroid dehydrogenase type 1: a tissue-specific regulator of glucocorticoid response. Endocr Rev. 2004;25:831–866. doi: 10.1210/er.2003-0031. [DOI] [PubMed] [Google Scholar]
- 2.Walker BR. Extra-adrenal regeneration of glucocorticoids by 11β-hydroxysteroid dehydrogenase type 1: physiological regulator and pharmacological target for energy partitioning. Proc Nutr Soc. 2007;66:1–8. doi: 10.1017/S002966510700523X. [DOI] [PubMed] [Google Scholar]
- 3.Seckl JR, Morton NM, Chapman KE, Walker BR. Glucocorticoids and 11β-hydroxysteroid dehydrogenase in adipose tissue. Recent Prog Horm Res. 2004;59:359–393. doi: 10.1210/rp.59.1.359. [DOI] [PubMed] [Google Scholar]
- 4.Michailidou Z, Jensen MD, Dumesic DA, Chapman KE, Seckl JR, Walker BR, Morton NM. Omental 11β-hydroxysteroid dehydrogenase 1 correlates with fat cell size independently of obesity. Obesity. 2007;15:1155–1163. doi: 10.1038/oby.2007.618. [DOI] [PubMed] [Google Scholar]
- 5.Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, Flier JS. A transgenic model of visceral obesity and the metabolic syndrome. Science. 2001;294:2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- 6.Masuzaki H, Yamamoto H, Kenyon CJ, Elmquist JK, Morton NM, Paterson JM, Shinyama H, Sharp MG, Fleming S, Mullins JJ, Seckl JR, Flier JS. Transgenic amplification of glucocorticoid action in adipose tissue causes high blood pressure in mice. J Clin Invest. 2003;112:83–90. doi: 10.1172/JCI17845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson JM, Morton NM, Fiévet C, Kenyon CJ, Holmes MC, Staels B, Seckl JR, Mullins JJ. Metabolic syndrome without obesity: Hepatic overexpression of 11β-hydroxysteroid dehydrogenase type 1 in transgenic mice. Proc Natl Acad Sci U.S.A. 2004;101:7088–7093. doi: 10.1073/pnas.0305524101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker BR, Connacher AA, Lindsay RM, Webb DJ, Edwards CRW. Carbenoxolone increases hepatic insulin sensitivity in man: a novel role for 11-oxosteroid reductase in enhancing glucocorticoid receptor activation. J Clin Endocrinol Metab. 1995;80:3155–3159. doi: 10.1210/jcem.80.11.7593419. [DOI] [PubMed] [Google Scholar]
- 9.Kotelevtsev Y, Holmes MC, Burchell A, Houston PM, Schmoll D, Jamieson P, Best R, Brown R, Edwards CRW, Seckl JR, Mullins JJ. 11β-hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid inducible responses and resist hyperglycaemia on obesity or stress. Proc Natl Acad Sci U.S.A. 1997;94:14924–14929. doi: 10.1073/pnas.94.26.14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morton NM, Holmes MC, Fiévet C, Staels B, Tailleux A, Mullins JJ, Seckl JR. Improved lipid and lipoprotein profile, hepatic insulin sensitivity, and glucose tolerance in 11β-hydroxysteroid dehydrogenase type 1 null mice. J Biol Chem. 2001;276:41293–41300. doi: 10.1074/jbc.M103676200. [DOI] [PubMed] [Google Scholar]
- 11.Alberts P, Engblom L, Edling N, Forsgren M, Klingstrom G, Larsson C, Ronquist-Nii Y, Ohman B, Abrahmsen L. Selective inhibition of 11β-hydroxysteroid dehydrogenase type 1 decreases blood glucose concentrations in hyperglycaemic mice. Diabetologia. 2002;45:1528–1532. doi: 10.1007/s00125-002-0959-6. [DOI] [PubMed] [Google Scholar]
- 12.Alberts P, Nilsson C, Selen G, Engblom LO, Edling NH, Norling S, Klingstrom G, Larsson C, Forsgren M, Ashkzari M, Nilsson CE, Fiedler M, Bergqvist E, Ohman B, Bjorkstrand E, Abrahmsen LB. Selective inhibition of 11β-hydroxysteroid dehydrogenase type 1 improves hepatic insulin sensitivity in hyperglycemic mice strains. Endocrinology. 2003;144:4755–4762. doi: 10.1210/en.2003-0344. [DOI] [PubMed] [Google Scholar]
- 13.Morton NM, Paterson JM, Masuzaki H, Holmes MC, Staels B, Fievet C, Walker BR, Flier JS, Mullins JJ, Seckl JR. Novel adipose tissue-mediated resistance to diet-induced visceral obesity in 11 β-hydroxysteroid dehydrogenase type 1-deficient mice. Diabetes. 2004;53:931–938. doi: 10.2337/diabetes.53.4.931. [DOI] [PubMed] [Google Scholar]
- 14.Draper N, Echwald SM, Lavery GG, Walker EA, Fraser R, Davies E, Sorensen TI, Astrup A, Adamski J, Hewison M, Connell JM, Pedersen O, Stewart PM. Association studies between microsatellite markers within the gene encoding human 11β-hydroxysteroid dehydrogenase type 1 and body mass index, waist to hip ratio, and glucocorticoid metabolism. J Clin Endocrinol Metab. 2002;87:4984–4990. doi: 10.1210/jc.2001-011375. [DOI] [PubMed] [Google Scholar]
- 15.Bruley C, Lyons V, Worsley AG, Wilde MD, Darlington GD, Morton NM, Seckl JR, Chapman KE. A novel promoter for the 11β-hydroxysteroid dehydrogenase type 1 gene is active in lung and is C/EBPα independent. Endocrinology. 2006;147:2879–2885. doi: 10.1210/en.2005-1621. [DOI] [PubMed] [Google Scholar]
- 16.Nair S, Lee YH, Lindsay RS, Walker BR, Tataranni PA, Bogardus C, Baier LJ, Permana PA. 11β-Hydroxysteroid dehydrogenase Type 1: genetic polymorphisms are associated with Type 2 diabetes in Pima Indians independently of obesity and expression in adipocyte and muscle. Diabetologia. 2004;47:1088–1095. doi: 10.1007/s00125-004-1407-6. [DOI] [PubMed] [Google Scholar]
- 17.Franks PW, Knowler WC, Nair S, Koska J, Lee YH, Lindsay RS, Walker BR, Looker HC, Permana PA, Tataranni PA, Hanson RL. Interaction between an 11βHSD1 gene variant and birth era modifies the risk of hypertension in Pima Indians. Hypertension. 2004;44:681–688. doi: 10.1161/01.HYP.0000144294.28985.d5. [DOI] [PubMed] [Google Scholar]
- 18.Morales MA, Carvajal CA, Ortiz E, Mosso LM, Artigas RA, Owen GI, Fardella CE. Possible pathogenetic role of 11β-hydroxysteroid dehydrogenase type 1 (11βHSD1) gene polymorphisms in arterial hypertension. Rev Med Chil. 2008;136:701–710. [PubMed] [Google Scholar]
- 19.Draper N, Walker EA, Bujalska IJ, Tomlinson JW, Chalder SM, Arlt W, Lavery GG, Bedendo O, Ray DW, Laing I, Malunowicz E, White PC, Hewison M, Mason PJ, Connell JM, Shackleton CH, Stewart PM. Mutations in the genes encoding 11β-hydroxysteroid dehydrogenase type 1 and hexose-6-phosphate dehydrogenase interact to cause cortisone reductase deficiency. Nat Genet. 2003;34:434–439. doi: 10.1038/ng1214. [DOI] [PubMed] [Google Scholar]
- 20.Gambineri A, Vicennati V, Genghini S, Tomassoni F, Pagotto U, Pasquali R, Walker BR. Genetic variation in 11β-hydroxysteroid dehydrogenase type 1 predicts adrenal hyperandrogenism among lean women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2006;91:2295–2302. doi: 10.1210/jc.2005-2222. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc Natl Acad Sci U S A. 1995;92:2662–2666. doi: 10.1073/pnas.92.7.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Wet JR, Wood KV, DeLuca M, Helsinki DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tannin GM, Agarwal AK, Monder C, New MI, White PC. The human gene for 11β-hydroxysteroid dehydrogenase. J Biol Chem. 1991;266:16653–16658. [PubMed] [Google Scholar]
- 25.Sai S, Esteves CL, Kelly V, Michailidou Z, Anderson K, Coll AP, Nakagawa Y, Ohzeki T, Seckl JR, Chapman KE. Glucocorticoid regulation of the promoter of 11β-hydroxysteroid dehydrogenase type 1 is indirect and requires C/EBPβ. Mol Endocrinol. 2008;22:2049–2060. doi: 10.1210/me.2007-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal AK, Monder C, Eckstein B, White PC. Cloning and expression of rat cDNA encoding corticosteroid 11β-dehydrogenase. J Biol Chem. 1989;264:18939–18943. [PubMed] [Google Scholar]
- 27.Bujalska IJ, Draper N, Michailidou Z, Tomlinson JW, White PC, Chapman KE, Walker EA, Stewart PM. Hexose-6-phosphate dehydrogenase confers oxo-reductase activity upon 11β-hydroxysteroid dehydrogenase type 1. J Mol Endocrinol. 2005;34:675–684. doi: 10.1677/jme.1.01718. [DOI] [PubMed] [Google Scholar]
- 28.ESHRE/ASMR-sponsored PCOS consensus workshop group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19:41–47. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 29.Nestler JE, Jakubowicz DJ, Evans WS, Pasquali R. Effects of metformin on spontaneous and clomiphene-induced ovulation in the polycystic ovary syndrome. N Engl J Med. 1998;338:1876–1880. doi: 10.1056/NEJM199806253382603. [DOI] [PubMed] [Google Scholar]
- 30.Hoogendoorn B, Owen MJ, Oefner PJ, Williams N, Austin J, O’Donovan MC. Genotyping single nucleotide polymorphisms by primer extension and high performance liquid chromatography. Hum Genet. 1999;104:89–93. doi: 10.1007/s004390050915. [DOI] [PubMed] [Google Scholar]
- 31.Odermatt A, Arnold P, Stauffer A, Frey BM, Frey FJ. The N-terminal anchor sequences of 11β-hydroxysteroid dehydrogenases determine their orientation in the endoplasmic reticulum membrane. J Biol Chem. 1999;274:28762–28770. doi: 10.1074/jbc.274.40.28762. [DOI] [PubMed] [Google Scholar]
- 32.Blum A, Martin HJ, Maser E. Human 11β-hydroxysteroid dehydrogenase type 1 is enzymatically active in its nonglycosylated form. Biochem Biophys Res Commun. 2000;276:428–434. doi: 10.1006/bbrc.2000.3491. [DOI] [PubMed] [Google Scholar]
- 33.Mercer W, Obeyesekere V, Smith R, Krozowski Z. Characterization of 11β-HSD1B gene expression and enzymatic activity. Mol Cell Endocrinol. 1993;92:247–251. doi: 10.1016/0303-7207(93)90015-c. [DOI] [PubMed] [Google Scholar]
- 34.Obeid J, Curnow KM, Aisenberg J, White PC. Transcripts originating in intron-1 of the HSD11 (11β-hydroxysteroid dehydrogenase) gene encode a truncated polypeptide that is enzymatically inactive. Mol Endocrinol. 1993;7:154–160. doi: 10.1210/mend.7.2.8469231. [DOI] [PubMed] [Google Scholar]