Abstract
In a bioinformatics based screen for chloroplast-localized protein kinases we noticed that available protein targeting predictors falsely predicted chloroplast localization. This seems to be due to interference with N-terminal protein acylation, which is of particular importance for protein kinases. Their N-myristoylation was found to be highly overrepresented in the proteome, whereas myristoylation motifs are almost absent in known chloroplast proteins. However, only abolishing their myristoylation was not sufficient to target those kinases to chloroplasts and resulted in nuclear accumulation instead. In contrast, inhibition of N-myristoylation of a calcium-dependent protein kinase was sufficient to alter its localization from the plasma membrane to chloroplasts and chloroplast localization of ferredoxin-NADP+ reductase and Rubisco activase could be efficiently suppressed by artificial introduction of myristoylation and palmitoylation sites.
Keywords: Protein kinase, Chloroplast targeting, Myristoylation, Palmitoylation, Calcium-dependent protein kinase
1. Introduction
The subcellular localization of proteins is crucial for their physiological function [1,2]. Accordingly, the correct assignment of protein localization is a prerequisite to understand its biological function. The rapidly increasing amount of sequenced genomes generated an increasing need to predict the subcellular localization of proteins from available sequence data. Protein sorting mechanisms are dependent on the presence of certain targeting sequences, mostly in the N-terminal parts of the protein, as well as on the general properties of the protein, for example its hydrophobicity [3,4]. Based on the physico-chemical properties of its targeting peptides, several algorithms have been developed to predict the subcellular localization of proteins [5].
It is estimated that ~30% of all cellular proteins are targeted to membranes [6], which can be achieved via hydrophobic transmembrane domains, electrostatic interaction with membrane components or lipid modifications [7]. The two major mechanisms of lipid modification are acylation and prenylation. While prenylation modifies C-terminal ends of proteins by covalent attachment of a farnesyl or geranylgeranyl moiety to a cysteine residue [8], protein acylation occurs mainly in the N-terminal part [9] and does therefore potentially interfere with protein sorting. Numerous proteins involved in signal transduction, are myristoylated and palmitoylated [9-11]. N-Myristoylation is the irreversible, co-translational attachment of myristic acid (C14:0) to an N-terminal glycine that is required at position 2 of a protein. Accordingly the mutation of this glycine (i.e. G2A) abolishes N-myristoylation of this protein. During translation, following the removal of the N-terminal methionine residue by a methionylaminopeptidase, myristic acid is linked to the N-terminal glycine via an amide bond by a N-myristoyltransferase (NMT) [12]. NMT recognizes a certain consensus motif – in many cases MGXXX(S/T) – which can be predicted by various programs [13-16]. In contrast, palmitoylation is the post-translational attachment of palmitic acid (C16:0) to N-terminal or internal cysteine residues of proteins via a reversible thioester bond catalyzed by a protein palmitoyltransferase (PPT). PPTs are much likely located at membranes, for example the ER or the Golgi apparatus [11], but the mechanism of their action is still unclear[17,18]. Internal palmitoylation of proteins is myristoylation-independent, whereas N-myristoylation is a prerequisite for N-terminal palmitoylation in most cases. Furthermore, palmitoylation is not restricted to the presence of a specific consensus motif [13].
Myristoylation facilitates only reversible membrane binding of proteins because the energy provided by myristate-lipid interaction alone is too low for a stable membrane attachment [19]. Palmitoylation in contrast is suggested to mediate a stable membrane anchoring, which corresponds to the fact that palmitoylated proteins are found almost exclusively in membrane preparations whereas myristoylated proteins are also present in soluble protein extracts [9,10]. Stable membrane attachment of myristoylated proteins can only be achieved by additional factors that support membrane binding such as palmitoylation, interaction of a polybasic amino acid stretch with acidic phospholipids or interaction with a membrane protein [18,20]. Acylation of proteins can influence their membrane targeting, their structure and activity or their interaction with other proteins [21].
The physiological relevance of protein N-acylation has already been demonstrated for a number of different examples, particularly for proteins involved in signal transduction and stress response[7,22,23]. For example, the plasma membrane Na+/H+ exchanger SOS1 is regulated in a calcium-dependent manner via the joint action of SOS3, a calcineurin B-like protein (CBL) and SOS2, a protein kinase in response to salt stress in Arabidopsis [24]. The salt-hypersensitive sos3-1 mutant exhibits impaired SOS1 activity, which could only be complemented by wild-type SOS3, but not by the non-myristoylatable SOS3 G2A mutant [23,24]. Similarly, cbll mutants are hypersensitive to salt, and again only wild-type CBL1 but not CBL1 G2A was able to partially complement the mutant. Furthermore, CBL1 C3S in which the cysteine on position 3 has been exchanged for serine to prevent its palmitoylation, was also not able to complement the salt-sensitive phenotype of the cbl1 mutant. This indicates that palmitoylation as well as N-myristoylation has strong effects on the physiological function of signaling components [7]. However, the interference of protein N- acylation and other targeting mechanisms has so far almost been overlooked, particularly in the plant field. In 2005 Colombo et al. [25] reported that N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to the ER and to mitochondrial outer membranes by a mechanism of kinetic partitioning, and here we show that protein N-acylation is able to override other targeting signals like chloroplast targeting peptides, which has strong implications particularly for protein kinases.
2. Materials and methods
2.1. Cloning of genes and production of mutants
The coding sequences of all investigated genes were amplified by PCR from Arabidopsis cDNA. All analyzed mutants were created by PCR mutagenesis using 5′ oligonucleotides carrying the indicated base changes. Subsequently, all constructs were sequenced and cloned into the vector pBAT [26], for the analysis of N-myristoylation, and into the vector pBIN-Basta, a derivate of pBIN 19 [27] carrying a C-terminal YFP fusion for localization studies.
2.2. In vitro myristoylation assays
Analysis of protein N-myristoylation was carried out exactly as previously described [28], using a cell free system (TNT Coupled Wheat Germ Extract System, Promega). In vitro translation was carried out either in the presence of 10 μCi of L-[35S] methionine (1175 Ci/mmol, Perkin–Elmer) for total protein labelling, or 50 μCi of [9,10-3H]-labelled myristic acid (60 Ci/mmol, American Radiolabeled Chemicals). Reaction products were separated on 12% (w/v) SDS-polyacrylamide gels and incubated with autoradiography intensifier (Amersham) before detection on X-ray film.
2.3. YFP localization studies
The localization of proteins fused to YFP was investigated by confocal laser scanning microscopy after agrobacterium-mediated transfection of Nicotiana tabacum epidermal leaf cells two days after transfection as described previously [28].
2.4. Western blotting
After microscopy, transfected leafs of N. tabacum expressing the YFP-fusion proteins were grinded in liquid nitrogen. Proteins were extracted in denaturing extraction buffer (0.175 M Tris–HCl pH 8.8, 5% SDS, 15% glycerol, 0.2 M DTT), precipitated with four volumes of cold acetone and resuspended in standard SDS–PAGE loading buffer. Western blot was carried out as described previously [29], and detection was carried out with anti-GFP primary antibody (1:1000 dilution, Roche) and anti-mouse secondary antibody (1:10000 dilution, GE healthcare). Western blots were developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce). Protein amounts were equalized in order to have similar signal strengths.
3. Results
3.1. N-Myristoylation affects particularly the subcellular localization of protein kinases
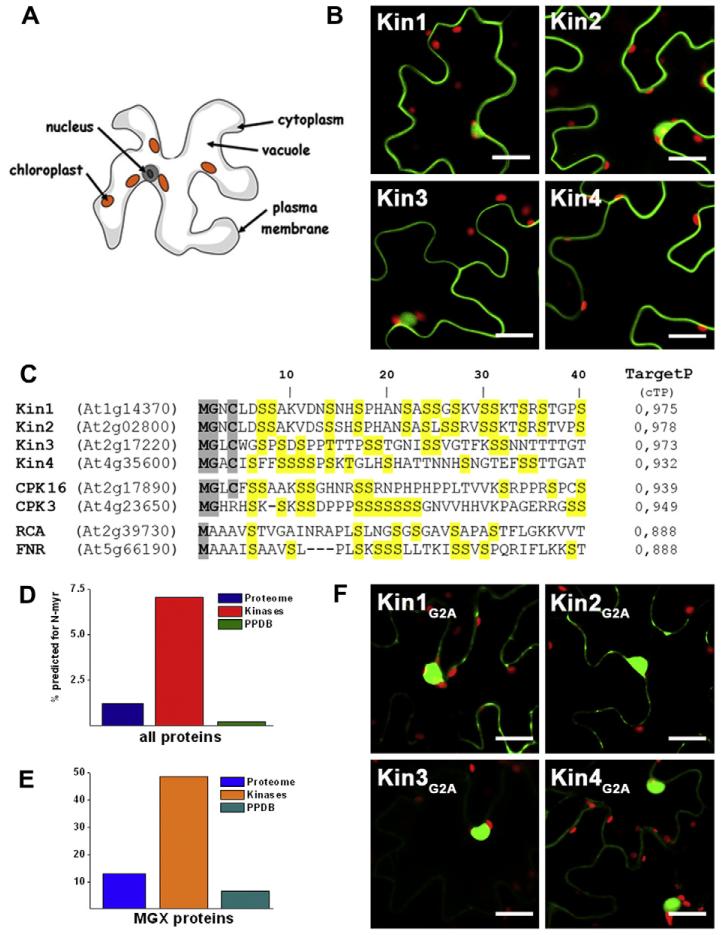
Initially we set out to identify chloroplast localized protein kinases in a bioinformatics based approach using multiple prediction methods. To experimentally test the in vivo localization of our selected candidate genes, we generated C-terminal YFP-fusion proteins and studied their localization by laser scanning microscopy after transient expression in tobacco leaves. Disapointingly, none out of the 10 protein kinases we tested appeared in chloroplasts, instead most of them (8/10) showed a predominant extra-plastidic membrane localization. Typical results are examplified in Fig. 1B for four serine/threonine-specific protein kinases: Kin1 (At1g14370), Kin2 (At2g02800), Kin3 (At2g17220), and Kin4 (At4g35600), which were all highly predicted to be targeted to chloroplasts by at least three different prediction methods. The N-terminal sequences of these kinases are shown in Fig. 1C including their TargetP score for chloroplast targeting. Considering that the known true positive rate of chloroplast prediction by TargetP in Arabidopsis is ~86%, whereas the false positive rate of prediction is ~35% [30] this was a very unexpected result. A similar observation has also been published by Schliebner et al. [31], who analyzed the localization of nine different protein kinases, which were also chloroplast predicted by several algorithms, and found only two of them localized in the chloroplast. As the overlap between the two studies is only one kinase (Kin3, At2g 17220), there is now a total number of 18 protein kinases with high prediction for chloroplast localization by different methods of which only two (11%) appeared in chloroplasts. This prompted us to have a closer look at the N-terminal sequences where we noticed that most (9/10) of the kinases we studied contained motifs for N-myristoylation and many (8/10) also cysteins for additional palmitoylation (Fig. 1C).
Fig. 1.
(A) Scheme of a leaf epidermal cell in which the vacuole fills most of the intracellular space. (B) Confocal microscopy images of tobacco epidermal leaf cells expressing YFP-fusions of Kin1-4 (kinases, green; chloroplasts, red; scale bar = 20 μm). (C) Alignment of the N-terminal 40 amino acids of Kin 1-4, CPK3 and 16, RCA and FNR. TargetP score indicates chloroplast prediction. Residues important for myristoylation and palmitoylation are shaded in grey and serines in yellow to illustrate their enrichment. (D) Comparison of myristoylation prediction for protein kinases compared to the entire Arabidopsis proteome and the chloroplast proteome (PPDB). (E) The same analysis as in (D) but considering only proteins starting with MGX. (F) Confocal microscopy images of tobacco epidermal leaf cells expressing YFP-fusions of the G2A versions of Kin1-4.
Therefore, we extended our analysis to the entire Arabidopsis proteome and asked whether N-myristoylation of protein kinases might be a more general phenomenon to regulate their subcellular localization. Strikingly, 7% of all 965 Arabidopsis protein kinases [32] but only 1.2% (320 proteins) of all other 26270 proteins (TAIR8 release) were predicted to be myristoylated using the myristoylation prediction program Myrist Predictor (http://plantsp.genomics.purdue.edu/html/myrist.html) [16]. On the other hand, only 0.2% (2 proteins) out of the ~1100 experimentally confirmed chloroplast proteins listed in the plant proteome database (PPDB) [33] are predicted to be myristoylated by the Myrist Predictor (Fig. 1D). When we looked more specifically at predicted chloroplast-localized protein kinases we found that 36.8% of those were predicted to be myristoylated whereas only 22.8% of all predicted chloroplast proteins were also predicted to be myristoylated. Looking at the experimentally identified proteins this difference becomes even more obvious: In total, 31 chloroplast annotated proteins in the PPDB database [30] have a glycine at position 2, and only two of them (i.e. At4g03415 and At2g25840) are predicted to be myristoylated. An alignment of the N-termini of 898 annotated chloroplast proteins revealed that clearly the penultimate position of these known chloroplast proteins is typically an Ala (56% of cases), Ser (10%) or Pro (7%) and not a Gly [30]. In comparison, 12.9% of all Arabidopsis proteins starting with a MGX sequence are predicted to be myristoylated compared to 48.6% of all protein kinases (Fig. 1E). The two experimentally identified chloroplast proteins that are predicted to be myristoylated are a protein phosphatase (At4g03415) and a tRNA synthetase (At2g25840) that has been shown to be dually targeted to chloroplasts and mitochondria [34]. However, it is still unclear whether these two proteins are really myristoylated in vivo. We could confirm that Kin1 and Kin3 are indeed N-myristoylated in vitro (Supplementary Fig. 1) and therefore continued to test the effect of N-myristoylation on the chloroplast predicted protein kinases Kin1, Kin2, Kin3, and Kin4 in vivo. We generated non-myristoylatable G2A mutants of those kinases and studied their localization in tobacco epidermal leaf cells. As expected, the localization of all four candidate protein kinases was altered, when myristoylation was abolished (Fig. 1F). The membrane localization was drastically reduced and the proteins accumulated strongly in the nucleus compared to the wild-type proteins but not in the chloroplast as we would have expected.
3.2. N-Acylation overrides chloroplast localization of a calcium-dependent protein kinase (CDPK)
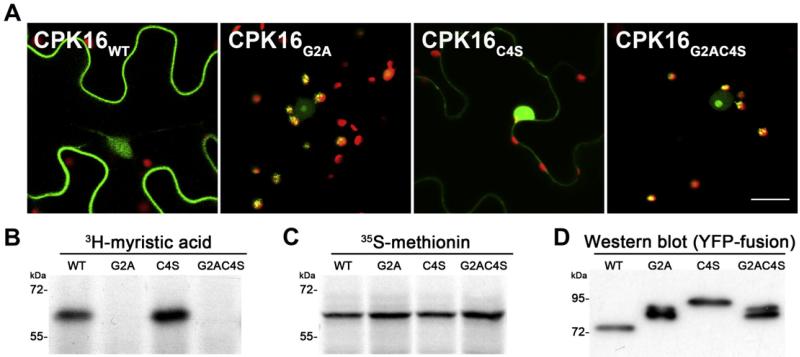
A much more striking effect of N-myristoylation on the subcellular localization of a protein kinase became obvious when analyzing the calcium-dependent protein kinase CPK16 (AT2G17890). CPK16 is also highly predicted to be localized in the chloroplast and harbors N-myristoylation and palmitoylation sites (Fig. 1C). Consistent with its predicted N-acylation, CPK16 appeared predominantly at the plasma membrane in the wild-type form (Fig. 2A). CPK16 is efficiently myristoylated in vitro (Fig. 2B and C), and intriguingly, CPK16 was relocated to chloroplasts when the glycine on position 2 was exchanged for an alanine (Fig 2A). This implies that myristoylation interferes with chloroplast localization. As CPK16 harbors both an N-terminal myristoylation and palmitoylation site, we set out to study the interference of these two modifications with chloroplast localization in more detail. Therefore, the CPK16 mutants C4S and G2AC4S were created in addition to the G2A mutant. They have the cysteine on position 4 exchanged for serine and thus cannot be palmitoylated anymore. Analysis of YFP-fusion proteins in infiltrated tobacco leaves revealed that the C4S mutant, which can still be myristoylated but not palmitoylated, was not targeted to chloroplasts but showed a much stronger nuclear accumulation instead (Fig. 2A). In contrast, the G2AC4S mutant was localized again in chloroplasts, thus suggesting that myristoylation alone inhibits chloroplast localization of CPK16 (Fig. 2A). As expected, only wild-type CPK16 and the C4S mutant could be myristoylated in vitro as shown by the incorporation of 3H-labeled myristic acid (Fig. 2B), while all proteins were translated with similar efficiencies (Fig. 2C). Notably the G2A and the G2AC4S mutant showed also a stronger accumulation in the nucleolus as compared to the wild-type. To rule out that the exchange of amino acids led to chloroplast import inhibition due to perturbation of the chloroplast transit peptide, additional control mutations were generated. A G2V version was created, as alanine is known to be the most frequently occurring amino acid on position 2 of chloroplast proteins, which might be a possible chloroplast targeting determinant [30,35]. Nevertheless, CPK16 G2V still localized to chloroplasts and showed also nuclear accumulation (Supplementary Fig. 2). Furthermore, we wanted to backup our microscopical studies with a biochemical assay. Therefore we extracted total protein extracts of the tobacco leaves expressing the YFP-fusion proteins and analyzed them by Western blotting using an antibody directed against GFP. The wild-type version of CPK16 appeared at a molecular mass of about 75 kDa (Fig. 2D), which was unexpected, but seemed not to be caused by chloroplast import-related processing. Comparison with in vitro translated CPK16-YFP (Supplementary Fig. 1C) revealed that the “full-length” (non-processed) protein appeared at a molecular mass of 93 kDa like the C4S mutant. Most importantly, the G2A and G2AC4S mutants of CPK16-YFP appeared at 85 kDa. TargetP or ChloroP (www.cbs.dtu.dk/services/) predicted a targeting peptide of 75 amino acids for CPK16, which would correspond to 8,3 kDa. Thus, the observed size difference of about 8 kDa would be in perfect agreement with the removal of the chloroplast transit peptide in these mutants after import into the chloroplast, demonstrating that CPK16 harbors a canonical targeting signal that is masked by N-myristoylation.
Fig. 2.
(A) Confocal microscopy images of CPK16 YFP-fusion proteins and mutant versions as indicated in image. The size of the scale bar is 20 μm. (B) Autoradiograph of myristoylation assays of CPK16 and mutants. (C) Autoradiograph of the translation controls. (D) Western blot of CPK16 YFP-fusion proteins and mutant versions from infiltrated tobacco leaves.
3.3. Artificial N-acylation of chloroplast localized proteins inhibits their import
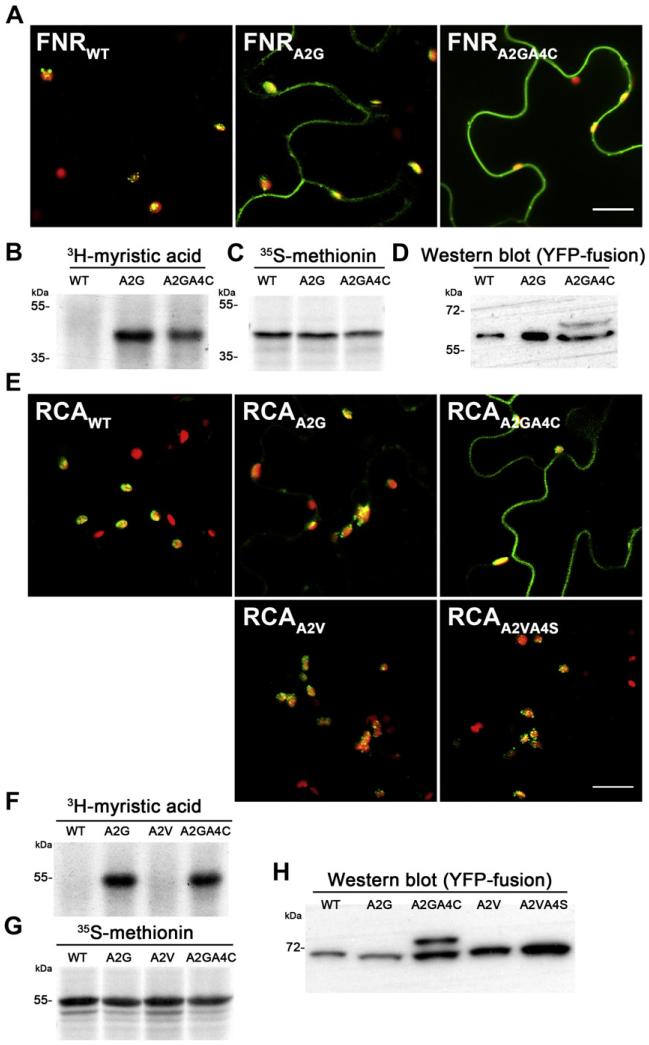
Based on these results we asked, whether it is possible to prevent import of canonical chloroplast proteins by the artificial introduction of N-myristoylation and palmitoylation. Therefore we selected the chloroplast proteins ferredoxin-NADP+ reductase (FNR) (At5g66190) and Rubisco activase (RCA) (At2g39730), which are both lacking a glycine on position 2 and therefore cannot be myristoylated per se. FNR and RCA show exclusive chloroplast localization as YFP-fusion proteins (Fig. 3A and E). However, according to the Myrist Predictor, the exchange of alanine on position 2 for a glycine results in the introduction of a strong myristoylation consensus motif in both proteins. Consequently, both FNR A2G and RCA A2G could be efficiently myristoylated in vitro (Fig. 3B, C and F, G). Interestingly, YFP-fusion proteins of FNR A2G and RCA A2G were still localized predominantly in chloroplasts and only a minor fraction appeared outside of chloroplasts (Fig. 3A and E). Therefore we introduced additional palmitoylation sites by generating A2GA4C mutants of FNR and RCA to test the effect of both N-terminal modifications on chloroplast import. Both mutants were still myristoylated in vitro, indicating that introduction of the cysteine did not eliminate the myristoylation consensus motif (Fig. 3B, C and F, G). But now the analysis of YFP-fusion proteins of FNR A2GA4C and RCA A2GA4C in tobacco leaves revealed that a great part of those mutants was not localized in the chloroplast but showed a strong membrane attachment instead (Fig. 3A and E). This indicates that introduction of myristoylation and palmitoylation impedes chloroplast localization in these cases. To exclude again that the observed changes in localization are caused by mutating the critical alanine at position 2 or by introducing a cysteine at position 4, we generated an A2V and A2VA4S mutant for RCA. Nevertheless, both mutants still showed chloroplast localization as shown in Fig. 3E. These results clearly indicated that chloroplast localization of FNR, and RCA could efficiently be inhibited by N-terminal acylation.
Fig. 3.
(A, E) Confocal microscopy images of FNR and RCA YFP-fusion proteins and mutant versions, respectively (as indicated in image). The scale bar indicates 20 μm. (B, F) Autoradiograph of myristoylation assays of FNR and RCA with their corresponding mutants. (C–G) Autoradiograph of translation controls of FNR and RCA with their corresponding mutants, respectively. (D, H) Western blot of FNR and RCA YFP-fusion proteins and the corresponding mutant versions from infiltrated tobacco leaves.
As already done for CPK16 we analyzed also protein extracts from infiltrated leaves by Western-blotting using the GFP antibody. These results confirmed the observations from fluorescence microscopy. We found that both A2G mutants were still perfectly processed and no unprocessed precursor was detectable in Western blots (Fig. 3D and H), which is in agreement with the observed predominant chloroplast localization. However, the introduction of an additional palmitoylation site, lead to the accumulation of unprocessed precursor in both A2GA4C mutants (Fig. 3D and H). This indicated that for FNR and RCA the N-terminal palmitoylation prevents their chloroplast localization.
4. Discussion
We noticed that currently all methods for prediction of subcellular protein localization do not consider N-terminal acylation. While this weakness seems to be of minor importance in the analysis of diverse protein sets, it becomes particularly important for protein kinases. At the proteome wide scale, they seem to be much more affected as other proteins, notably 48.6% of all protein kinases starting with MGX are predicted to be N-myristoylated. On the other hand, only two proteins with predicted N-myristoylation have been experimentally identified in chloroplasts. Our analysis of predicted chloroplast-localized protein kinases and their G2A mutants clearly showed that N-myristoylation strongly affects their localization, leading to a predominant extra-plastidic membrane attachment of the (wild-type) proteins. Moreover, YFP localization studies on CPK16, FNR, RCA and their acylation mutants revealed that myristoylation as well as palmitoylation is able to interfere with chloroplast localization. However, it seems that this is not a general mechanism but has to be analyzed for each protein separately. In the case of CPK16 abolishing myristoylation in the G2A mutant led to its accumulation in chloroplasts, but removal of only the palmitoylation site in the C4S mutant had no effect on the chloroplast localization and rather affected nuclear accumulation. Thus, we concluded that myristoylation alone did inhibit chloroplast import of CPK16 in vivo. In contrast, abolishing N-myristoylation of CPK3 (At4g23650), another myristoylated CDPK which is also highly predicted to be targeted to chloroplasts (Fig. 1C), did not lead to chloroplast localization as we have shown previously [29].
The artificial introduction of N-myristoylation sites in FNR and RCA in the A2G mutants did only slightly influence chloroplast targeting. Only the additional introduction of palmitoylation sites in the A2GA4C mutants led to a strong accumulation of the proteins outside the chloroplast and appearance of the unprocessed precursor in Western blots from leaf extracts. It seems that in these cases inhibition of chloroplast localization must primarily be attributed to palmitoylation. An explanation therefore would be that acylation does not inhibit the passage of proteins through the chloroplastic TOC-TIC apparatus per se. Acylation may rather direct proteins to different compartments before they can be recognized by chloroplast import components in vivo. For example it is possible that myristoylated CPK16 is recognized by SRP, co-translationally targeted to the ER were it is palmitoylated and subsequently transported to the plasma membrane. In contrast, non-myristoylated CPK16 G2A would not be recognized by SRP and therefore, after completed translation, CPK16 G2A would be available for components of the chloroplast import machinery. Altogether, our studies show that N-terminal protein acylation offers an additional layer to regulate protein targeting, which is of particular importance for protein kinases and needs clearly to be considered in the context of potential organellar targeting.
Supplementary Material
Acknowledgements
We thank Helga Waltenberger for excellent technical support. This work has been funded by the Austrian GEN-AU program in the ERA-PG project CROPP (Project No. 818514), and by the EU in the Marie-Curie ITN COSI (GA 215-174).
Abbreviations
- CDPK
calcium-dependent protein kinase
- FNR
ferredoxin-NADP+ reductase
- PPDB
plant protein database
- RCA
Rubisco activase
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2011.01.001.
References
- [1].Scott MS, Calafell SJ, Thomas DY, Hallett MT. Refining protein subcellular localization. PLoS Comput. Biol. 2005;1:e66. doi: 10.1371/journal.pcbi.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lunn JE. Compartmentation in plant metabolism. J. Exp. Bot. 2007;58:35–47. doi: 10.1093/jxb/erl134. [DOI] [PubMed] [Google Scholar]
- [3].Blobel G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].von Heijne G. On the hydrophobic nature of signal sequences. Eur. J. Biochem. 1981;116:419–422. doi: 10.1111/j.1432-1033.1981.tb05351.x. [DOI] [PubMed] [Google Scholar]
- [5].Emanuelsson O, von Heijne G. Prediction of organellar targeting signals. Biochim. Biophys. Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- [6].Kleinschmidt JH. Membrane proteins – introduction. Cell. Mol. Life Sci. 2003;60:1527–1528. doi: 10.1007/s00018-003-3170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Batistic O, Sorek N, Schultke S, Yalovsky S, Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell. 2008;20:1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- [9].Towler DA, Gordon JI, Adams SP, Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu. Rev. Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- [10].Taniguchi H. Protein myristoylation in protein-lipid and protein-protein interactions. Biophys. Chem. 1999;82:129–137. doi: 10.1016/s0301-4622(99)00112-x. [DOI] [PubMed] [Google Scholar]
- [11].Iwanaga T, Tsutsumi R, Noritake J, Fukata Y, Fukata M. Dynamic protein palmitoylation in cellular signaling. Prog. Lipid Res. 2009;48:117–127. doi: 10.1016/j.plipres.2009.02.001. [DOI] [PubMed] [Google Scholar]
- [12].Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- [13].Sorek N, Bloch D, Yalovsky S. Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 2009;12:714–720. doi: 10.1016/j.pbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- [14].Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-Terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- [15].Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- [16].Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yalovsky S, Rodr Guez-Concepcion M, Gruissem W. Lipid modifications of proteins – slipping in and out of membranes. Trends Plant Sci. 1999;4:439–445. doi: 10.1016/s1360-1385(99)01492-2. [DOI] [PubMed] [Google Scholar]
- [18].Weber CN. Molekulare determinanten fur die palmitoylierung integraler membranproteine. In: Institut fur Immunologie und Molekularbiologie, editor. Ph.D. thesis. Freie Universitat Berlin; Berlin: 2006. [Google Scholar]
- [19].Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- [20].Murray D, Ben-Tal N, Honig B, McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure. 1997;5:985–989. doi: 10.1016/s0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- [21].Beven L, Adenier H, Kichenama R, Homand J, Redeker V, Le Caer JP, Ladant D, Chopineau J. Ca2+-myristoyl switch and membrane binding of chemically acylated neurocalcins. Biochemistry. 2001;40:8152–8160. doi: 10.1021/bi010188e. [DOI] [PubMed] [Google Scholar]
- [22].Pierre M, Traverso JA, Boisson B, Domenichini S, Bouchez D, Giglione C, Meinnel T. N-Myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell. 2007;19:2804–2821. doi: 10.1105/tpc.107.051870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1678. [Google Scholar]
- [24].Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, Borgese N. N-Myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 2005;168:735–745. doi: 10.1083/jcb.200407082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Annweiler A, Hipskind RA, Wirth T. A strategy for efficient in vitro translation of cDNAs using the rabbit beta-globin leader sequence. Nucleic Acids Res. 1991;19:3750. doi: 10.1093/nar/19.13.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Benetka W, et al. Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle. 2008;7:3709–3719. doi: 10.4161/cc.7.23.7176. [DOI] [PubMed] [Google Scholar]
- [29].Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010;63:484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schliebner I, Pribil M, Zuhlke J, Dietzmann A, Leister D. A survey of chloroplast protein kinases and phosphatases in Arabidopsis thaliana. Curr. Genomics. 2008;9:184–190. doi: 10.2174/138920208784340740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gribskov M, Fana F, Harper J, Hope DA, Harmon AC, Smith DW, Tax FE, Zhang G. PlantsP: a functional genomics database for plant phosphorylation. Nucleic Acids Res. 2001;29:111–113. doi: 10.1093/nar/29.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. PPDB, the plant proteomics database at cornell. Nucleic Acids Res. 2009;37:D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Duchene AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pujol C, Marechal-Drouard L, Duchene AM. How can organellar protein N-terminal sequences be dual targeting signals? In silico analysis and mutagenesis approach. J. Mol. Biol. 2007;369:356–367. doi: 10.1016/j.jmb.2007.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.