Abstract
Cancer cells remodel their metabolic programmes to meet the requirements of rapid proliferation. Glutaminase (GLS1) is a mitochondrial enzyme that converts glutamine to glutamate. Our aim was to investigate, for the first time, GLS1 protein expression in colorectal cancer and to evaluate its clinical significance. Immunohistochemical analysis was performed on tissue microarrays containing pairs of cancer and adjacent normal tissues from colorectal cancer patients (n=257). The expression of GLS1 protein in normal colonic tissues and colorectal cancer was measured by western blotting. Proliferation and cell death were evaluated in colorectal cancer cell lines after GLS1 inhibitor treatment. Compared with normal tissues (18.15%), we observed that the expression of GLS1 increased significantly in colorectal cancer (80.24%; P<0.0001) by immunohistochemical analysis, and the elevation of GLS1 protein expression levels in fresh colorectal cancer samples versus normal colonic tissues were also observed by western blotting. Furthermore, GLS1 expression levels were significantly associated with deeper tumour infiltration (P=0.0002), and the pathological pattern of tubular adenocarcinoma (p=0.0008). In addition, treatment with the GLS1 inhibitor suppressed proliferation and induced apoptosis in HT29 and SW480 cell lines. These results suggest that the expression of GLS1 is upregulated and correlates with clinicopathological factors in colorectal cancer. GLS1 exhibits functional importance in colon cancer tumorigenesis. Moreover, GLS1 may serve as a target for colorectal cancer therapy.
Keywords: Colorectal cancer, glutaminase, tumorigenesis
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide. Many studies have demonstrated that altered metabolism is a hallmark of cancer cells [1], and both glutamine uptake and the rate of glutaminolysis are increased in tumours [2]. This relationship suggests that we could utilise the specificity of tumour metabolism to the benefit of cancer patients.
Glutamine is considered to be non-essential under normal condition but becomes semi-essential in critically ill patients [3] and essential when culturing cells in vitro [2,4,5]. Some types of cancer cells cannot survive without exogenous glutamine, which is termed “glutamine addiction” [6]. This dependence is due to the diverse effects of glutamine metabolism in cancer cells. Glutamine plays a vital role in multiple metabolic processes, including the biosynthesis of nucleotides, hexosamine, glutathione and reducing equivalents, serving as a nonessential amino acid and respiratory substrate, and participating in ammoniagenesis and glycosylation reactions. Additionally, oncogenes, such as Myc, Ras, Rho GTPases and p53, have been implicated in the regulation of glutamine metabolism [7]. Myc, as a proto-oncogene, stimulates the uptake and catabolism of glutamine [8]. Activation of the Rho GTPase signalling pathway can up-regulate glutaminase activity in an NF-kB-dependent manner in cancer cells [9]. Utilisation of glutamine is increased in support of both the TCA cycle and biosynthesis in K-ras-transformed cells [10]. Glutamine metabolism is also influenced by the mammalian target of rapamycin (mTOR) pathway [11] and the extracellular signal-regulated protein kinase (ERK) signalling pathway [12,13], which were known to be involved in tumour growth.
In glutamine metabolism, mitochondrial glutaminase (GLS) is the key enzyme in the conversion of glutamine to glutamate and is highly expressed in many tumours and cancer cell lines. In mammals, there are two different phosphate-activated GLS isoforms: GLS1 (kidney-type) and GLS2 (liver-type), which are encoded by separate genes on different chromosomes. Altered expression of GLS has been reported in cancer cells of different origins, although it is unknown whether the altered expression is a cause or an effect of neoplastic transformation. GLS1 is the main isoform in hepatoma cells [14-16], Ehrlich ascites tumour cells (EATCs) [17], breast cancer cell lines [16], and KU812F human myeloid leukaemia cells. Furthermore, GLS1 is up-regulated compared with cells from normal tissues.
Expression of GLS is up-regulated in various cancers, including colorectal cancer cell lines [18]. GLS activity in the carcinoma cells was lower than in adenoma cells in colorectal cancer cell lines [5]. However, the alteration of GLS1 in solid colorectal tumours and its clinical significance remain unclear. In this study, we focused on the role of GLS1 in the colorectal cancer and its potential association with clinical features.
Materials and methods
Patients and tissue microarray (TMA)
Fourteen pairs of fresh tumour and paraneoplastic specimens from colorectal cancer patients were collected from the Union Hospital of Tongji Medical College of Huazhong University of Science and Technology (Hubei, China) as approved by the Human Ethics Review Board. All patients were informed and agreed with the sample collection. Forty-seven pairs of colorectal cancer and paraneoplastic normal specimens section were also obtained from Union Hospital. In addition, three colorectal cancer TMAs were used, which included 210 pairs of tumour and matched normal colonic tissue in total. The diagnosis of specimens was confirmed by immunohistochemistry, and all of the patients were staged according to the 7th AJCC stages.
Cell culture
HT29 and SW480 cells were cultured in DMEM with 4.5 g/L glucose, supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in 5% CO2.
Immunostaining for GLS1
Paraffin-embedded sections were deparaffinised in xylene and rehydrated in graded alcohol. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 10 min. Antigen retrieval was performed by autoclave sterilisation in sodium citrate buffer for 3 min, and slides were then incubated with 10% normal goat serum solution for 20 min. The rabbit polyclonal antibody directed against GLS1 (ab93434, 1:100; Abcam, USA) was added to sections and incubated at 4°C overnight. HRP-conjugated secondary antibody was used according to the manufacturer’s instructions. The slides were then incubated with DAB to visualise GLS1 expression, followed by hematoxylin counterstaining. PBS was used rather than the primary antibody as a negative control. The images were captured using a RGB JVC solid-state camera connected to an Olympus BH2 microscope. Staining results were assessed independently and blindly by two pathologists. The intensity of GLS1 staining was scored 0-3 (0, negative; 1, weak; 2, moderate; and 3, strong).
Determination of GLS1 protein by western blot analysis
Tissues were ground completely in liquid nitrogen and lysed in a radio immunoprecipitation assay buffer with a protease inhibitor cocktail for protein extraction. After incubation on ice for 45 min, the solution was centrifuged at 12000 g for 15 min at 4°C. The supernatant was collected, and the total protein concentration was detected by Bradford assay. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. After blocking in 5% non-fat milk in PBST, the membranes were incubated with primary antibody against GLS1 (1:500, Abcam, USA). A peroxidase-conjugated secondary antibody was used, and the bands were detected by ECL (Pierce). β-actin was used as an internal reference. The Gel-Pro analyser (Exon-Intron, Inc., Bob Farrell, USA) was used to quantify the bands.
Cell proliferation and viability assays
Approximately 5×10³ cells in 100 μl of DMEM were seeded into a 96-well culture plate and cultured for 24 h. After treatments, the cell number was evaluated by MTT assay. Ten microliters of MTT (5 mg/ml) was added to each well, and the cells were incubated at 37°C for 4 h. DMSO was used to dissolve the formazan, and the absorbance at 490 nm was measured using a microplate reader.
Immunofluorescence microscopy
After growing to 70% confluence, cells were treated by 6-diazo-5-oxo-L-norleucine (DON) and fixed with freshly prepared 3.7% formaldehyde at 37°C for 15 minutes. The cells were washed twice using PBS and then incubated with Hoechst (10 μg/ml) for 20 min to stain the nucleus. After washing, typical apoptotic cells with condensed nuclei were observed.
Statistical analysis
Data are expressed as the mean ± SD. Statistically significant differences were performed using Student’s t-test or chi-square test, and a P-value<0.05 was considered significant. Statistical analyses were performed by GraphPad Prism 4.02 version (GraphPad, San Diego, CA, USA).
Results
Expression of GLS1 was increased in human colorectal cancer tissues by immunohistochemistry
GLS1 expression has previously been analysed in some human tumours, but the only studies of GLS1 expression in colorectal cancer have been limited to human colon cancer cell lines. Initially, we examined GLS1 expression by immunohistochemistry of 257 colorectal cancer tissues. Representative staining is presented in Figure 1A, and most of the cancer tissues exhibited strong cytoplasmic immunoreactivity for GLS1. Furthermore, we found that GLS1 staining intensity is higher in the tumour tissues than in the normal tissues (as in Figure 1B). After excluding the falling tissues, we scored for GLS1 staining, and positivity was defined as an IHC score >0. Statistics were calculated for the GLS1 staining of remaining 248 pairs of tissue sections (as in Table 1). The result suggested that 80.24% of tumours were positive for GLS1 (n=199/248), and only 19.76% (49/248) showed negative staining. By contrast, in the adjacent normal tissues, 81.85% (203/248) exhibited a negative immune response, and 18.15% (45/248) exhibited positive reactivity. Therefore, the up-regulation of GLS1 was obvious in colorectal tumour tissues compared with normal tissues (P<0.0001).
Figure 1.
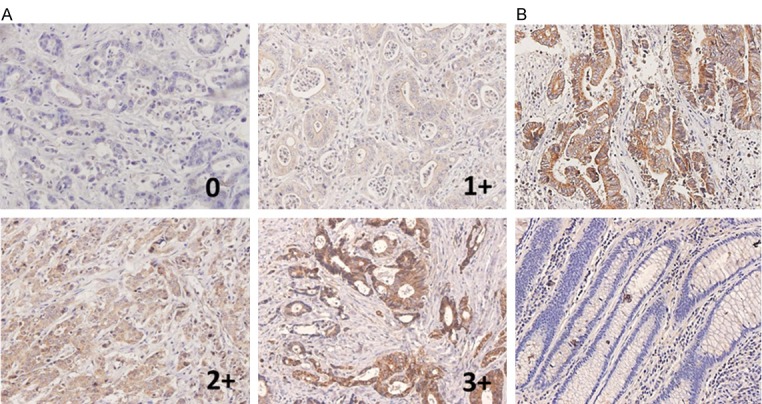
The expression of GLS1 in colorectal tumour tissues and normal adjacent tissues by immunohistochemistry. A. Representative staining of colorectal cancer sections exhibiting negative, low, moderate and high immunostaining for GLS1. B. Positive immunostaining of GLS1 in tumour (upper panel) and negative immunostaining of GLS1 in corresponding normal adjacent issues (inferior panel).
Table 1.
Analysis of GLS1 expression in colorectal tumour and normal tissues. Statistics of immunostaining for the expression of GLS1 in 248 pairs of colorectal tumour and normal tissues. The Pearson’s χ2 test was used to assess the statistical significance, and differences were significant with a P-value<0.05
| GLS1 expression | Cancer | Normal | P value |
|---|---|---|---|
| Negative | 49 (19.76%) | 203 (81.85%) | <0.0001 |
| Positive | 199 (80.24%) | 45 (18.15%) |
Expression of GLS1 protein was increased in human colorectal cancer tissues by western blotting analysis
Following analysis of GLS1 immunostaining in 257 colorectal cancer tissues, we proceeded to analyse 14 fresh pairs of colorectal cancer and adjacent normal tissues for GLS1 protein expression by western blotting. Figure 2A presents the general pattern of increased GLS1 protein expression in tumour tissues compared with adjacent normal tissues. The up-regulation of GLS1 expression was observed in most of the tumour tissues examined (n=13/14). Figure 2B demonstrates that the average GLS1 protein level in the tumour tissues was three times more than that in adjacent normal tissues (P=0.0015). Thus, this result was consistent with increased levels of GLS1 in colorectal cancer tissues by immunohistochemistry.
Figure 2.
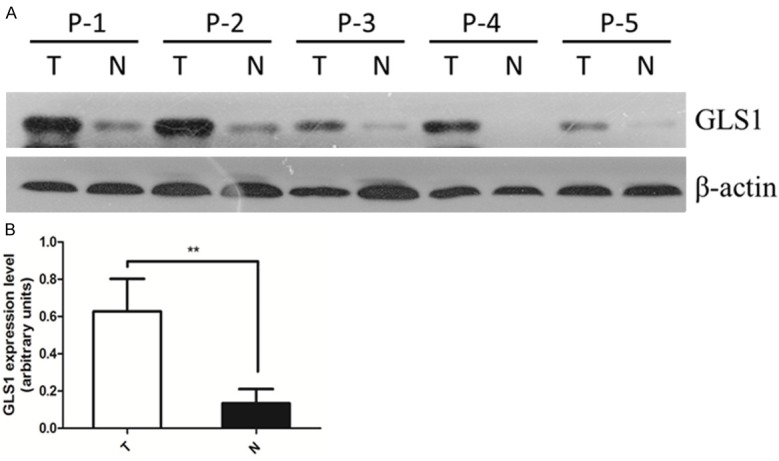
GLS1 protein expression is increased in colorectal cancer compared with normal tissues by western blotting. A. Fourteen pairs of tumour and adjacent normal tissues were analysed for GLS1 protein expression by western blotting. Five representative pairs of tumour (T) and normal (N) tissues are shown. β-actin was used as an internal loading control. B. Statistics of average GLS1 expression in 14 pairs of tumour (T) and normal (N) tissues are shown, P=0.0015**.
Correlations between the expression of GLS1 and clinicopathological factors
To determine the clinical significance of elevated GLS1 expression in colorectal cancer patients, we further examined the correlation between GLS1 protein expression and clinicopathological factors, including gender, age, clinical staging, tumour stage, lymphatic metastasis, distant metastasis and histological type of the colorectal cancer patients. After excluding the failing tissues and the cases with incomplete information, statistics and analyses were calculated for the correlations between GLS1 protein expression of remaining 237 pairs of tissue sections and the clinicopathological parameters (as in Table 2). The patients’ characteristics are presented in Table 3.
Table 2.
The relationships between the expression of GLS1 and clinicopathological factors. Statistics of immunostaining for the expression of GLS1 in 237 colorectal tumour tissues. The statistical analysis was performed using the Pearson’s χ2 test; P-value<0.05 was considered significant
| Variable0 | GLS1 expression | P value | ||
|---|---|---|---|---|
|
| ||||
| None/Low | moderate | strong | ||
| Gender | 0.1401 | |||
| Male | 23 | 40 | 61 | |
| Female | 33 | 29 | 51 | |
| Age, Y | 0.1009 | |||
| ≤59 | 25 | 31 | 35 | |
| ≥60 | 31 | 38 | 77 | |
| TNM Stage | 0.0309 | |||
| I | 7 | 11 | 5 | |
| II | 15 | 29 | 55 | |
| III | 23 | 21 | 39 | |
| IV | 11 | 8 | 13 | |
| T stage | 0.0002 | |||
| T1/2 | 10 | 14 | 7 | |
| T3 | 18 | 41 | 69 | |
| T4 | 28 | 14 | 36 | |
| N stage | 0.1291 | |||
| N0 | 25 | 43 | 64 | |
| N1/2 | 31 | 26 | 48 | |
| M stage | 0.0259 | |||
| M0 | 40 | 57 | 98 | |
| M1 | 16 | 13 | 13 | |
| Histological type | ||||
| Tubular adenocarcinoma | 38 | 57 | 102 | 0.0008 |
| Others | 18 | 12 | 10 | |
Table 3.
Patient characteristics
| Variables | Results |
|---|---|
| NO. of patients | 237 |
| Age, y (median, range) | 63, 24-90 |
| Sex (male/female) | 124/113 (52.3%/47.7%) |
| TNM stage (I+II+III+IV) | 23/99/83/32 (9.7%/41.8%/35%/13.5%) |
| T stage (T1+T2/T3/T4) | 31/128/78 (13.1%/54%/32.9%) |
| N stage (N0/N1+N2) | 132/105 (55.7%/44.3%) |
| M stage (M0/M1) | 195/42 (82.3%/17.7%) |
| Pathology (Tubular adenocarcinoma/Others) | 197/40 (83.1%/16.9%) |
The result demonstrated that the expression of GLS1 was associated with T staging, distant metastasis, TNM staging and the histological type of colorectal cancer. We found that the expression of GLS1 was significantly higher in tumour tissues of T3/T4 than in T1/T2 (P=0.0002). We also observed that GLS1 staining of tumours with distant metastases was weaker than tumours without distant metastasis (P=0.0259). In addition, the expression of GLS1 in colorectal cancer patients of TNM stage II-IV was higher than that of TNM staging I (P=0.0309). We lastly observed that the expression of GLS1 was significantly higher in tubular adenocarcinoma than other histological types of colorectal cancer (p=0.0008), such as signet-ring cell carcinoma or mucinous adenocarcinoma. Conversely, we found that GLS1 levels in colorectal cancer tissues did not correlate with gender, age or lymph node metastasis of the patients.
GLS1 plays a role in colon cancer cell proliferation and apoptosis
Many studies have demonstrated that GLS1 is overexpressed in various kinds of cancer cells and is plays an important role in cell survival and proliferation [19,20]. To determine whether such phenotypes could also be observed in colorectal cancer cells, we tested the effect of GLS1 activity inhibition induced by DON (6-diazo-5-oxo-L-norleucine, Sigma-Aldrich) on HT29 and SW480 colon cancer cell lines. We observed that GLS1 exhibits functional importance in colon cancer cell proliferation and apoptosis.
For glutaminase inhibition, DON was administered to cells in a concentration gradient of 0, 10, 20, 40 and 80 μmol for 24 and 48 hours. After treatment, cell growth was tested by MTT assay, and the apoptotic effect was assessed by immunofluorescent assay of karyopyknosis or nuclear fragmentation, which is a morphological hallmark of apoptosis. We found that HT29 and SW480 cells exhibited significantly reduced growth rates after treatment with different concentrations of DON for 24 h (p<0.0001), as in Figure 3A, 3B. When the treating time was prolonged, the growth rates of HT29 and SW480 declined further (Figure 3C). These results suggest that the growth inhibitory effects induced by DON were dose- and time-dependent. In addition, GLS1 activity inhibition by DON led to prominent karyopyknosis and nuclear fragmentation (Figure 3D).
Figure 3.
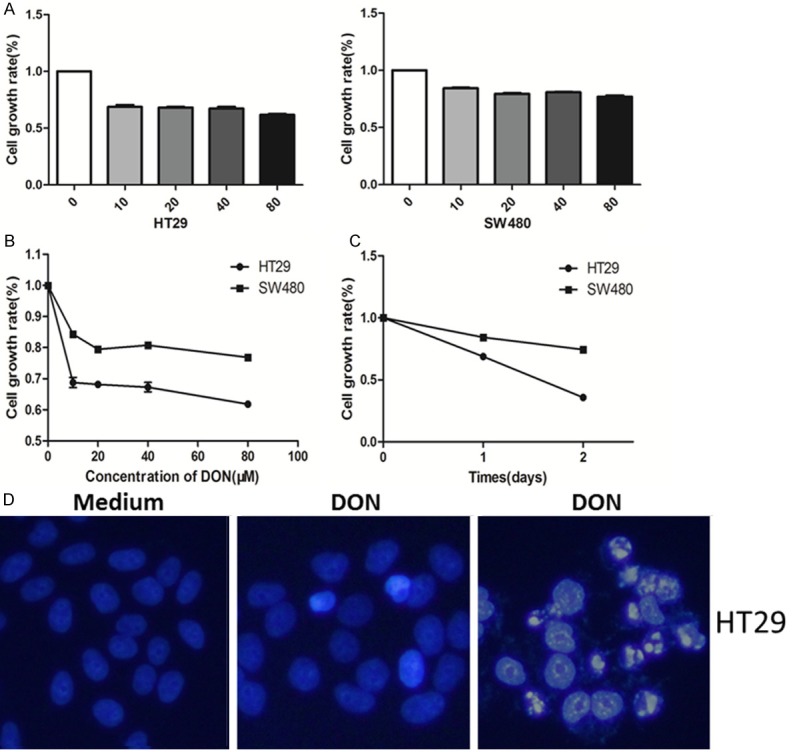
GLS1 activity inhibition can promote apoptosis and suppress proliferation in colon cancer cell lines. A, B. The normalised cell growth (MTT assay) of HT29 and SW480 cell lines after different 24 h treatment at DON concentrations (both P values<0.0001, ***). C. The normalised cell growth (MTT assay) of HT29 and SW480 cell lines after 24 h and 48 h treatment with 10 μM DON. D. Representative apoptosis induced by DON in colon cancer cells. Hoechst staining revealed karyopyknosis (middle panel) and nucleus fragmentation (right panel) of HT29 cells. SW480 and HT29 cells were treated with 10 μM DON for 48 h.
Discussion
Since 1920s, it has been established that cancer cell metabolism is altered compared with normal tissue, which contributes to the initiation, proliferation and development of tumours. In addition to elevated aerobic glycolysis, cancer cells exhibit increased dependence on glutamine for growth and proliferation [2,15,21-23].
GLS1 plays a critical role in catalysing glutaminolysis, and its expression is often increased in tumours and rapidly dividing cells [16]. Some recent studies have stressed the importance of glutaminolysis in maintaining the malignant phenotype [8]. Our study demonstrated for the first time elevated GLS1 expression in colorectal cancer patients compared with adjacent normal tissue, which suggested that GLS1 may correlate with the initiation and progression of colorectal cancer.
We also observed the correlation between GLS1 protein expression and clinicopathological factors of colorectal cancer patients. The data demonstrated that the expression of GLS1 was associated with T staging, distant metastasis, TNM staging and the histological type of colorectal cancer. The expression of GLS1 was higher in T3/T4 than in T1/T2 tumours, so the deeper the tumour infiltrates, the higher the expression of GLS1. This suggests that GLS1 may be associated with the initiation and progression of colorectal cancer. The study also demonstrated that GLS1 was less obviously increased in tumours with distant metastasis, possibly because of the statistical bias due to the small sample sizes of distant metastasis. Moreover, tubular adenocarcinomas expressed higher levels of GLS1 than other histological types of colorectal cancer, such as signet-ring cell carcinoma and mucinous adenocarcinoma. As rare and special types of colorectal cancer, signet-ring cell carcinoma and mucinous adenocarcinoma are often misdiagnosed and exhibit poor prognosis. Positron emission tomography (PET) technology is widely used for diagnosing carcinomas and evaluating the tumour stage by detecting glucose uptake. PET based on fluorodeoxyglucose (FDG) is the most widely used but cannot detect some poorly differentiated carcinomas, such as neuroglia, signetring cell carcinoma and mucinous adenocarcinoma. Today, a new type of PET based on the metabolism of glutamine is regarded as a better choice for tumours with low glucose metabolism due to the technique’s sensitivity for poorly differentiated carcinomas [24,25]. However, our study found that signet-ring cell carcinoma and mucinous adenocarcinoma exhibit low or even absent expression of GLS1. Thus, these diagnoses should be taken into consideration when the PET is negative.
The fact that GLS1 is involved in many metabolic procedures of tumour cells suggests that GLS1 might be a potential target in tumour therapy. In light of this, research has demonstrated that blocking the activation of GLS1 by compound 968 resulted in inhibition of Rho GTPase-induced transformation in fibroblasts, and that compound 968 strongly inhibited the proliferation of breast cancer cell. Additionally, the inhibition of GLS1 led to suppressed oncogenic transformation without affecting normal cells, which suggested a promising role of GLS1 in cancer therapy [9]. In our study, we found that the inhibition of GLS1 activity in HT29 and SW480 cells led to a significant reduction in growth as well as an increase of apoptosis, thus inhibiting tumour progression. To our knowledge, there is no study on the inhibition of GLS1 in colorectal cancer cells, and our results suggest that GLS1 is associated with tumorigenesis of colorectal cancer and that it may be used as an early diagnostic and a potential therapeutic target in colorectal cancer.
Despite increasing number of studies focused on the expression and role of GLS1 in cancer cells, further studies are needed. The mechanisms of GLS1 regulation are incompletely understood, and the interaction between GLS1 and neoplastic transformation is unclear. In addition, more detailed studies of the molecular mechanisms of the functional importance of GLS1 in tumour cells are needed.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (No. 81101507) to Fang Huang.
Disclosure of conflict of interest
None.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 3.Lacey JM, Wilmore DW. Is glutamine a conditionally essential amino acid. Nutr Rev. 1990;48:297–309. doi: 10.1111/j.1753-4887.1990.tb02967.x. [DOI] [PubMed] [Google Scholar]
- 4.Kovacevic Z, Morris HP. The role of glutamine in the oxidative metabolism of malignant cells. Cancer Res. 1972;32:326–333. [PubMed] [Google Scholar]
- 5.Turner A, McGivan JD. Glutaminase isoform expression in cell lines derived from human colorectal adenomas and carcinomas. Biochem J. 2003;370:403–408. doi: 10.1042/BJ20021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobo C, Ruiz-Bellido MA, Aledo JC, Marquez J, Nunez De Castro I, Alonso FJ. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells. Biochem J. 2000;348 Pt 2:257–261. [PMC free article] [PubMed] [Google Scholar]
- 11.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhoads JM, Argenzio RA, Chen W, Rippe RA, Westwick JK, Cox AD, Berschneider HM, Brenner DA. L-glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am J Physiol. 1997;272:G943–953. doi: 10.1152/ajpgi.1997.272.5.G943. [DOI] [PubMed] [Google Scholar]
- 13.Larson SD, Li J, Chung DH, Evers BM. Molecular mechanisms contributing to glutamine-mediated intestinal cell survival. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1262–1271. doi: 10.1152/ajpgi.00254.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder-Horowitz M, Knox WE, Morris HP. Glutaminase activities and growth rates of rat hepatomas. Cancer Res. 1969;29:1195–1199. [PubMed] [Google Scholar]
- 15.Matsuno T, Goto I. Glutaminase and glutamine synthetase activities in human cirrhotic liver and hepatocellular carcinoma. Cancer Res. 1992;52:1192–1194. [PubMed] [Google Scholar]
- 16.Perez-Gomez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sanchez P, Gonzalez ME, Marquez J, Mates JM. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–542. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quesada AR, Sanchez-Jimenez F, Perez-Rodriguez J, Marquez J, Medina MA, Nunez de Castro I. Purification of phosphate-dependent glutaminase from isolated mitochondria of Ehrlich ascites-tumour cells. Biochem J. 1988;255:1031–1035. doi: 10.1042/bj2551031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regulation of renal ammoniagenesis. Purification and characterization of phosphate-dependent glutaminase from rat kidney. Arch Biochem Biophys. 1976;174:82–89. [PubMed] [Google Scholar]
- 19.Segura JA, Ruiz-Bellido MA, Arenas M, Lobo C, Marquez J, Alonso FJ. Ehrlich ascites tumor cells expressing anti-sense glutaminase mRNA lose their capacity to evade the mouse immune system. Int J Cancer. 2001;91:379–384. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1046>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Rufian M, Segura JA, Lobo C, Mates JM, Marquez J, Alonso FJ. Identification of genes downregulated in tumor cells expressing antisense glutaminase mRNA by differential display. Cancer Biol Ther. 2006;5:54–58. doi: 10.4161/cbt.5.1.2238. [DOI] [PubMed] [Google Scholar]
- 21.Dranoff G, Elion GB, Friedman HS, Campbell GL, Bigner DD. Influence of glutamine on the growth of human glioma and medulloblastoma in culture. Cancer Res. 1985;45:4077–4081. [PubMed] [Google Scholar]
- 22.Matsuno T, Hirai H. Glutamine synthetase and glutaminase activities in various hepatoma cells. Biochem Int. 1989;19:219–225. [PubMed] [Google Scholar]
- 23.Martin M, Beauvoit B, Voisin PJ, Canioni P, Guerin B, Rigoulet M. Energetic and morphological plasticity of C6 glioma cells grown on 3-D support; effect of transient glutamine deprivation. J Bioenerg Biomembr. 1998;30:565–578. doi: 10.1023/a:1020584517588. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman BP, Ploessl K, Wang L, Qu W, Zha Z, Wise DR, Chodosh LA, Belka G, Thompson CB, Kung HF. PET imaging of glutaminolysis in tumors by 18F-(2S,4R)4-fluoroglutamine. J Nucl Med. 2011;52:1947–1955. doi: 10.2967/jnumed.111.093815. [DOI] [PubMed] [Google Scholar]
- 25.Qu W, Zha Z, Ploessl K, Lieberman BP, Zhu L, Wise DR, Thompson CB, Kung HF. Synthesis of optically pure 4-fluoro-glutamines as potential metabolic imaging agents for tumors. J Am Chem Soc. 2011;133:1122–1133. doi: 10.1021/ja109203d. [DOI] [PubMed] [Google Scholar]


