Abstract
Recent studies have demonstrated that cathepsin K seems to be a powerful marker in identifying the microphthalmia associated transcription factor (MITF) family tumors such as renal perivascular epithelioid cell neoplasms (PEComas), alveolar soft part sarcoma, and translocation-associated renal cell carcinomas. However, the expression of cathepsin K in melanocytic lesions has not been well characterized. Our aim was to investigate the expression of cathepsin K in a wide histological spectrum of melanocytic lesions and to evaluate its potential diagnostic and molecular target therapy usefulness in comparison with other commonly used markers. 143 consecutive melanocytic lesions were selected for study including 56 primary malignant melanomas, 62 metastatic melanomas, and 25 benign nevi (16 intradermal melanocytic nevi and 9 compound melanocytic nevi). 107 of the 118 (91%) primary and metastatic melanomas displayed a high percentage of cells with moderately to strongly positive reactions for cathepsin K (mean 82%; range 0-95%). MITF, HMB45, Melan-A, and S100 were expressed in 85, 76, 78 and 96% of cases, respectively, with various percentages of positive cells (mean, 63, 49, 55 and 86%; range 0-90, 0-80, 0-90 and 0-95%). Among the benign nevi, cathepsin K, MITF, HMB45, Melan-A, and S100 were expressed in 88, 80, 36, 68 and 100% of cases, respectively. Cathepsin K appears to be consistently and strongly expressed in melanocytic lesions and valuable in distinguishing malignant melanomas from the majority of human cancers.
Keywords: Melanoma, melanocytic lesions, immunohistochemistry, cathepsin K, nevi, MITF
Introduction
The diagnosis of metastatic malignant melanoma may be difficult in surgical pathology because of its extremely variable morphology. Histologically, the tumor cells may have diverse features including epithelioid cell, round cell, spindle cell, rhabdoid cell, and so on. Therefore, it may be misdiagnosed as other tumors such as poor differentiated carcinomas, sarcomas, and large cell lymphomas. Based on these considerations, the diagnosis of such lesions should be based not only on morphology itself, but also on immunophenotype. Although some immunohistochemical markers such as S100, HMB45, Melan-A, and MITF are often used and shown to be helpful in the detection of these tumors, the limitations in the sensitivity and /or specificity of these available melanocytic markers still complicate this problem [1-7].
Recent researches have implicated that microphthalmia associated transcription factor (MITF) is required for the growth of melanocytes and melanoma [8-10]. Approximately 30% to 40% of melanomas harbor amplifications of MITF, although single nucleotide somatic mutations within its coding region have also been observed in human melanomas, making MITF the major melanoma oncogene [11-13]. MITF is one of four members of the MITF family, which includes MITF, TFEB, TFEC and TFE3 [9,10]. All family members share a homologous basic helix-loop-helix DNA binding domain and have overlapping transcriptional targets [9,10]. Several distinct tumors are associated with the overexpression of this gene family, including melanoma, clear cell sarcoma, angiomyolipoma, perivascular epithelioid cell neoplasms (PEComas), alveolar soft part sarcoma, and translocation-associated renal cell carcinomas [9,10,14-22]. All these tumors have been considered as a member of the MITF family of tumors, owing to their histological, immunochemical and molecular genetic similarity [9,10].
Cathepsin K is a cysteine protease from the papain family of proteases, which plays an important role in osteoclasts function and the overexpression of cathepsin K in osteoclasts is regulated by MITF [23]. In recent studies cathepsin K was identified as a transcriptional target of the MITF family, and was expressed consistently and strongly in TFEB translocation renal cell carcinomas, a wide spectrum of PEComas, and some cases of TFE3 translocation renal cell carcinomas [19,21,23-25]. However, the expression of cathepsin K in melanocytic lesions has not been well characterized. We hypothesized that the overexpression of cathepsin K may have the same effect in melanocytic lesions. The aim of this study was to investigate the expression of cathepsin K in melanocytic lesions and to evaluate its potential diagnostic and molecular target therapy usefulness in comparison with other commonly used markers.
Materials and methods
Case selection
A total of 143 consecutive melanocytic lesions were selected in the archives of the Department of Pathology at Nanjing Jinling Hospital. The melanocytic cases include 56 primary malignant melanomas, 62 metastatic melanomas, and 25 benign nevi (16 intradermal melanocytic nevi and 9 compound melanocytic nevi). The metastases were located in the lymph nodes (n=13), deep soft tissues (n=15), lung (n=5), gastrointestinal tract (n=4), and bone (n=3).
To determine the prevalence of immunoreactivity for cathepsin K in control neoplasms, both tissue microarray (TMA) sections and conventional unstained whole tissue sections were used in the study. Organ-specific TMAs constructed using a manual tissue arrayer (Beecher Instruments Inc, Sun Prarie, WI, USA) contained 50-99 spots of tumor with a diameter of 2 mm. The control TMAs included 70 breast carcinomas, 99 non-small lung carcinomas (70 lung adenocarcinomas and 29 lung squamous cell carcinomas), 80 gastric carcinomas, and 50 colorectal adenocarcinomas. To analyze certain tumors for which TMAs were not available to us, we obtained unstained whole tissue sections including 20 prostate adenocarcinoma, 30 bladder urothelial carcinomas, 30 clear cell renal cell carcinomas, 20 gallbladder carcinomas, 30 hepatocellular carcinomas, 15 uterine leiomyomas, 10 schwannomas, and 15 anaplastic large cell lymphomas.
Immunohistochemistry
Tissues were fixed in 10% formalin and embedded in paraffin. Sections 3 mm thick were stained with immunohistochemistry. The following antibodies for immunohistochemistry were used: cathepsin K (3F9, Abcam, 1:300), MITF (D5, Dako, 1:100), HMB45 (HMB45, Dako, 1:500), Melan-A (A103/M2-72, Neomarkers, 1:100), and S100 (Polyclonal, Dako, 1:2000). Immunoreaction was performed using the labelled streptavidin-biotin method and overnight incubation as previously described and evaluated in a semiquantitative way assessing both staining intensity and percentage of positive cells as previously described [23,26,27]. For all antibodies, the resulting score was calculated by multiplying the staining intensity (0=no staining, 1=mild staining, 2=moderate staining, and 3=strong staining) by the percentage of immunoreactive tumor cells (0 to 100). The immunostaining was considered 0 or negative when the score was <25; 1+ or weak, 26 to 100; 2+ or moderate, 101 to 200; and 3+ or strong, 201 to 300.
Result
Primary and metastatic melanomas
The results of immunohistochemical findings in primary and metastatic melanomas were listed in Table 1. Overall, 107 of the 118 (91%) primary and metastatic melanomas with different types of tumors cells including rhabdoid cell, epithelioid cell, round cell and spindle cell displayed a high percentage of cells with moderately to strongly positive reactions for cathepsin K (mean 82%; range 0-95%). In contrast, the adjacent normal squamous epithelium, sweat glands, and the walls of the vessels were completely negative for cathepsin K. The MITF, HMB45, Melan-A, and S100 were expressed in melanomas as previously described, which were focally (1+) to strongly (3+) positive in 85% (100 of 118), 76% (90 of 118) and 78% (92 of 118) of melanomas, respectively, with various percentages of positive cells (MITF: mean 63%; range 0-90%; HMB45: mean 49%; range 0-80%; Melan-A: mean 55%; range 0-90%; S100: mean 86%; range 0-95%). If only moderate or strong positive staining was taken into account, cathepsin K, MITF, HMB45, Melan-A, and S100 staining were found to be moderately to strongly positive in 81%, 71%, 62%, 65%, and 87% of the melanomas (Figure 1).
Table 1.
Immunohistochemical findings in primary and metastatic melanomas
| Cathepsin K | MITF | HMB45 | Melan-A | S100 | |
|---|---|---|---|---|---|
| Strong (+++) | 61 | 52 | 41 | 48 | 62 |
| Moderate (++) | 34 | 32 | 32 | 29 | 41 |
| weak (+) | 12 | 16 | 17 | 15 | 10 |
| negative (-) | 11 | 18 | 28 | 26 | 5 |
Figure 1.
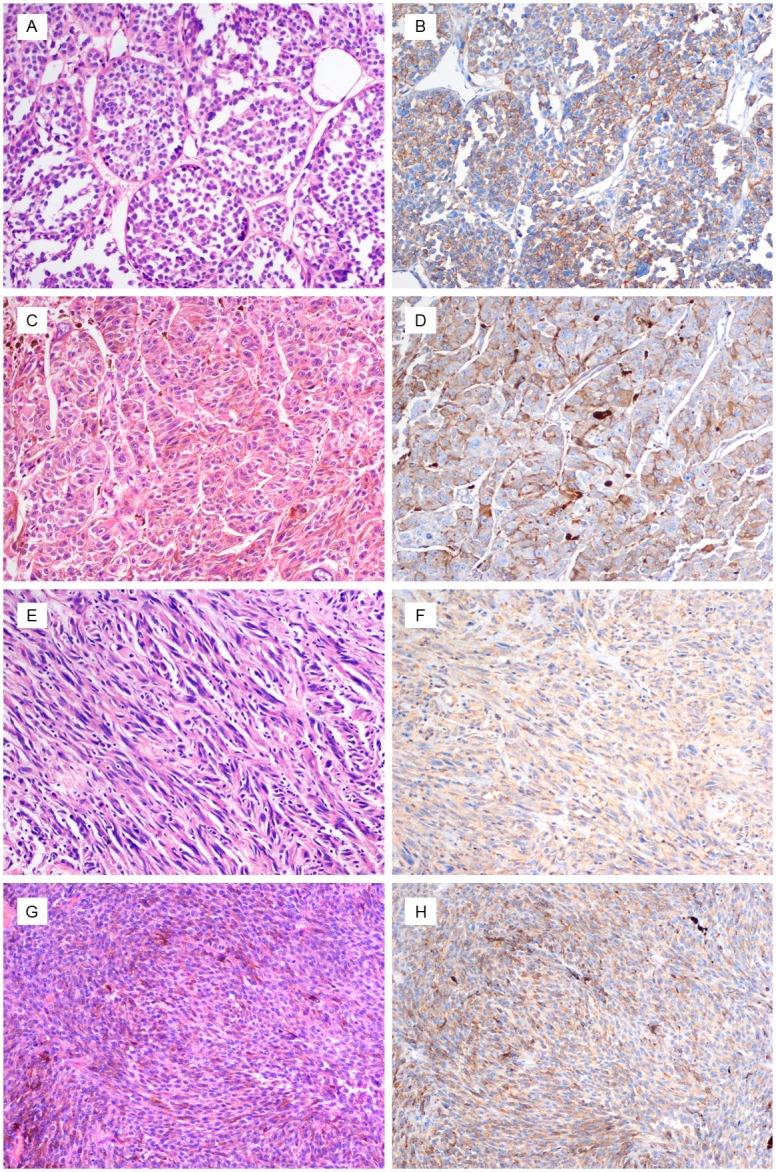
Melanoma with epithelioid (A and C) and spindle cell (E and G) morphology. (B, D, F and H) The tumor strongly expresses cathepsin K. (original magnification, ×200).
Cathepsin K positivity in primary and metastatic melanomas that were negative for other markers
Among the MITF-negative melanomas, 7 of 21 (33%) were positive for cathepsin K. 15 of the 28 melanomas (54%) that were HMB45 negative were positive for cathepsin K. Of the Melan-A-negative melanomas, 10 of 26 (38%) were cathepsin K-positive. Similarly, 2 of 5 S100-negative melanomas (40%) were cathepsin K-positive (Figure 2A-D).
Figure 2.

(A) Melanoma with rhabdoid morphology. (B) The tumour strongly expresses cathepsin K and is negative for HMB45 (C). (D) Melan-A is weakly expressed. (E) The histologic features of benign nevi. (F) Cathepsin K is diffusely expressed. (original magnification, ×200).
Benign nevi
Moderate to strong cathepsin K staining was seen in 22 of 25 (88%) cases of nevi (intradermal melanocytic nevi and compound melanocytic nevi). This trend was similar to the staining pattern of S100, which was positive in all nevi with diffuse (3+) staining pattern. 20 of the 25 (80%) nevi, 9 of the 25 (36%) nevi, and 17 of the 25 (68%) nevi showed focal (1+) to diffuse (3+) positivity with MITF, HMB45, and Melan-A respectively (Figure 2E and 2F).
Control tissues
None of the breast carcinomas, non-small lung carcinomas, gastric carcinomas, colorectal adenocarcinomas, prostate adenocarcinoma, bladder urothelial carcinomas, clear cell renal cell carcinomas, gallbladder carcinomas, hepatocellular carcinomas, uterine leiomyomas, schwannomas, and anaplastic large cell lymphoma was positive for cathepsin K, except for 1 breast carcinoma and 1lung adenocarcinoma that showed moderate immunoreactivity for cathepsin K.
Discussion
In this study, we showed that cathepsin K was expressed in 107 of the 118 (91%) primary and metastatic melanomas with different types of tumors cells including rhabdoid cell, epithelioid cell, round cell and spindle cell and in 22 of 25 (88%) cases of nevi including 16 intradermal melanocytic nevi and 9 compound melanocytic nevi, most of them in a high number of tumor cells with moderate or strong positive reactions for cathepsin K. If only moderate or strong positive staining was taken into account, cathepsin K, MITF, HMB45, Melan-A, and S100 staining were found to be moderately to strongly positive in 81%, 71%, 62%, 65%, and 87% of the melanomas. The tumors displayed a higher percentage of positive reaction cells for cathepsin K than for MITF, HMB45, and Melan-A. In some melanomas for which other markers were negative, more than half of the HMB45-negative melanomas, many MITF- and Melan-A-negative melanomas, and a few S100-negative melanomas were cathepsin K-positive. In the control group, except for moderate immunoreactivity for cathepsin K in one breast carcinoma and one lung adenocarcinoma, none of the various other neoplasms used as controls was positive for cathepsin K. These findings therefore suggest that cathepsin K is a useful addition to the diagnostic panel used in possible cases of melanomas and that cathepsin K can be used as a relatively specific marker to distinguish melanomas from the majority of human cancers.
Cathepsin K is a lysosomal papain-like cystine proteinase with strong collagenolytic and elastolytic activity, which plays an important role in osteoclast function and is regulated by MITF by increasing mRNA and protein level of this papain-like cysteine protease [8]. As recent studies have demonstrated cathepsin K to be a transcriptional target of the microphthalmia-associated transcription factor family, immunohistochemistry antibody to cathepsin K has been utilized in the diagnosis of several microphthalmia-associated transcription factor family of tumors such as translocation-associated renal cell carcinomas, PEComas, and alveolar soft part sarcoma [19,21,23-25,28]. In addition, cathepsin K is also expressed in reactive activated macrophages, but not in resident macrophages [8,29,30]. Consistent overexpression of cathepsin K has been demonstrated in granulomatous disorders including hypersensitivity pneumonitis, sarcoidosis, Wegener granulomatosis, berylliosis, and tuberculosis [8,29,30]. Our results confirmed the same effect of cathepsin K in melanocytic lesions and expanded the immunohistochemistry application of the antibody.
Recent studies have demonstrated that mammalian target of rapamycin (MTOR) is activated in the majority of malignant melanomas and may be targets for melanoma therapy [31-33]. In the study of Kneissel et al. MTOR inhibitors have efficacy in limiting the bone resorption mediated by osteoclastic cathepsin K [34]. These findings make the possibility to hypothesized that MTOR inhibitors can exert part of their activity by limiting the expression of cathepsin K [25]. In addition, cathepsin K inhibitors are being developed for cancer therapy in bone metastases, for rendering the bone a less favorable microenvironment for tumor growth by inhibiting osteoclast-mediated bone resorption [35,36]. All these findings make novel interventional approaches with cathepsin K worth further investigation.
In conclusion, cathepsin K appears to be consistently and strongly expressed in melanocytic lesions and valuable in distinguishing malignant melanomas from the majority of human cancers. The potential target of cathepsin K for melanoma therapy remains to be clarified.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81101933; Q Rao); and (81372743; Xiao-Jun Zhou); Natural Science Foundation of Jiangsu Province, China (BK2010463; Q Rao); and Maixin fund (m1203; Shan-Shan Shi).
Disclosure of conflict of interest
None.
References
- 1.Busam KJ, Iversen K, Coplan KC, Jungbluth AA. Analysis of microphthalmia transcription factor expression in normal tissues and tumors, and comparison of its expression with S-100 protein, gp100, and tyrosinase in desmoplastic malignant melanoma. Am J Surg Pathol. 2001;25:197–204. doi: 10.1097/00000478-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Clarkson KS, Sturdgess IC, Molyneux AJ. The usefulness of tyrosinase in the immunohistochemical assessment of melanocytic lesions: a comparison of the novel T311 antibody (anti-tyrosinase) with S-100, HMB45, and A103 (anti-melan-A) J Clin Pathol. 2001;54:196–200. doi: 10.1136/jcp.54.3.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King R, Googe PB, Weilbaecher KN, Mihm MC Jr, Fisher DE. Microphthalmia transcription factor expression in cutaneous benign, malignant melanocytic, and nonmelanocytic tumors. Am J Surg Pathol. 2001;25:51–57. doi: 10.1097/00000478-200101000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Miettinen M, Fernandez M, Franssila K, Gatalica Z, Lasota J, Sarlomo-Rikala M. Microphthalmia transcription factor in the immunohistochemical diagnosis of metastatic melanoma: comparison with four other melanoma markers. Am J Surg Pathol. 2001;25:205–211. doi: 10.1097/00000478-200102000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Chu AY, Pasha TL, Elder DE, Zhang PJ. Immunoprofile of MITF, tyrosinase, melan-A, and MAGE-1 in HMB45-negative melanomas. Am J Surg Pathol. 2002;26:82–87. doi: 10.1097/00000478-200201000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Krishna M. Diagnosis of metastatic neoplasms: an immunohistochemical approach. Arch Pathol Lab Med. 2010;134:207–215. doi: 10.5858/134.2.207. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen PS, Riber-Hansen R, Steiniche T. Immunohistochemical double stains against Ki67/MART1 and HMB45/MITF: promising diagnostic tools in melanocytic lesions. Am J Dermatopathol. 2011;33:361–370. doi: 10.1097/DAD.0b013e3182120173. [DOI] [PubMed] [Google Scholar]
- 8.Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, Fisher DE. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci U S A. 2001;98:5798–5803. doi: 10.1073/pnas.091479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis IJ, Fisher DE. MiT transcription factor associated malignancies in man. Cell Cycle. 2007;6:1724–1729. doi: 10.4161/cc.6.14.4484. [DOI] [PubMed] [Google Scholar]
- 10.Haq R, Fisher DE. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. J. Clin. Oncol. 2011;29:3474–3482. doi: 10.1200/JCO.2010.32.6223. [DOI] [PubMed] [Google Scholar]
- 11.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 12.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 13.Cronin JC, Wunderlich J, Loftus SK, Prickett TD, Wei X, Ridd K, Vemula S, Burrell AS, Agrawal NS, Lin JC, Banister CE, Buckhaults P, Rosenberg SA, Bastian BC, Pavan WJ, Samuels Y. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22:435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Righi A, Dimosthenous K, Rosai J. PEComa: another member of the MiT tumor family? Int J Surg Pathol. 2008;16:16–20. doi: 10.1177/1066896907309733. [DOI] [PubMed] [Google Scholar]
- 15.Argani P, Aulmann S, Illei PB, Netto GJ, Ro J, Cho HY, Dogan S, Ladanyi M, Martignoni G, Goldblum JR, Weiss SW. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol. 2010;34:1395–1406. doi: 10.1097/PAS.0b013e3181f17ac0. [DOI] [PubMed] [Google Scholar]
- 16.Dickson BC, Brooks JS, Pasha TL, Zhang PJ. TFE3 Expression in Tumors of the Microphthalmia-Associated Transcription Factor (MiTF) Family. Int J Surg Pathol. 2011;19:26–30. doi: 10.1177/1066896909352861. [DOI] [PubMed] [Google Scholar]
- 17.Rao Q, Chen JY, Wang JD, Ma HH, Zhou HB, Lu ZF, Zhou XJ. Renal cell carcinoma in children and young adults: clinicopathological, immunohistochemical, and VHL gene analysis of 46 cases with follow-up. Int J Surg Pathol. 2011;19:170–179. doi: 10.1177/1066896909354337. [DOI] [PubMed] [Google Scholar]
- 18.Inamura K, Fujiwara M, Togashi Y, Nomura K, Mukai H, Fujii Y, Yamamoto S, Yonese J, Fukui I, Ishikawa Y. Diverse fusion patterns and heterogeneous clinicopathologic features of renal cell carcinoma with t(6;11) translocation. Am J Surg Pathol. 2012;36:35–42. doi: 10.1097/PAS.0b013e3182293ec3. [DOI] [PubMed] [Google Scholar]
- 19.Rao Q, Liu B, Cheng L, Zhu Y, Shi QL, Wu B, Jiang SJ, Wang Y, Wang X, Yu B, Zhang RS, Ma HH, Lu ZF, Tu P, Wang JD, Zhou XJ. Renal cell carcinomas with t(6;11)(p21;q12): A clinicopathologic study emphasizing unusual morphology, novel alpha-TFEB gene fusion point, immunobiomarkers, and ultrastructural features, as well as detection of the gene fusion by fluorescence in situ hybridization. Am J Surg Pathol. 2012;36:1327–1338. doi: 10.1097/PAS.0b013e31825aafb5. [DOI] [PubMed] [Google Scholar]
- 20.Argani P, Yonescu R, Morsberger L, Morris K, Netto GJ, Smith N, Gonzalez N, Illei PB, Ladanyi M, Griffin CA. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am J Surg Pathol. 2012;36:1516–1526. doi: 10.1097/PAS.0b013e3182613d8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao Q, Williamson SR, Zhang S, Eble JN, Grignon DJ, Wang M, Zhou XJ, Huang W, Tan PH, Maclennan GT, Cheng L. TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol. 2013;37:804–815. doi: 10.1097/PAS.0b013e31827e17cb. [DOI] [PubMed] [Google Scholar]
- 22.Williamson SR, Bunde PJ, Montironi R, Lopez-Beltran A, Zhang S, Wang M, Maclennan GT, Cheng L. Malignant Perivascular Epithelioid Cell Neoplasm (PEComa) of the Urinary Bladder With TFE3 Gene Rearrangement: Clinicopathologic, Immunohistochemical, and Molecular Features. Am J Surg Pathol. 2013;37:1619–1626. doi: 10.1097/PAS.0b013e318293729d. [DOI] [PubMed] [Google Scholar]
- 23.Rao Q, Cheng L, Xia QY, Liu B, Li L, Shi QL, Shi SS, Yu B, Zhang RS, Ma HH, Lu ZF, Tu P, Zhou XJ. Cathepsin K expression in a wide spectrum of perivascular epithelioid cell neoplasms (PEComas): a clinicopathological study emphasizing extrarenal PEComas. Histopathology. 2013;62:642–650. doi: 10.1111/his.12059. [DOI] [PubMed] [Google Scholar]
- 24.Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, Pan CC, Netto G, Doglioni C, Hes O, Argani P, Chilosi M. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–1022. doi: 10.1038/modpathol.2009.58. [DOI] [PubMed] [Google Scholar]
- 25.Martignoni G, Bonetti F, Chilosi M, Brunelli M, Segala D, Amin MB, Argani P, Eble JN, Gobbo S, Pea M. Cathepsin K expression in the spectrum of perivascular epithelioid cell (PEC) lesions of the kidney. Mod Pathol. 2012;25:100–111. doi: 10.1038/modpathol.2011.136. [DOI] [PubMed] [Google Scholar]
- 26.Rao Q, Zhang XM, Tu P, Xia QY, Shen Q, Zhou XJ, Shi QL. Renal cell carcinomas with t(6;11)(p21;q12) presenting with tubulocystic renal cell carcinoma-like features. Int J Clin Exp Pathol. 2013;6:1452–1457. [PMC free article] [PubMed] [Google Scholar]
- 27.Xia QY, Rao Q, Shen Q, Shi SS, Li L, Liu B, Zhang J, Wang YF, Shi QL, Wang JD, Ma HH, Lu ZF, Yu B, Zhang RS, Zhou XJ. Oncocytic papillary renal cell carcinoma: a clinicopathological study emphasizing distinct morphology, extended immunohistochemical profile and cytogenetic features. Int J Clin Exp Pathol. 2013;6:1392–1399. [PMC free article] [PubMed] [Google Scholar]
- 28.Martignoni G, Gobbo S, Camparo P, Brunelli M, Munari E, Segala D, Pea M, Bonetti F, Illei PB, Netto GJ, Ladanyi M, Chilosi M, Argani P. Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod Pathol. 2011;24:1313–1319. doi: 10.1038/modpathol.2011.93. [DOI] [PubMed] [Google Scholar]
- 29.Buhling F, Reisenauer A, Gerber A, Kruger S, Weber E, Bromme D, Roessner A, Ansorge S, Welte T, Rocken C. Cathepsin K--a marker of macrophage differentiation? J Pathol. 2001;195:375–382. doi: 10.1002/path.959. [DOI] [PubMed] [Google Scholar]
- 30.Reghellin D, Poletti V, Tomassett S, Dubini A, Cavazza A, Rossi G, Lestani M, Pedron S, Daniele I, Montagna L, Murer B, Chilos M. Cathepsin-K is a sensitive immunohistochemical marker for detection of micro-granulomas in hypersensitivity pneumonitis. Sarcoidosis Vasc Diffuse Lung Dis. 2010;27:57–63. [PubMed] [Google Scholar]
- 31.Bundscherer A, Hafner C, Maisch T, Becker B, Landthaler M, Vogt T. Antiproliferative and proapoptotic effects of rapamycin and celecoxib in malignant melanoma cell lines. Oncol Rep. 2008;19:547–553. [PubMed] [Google Scholar]
- 32.Karbowniczek M, Spittle CS, Morrison T, Wu H, Henske EP. mTOR is activated in the majority of malignant melanomas. J Invest Dermatol. 2008;128:980–987. doi: 10.1038/sj.jid.5701074. [DOI] [PubMed] [Google Scholar]
- 33.Soon CW, Algazi AP, Cha EN, Daud AI. New horizons in melanoma treatment: targeting molecular pathways. Ochsner J. 2010;10:93–98. [PMC free article] [PubMed] [Google Scholar]
- 34.Kneissel M, Luong-Nguyen NH, Baptist M, Cortesi R, Zumstein-Mecker S, Kossida S, O’Reilly T, Lane H, Susa M. Everolimus suppresses cancellous bone loss, bone resorption, and cathepsin K expression by osteoclasts. Bone. 2004;35:1144–1156. doi: 10.1016/j.bone.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Le Gall C, Bonnelye E, Clezardin P. Cathepsin K inhibitors as treatment of bone metastasis. Curr Opin Support Palliat Care. 2008;2:218–222. doi: 10.1097/SPC.0b013e32830baea9. [DOI] [PubMed] [Google Scholar]
- 36.Clezardin P. Therapeutic targets for bone metastases in breast cancer. Breast Cancer Res. 2011;13:207. doi: 10.1186/bcr2835. [DOI] [PMC free article] [PubMed] [Google Scholar]


