Abstract
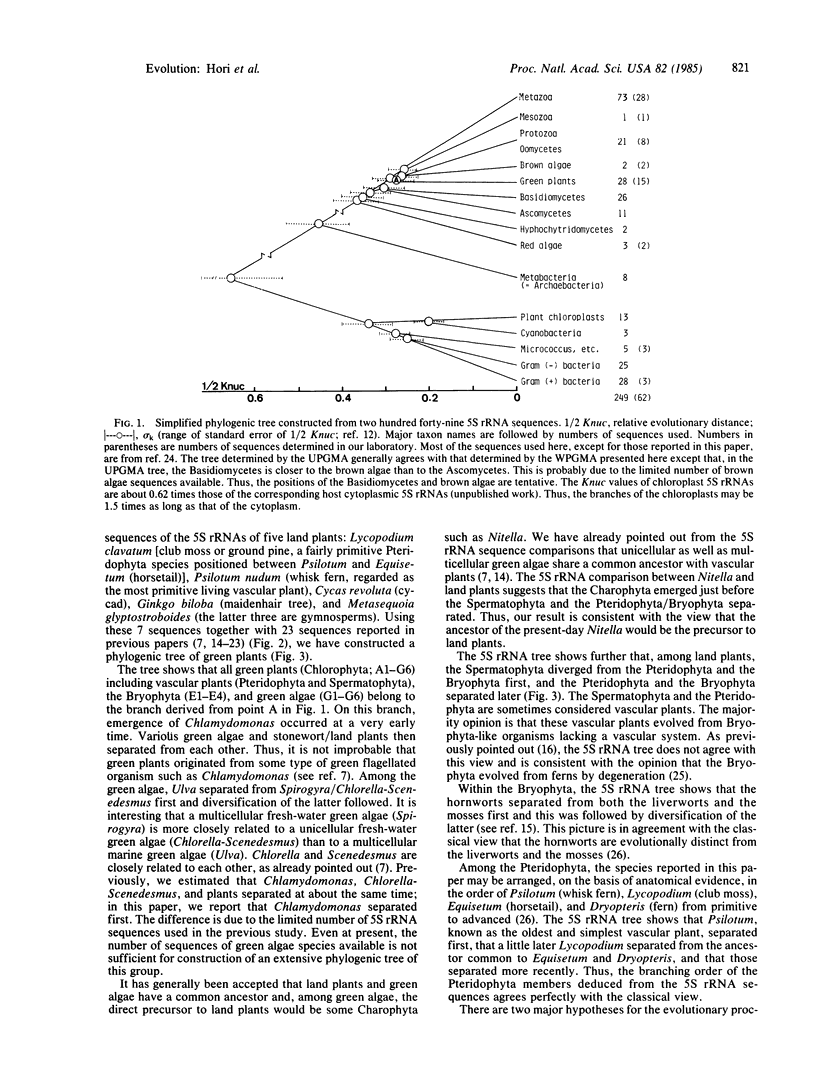
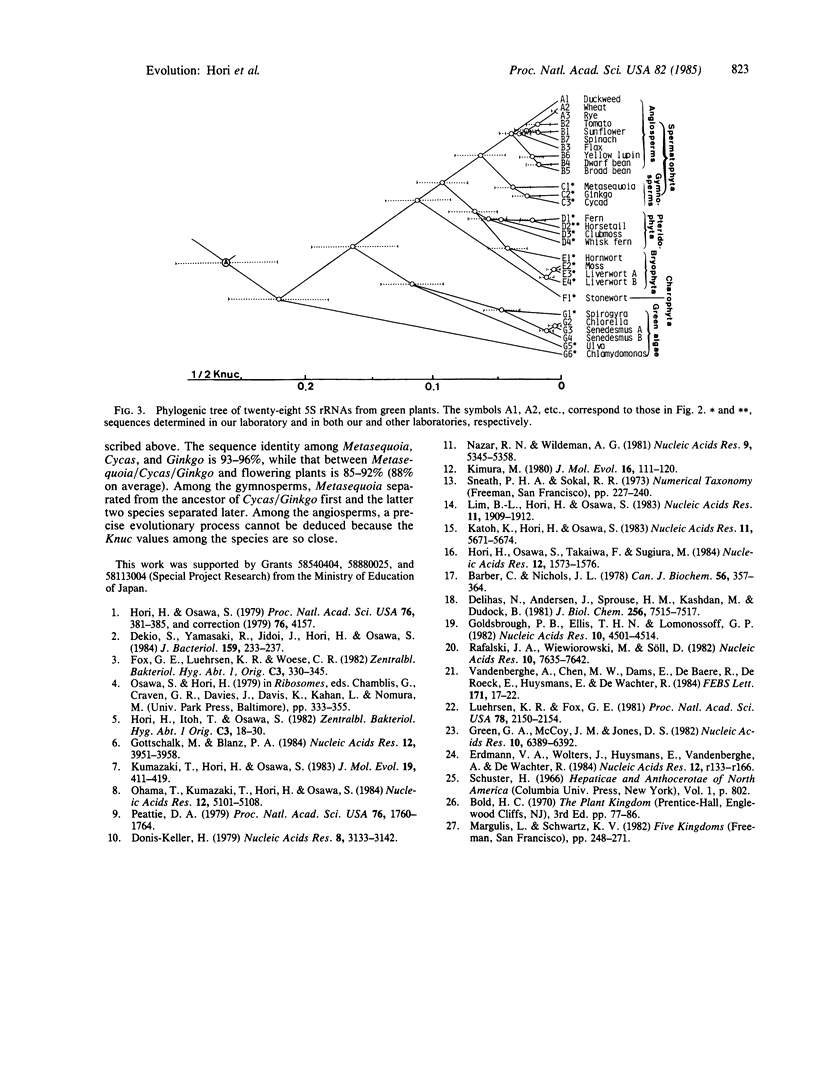
We have constructed a phylogenic tree for green plants by comparing 5S rRNA sequences. The tree suggests that the emergence of most of the uni- and multicellular green algae such as Chlamydomonas, Spirogyra, Ulva, and Chlorella occurred in the early stage of green plant evolution. The branching point of Nitella is a little earlier than that of land plants and much later than that of the above green algae, supporting the view that Nitella-like green algae may be the direct precursor to land plants. The Bryophyta and the Pteridophyta separated from each other after emergence of the Spermatophyta. The result is consistent with the view that the Bryophyta evolved from ferns by degeneration. In the Pteridophyta, Psilotum (whisk fern) separated first, and a little later Lycopodium (club moss) separated from the ancestor common to Equisetum (horsetail) and Dryopteris (fern). This order is in accordance with the classical view. During the Spermatophyta evolution, the gymnosperms (Cycas, Ginkgo, and Metasequoia have been studied here) and the angiosperms (flowering plants) separated, and this was followed by the separation of Metasequoia and Cycas (cycad)/Ginkgo (maidenhair tree) on one branch and various flowering plants on the other.
Keywords: 5S rRNA sequencing, phylogenetic tree
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barber C., Nichols J. L. Conformational studies on wheat embryo 5S RNA using nuclease S1 as a probe. Can J Biochem. 1978 May;56(5):357–364. doi: 10.1139/o78-057. [DOI] [PubMed] [Google Scholar]
- Dekio S., Yamasaki R., Jidoi J., Hori H., Osawa S. Secondary structure and phylogeny of Staphylococcus and Micrococcus 5S rRNAs. J Bacteriol. 1984 Jul;159(1):233–237. doi: 10.1128/jb.159.1.233-237.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N., Andersen J., Sprouse H. M., Kashdan M., Dudock B. The nucleotide sequence of spinach cytoplasmic 5 S ribosomal RNA. J Biol Chem. 1981 Jul 25;256(14):7515–7517. [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Wolters J., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r133–r166. doi: 10.1093/nar/12.suppl.r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough P. B., Ellis T. H., Lomonossoff G. P. Sequence variation and methylation of the flax 5S RNA genes. Nucleic Acids Res. 1982 Aug 11;10(15):4501–4514. doi: 10.1093/nar/10.15.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk M., Blanz P. A. Highly conserved 5S ribosomal RNA sequences in four rust fungi and atypical 5S rRNA secondary structure in Microstroma juglandis. Nucleic Acids Res. 1984 May 11;12(9):3951–3958. doi: 10.1093/nar/12.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green G. A., McCoy J. M., Jones D. S. The nucleotide sequence of 5S rRNA from Scenedesmus obliquus. Nucleic Acids Res. 1982 Oct 25;10(20):6389–6392. doi: 10.1093/nar/10.20.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S., Takaiwa F., Sugiura M. The nucleotide sequences of 5S rRNAs from a fern Dryopteris acuminata and a horsetail Equisetum arvense. Nucleic Acids Res. 1984 Feb 10;12(3):1573–1576. doi: 10.1093/nar/12.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Hori H., Osawa S. The nucleotide sequences of 5S ribosomal RNAs from four Bryophyta-species. Nucleic Acids Res. 1983 Aug 25;11(16):5671–5674. doi: 10.1093/nar/11.16.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S. Phylogeny of protozoa deduced from 5S rRNA sequences. J Mol Evol. 1983;19(6):411–419. doi: 10.1007/BF02102316. [DOI] [PubMed] [Google Scholar]
- Lim B. L., Hori H., Osawa S. The nucleotide sequences of 5S rRNAs from a multicellular green alga, Ulva pertusa, and two brown algae, Eisenia bicyclis and Sargassum fulvellum. Nucleic Acids Res. 1983 Mar 25;11(6):1909–1912. doi: 10.1093/nar/11.6.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Wildeman A. G. Altered features in the secondary structure of Vicia faba 5.8s rRNA. Nucleic Acids Res. 1981 Oct 24;9(20):5345–5358. doi: 10.1093/nar/9.20.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama T., Kumazaki T., Hori H., Osawa S. Evolution of multicellular animals as deduced from 5S rRNA sequences: a possible early emergence of the Mesozoa. Nucleic Acids Res. 1984 Jun 25;12(12):5101–5108. doi: 10.1093/nar/12.12.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski J. A., Wiewiorowski M., Söll D. Organization and nucleotide sequence of nuclear 5S rRNA genes in yellow lupin (Lupinus luteus). Nucleic Acids Res. 1982 Dec 11;10(23):7635–7642. doi: 10.1093/nar/10.23.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]



