Abstract
The classic paradigm of G protein-coupled receptor (GPCR) activation was based on the understanding that agonist binding to a receptor induces or stabilizes a conformational change to an “active” conformation. In the past decade, however, it has been appreciated that ligands can induce distinct “active” receptor conformations with unique downstream functional signaling profiles. Building on the initial recognition of the existence of such “biased ligands”, recent years have witnessed significant developments in several areas of GPCR biology. These include increased understanding of structural and biophysical mechanisms underlying biased agonism, improvements in characterization and quantification of ligand efficacy, as well as clinical development of these novel ligands. Here we review recent major developments in these areas over the past several years.
Introduction
That a given G protein coupled receptor (GPCR) can functionally couple to more than one heterotrimeric G protein has been known for many years. However, it was quite surprising when it was first noted in the mid 90’s that, at a single GPCR, different ligands could be “biased” or “functionally selective” toward one or another of these G proteins. Even more surprising were the discoveries a few years later that GPCRs could also signal through β-arrestins and that ligands could be biased towards either a G protein or β-arrestin-mediated pathways. The study of this important and potentially therapeutically relevant phenomenon has exploded over the last several years. Here we review some of the most important recent developments.
In recent years the list of known biased ligands for GPCRs has grown substantially. While the majority of the ligands identified target the binding site of the endogenous ligand for a given receptor (known as orthosteric ligands), recent work has identified a new class of biased ligands, biased allosteric modulators, which bind non-traditional ligand binding sites topographically distinct from the orthosteric binding site [1]. Biased allosteric modulators are characterized by the ability to modulate agonist affinity and/or efficacy towards a biased receptor conformation without affecting receptor activity on their own. In addition to the discovery of a large number of biased ligands acting on multiple receptor types, several major advances have been made regarding the mechanisms underlying biased agonism.
Mechanistic insights into biased agonism
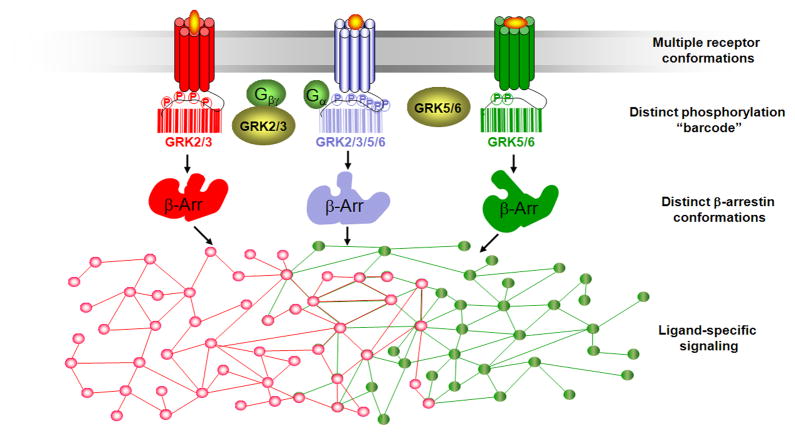
Different receptor states may vary in their ability to activate specific transducers such as G proteins or β-arrestins, as well as to affect transducer functionality in a selective manner. This is supported by the observation that β-arrestin function is dependent on the phosphorylation pattern or “barcode” of the receptor to which it is recruited (figure 1) [2–5]. Nobles et al. [3] demonstrated that β-arrestin recruited to the β2-adrenergic receptor (β2AR) phosphorylated by either G protein receptor kinase (GRK) 2 or GRK6 results in desensitization of receptor signaling and/or receptor internalization whereas only GRK6-phosphorylated β2AR induced recruitment of β-arrestin involved in extracellular-signal regulated kinase 1 and 2 (ERK1/2) activation. This is consistent with previous work on multiple different receptors showing a requirement for GRK2 and GRK3-mediated receptor phosphorylation for receptor internalization, whereas GRK5 and GRK6 mediated receptor phosphorylation is necessary for β-arrestin-dependent ERK1/2 signaling [2, 5–9].
Figure 1. Bar code hypothesis to explain differential functions of β-arrestin.
At the level of the receptor, biased ligands stabilize active receptor conformations structurally distinct from active conformations stabilized by balanced ligands. These unique conformations, in turn, recruit unique subsets of G protein receptor kinases and as a consequence, differential phosphorylation patterns or “bar codes” are generated on the receptor. At the level of the transducer, in this case β-arrestin, phosphorylation on the receptor promotes its recruitment and binding to the receptor. However, different phosphorylation “bar codes” may stabilize distinct, active conformations of transducers with resulting unique functional profiles. These ligand-specific functional profiles promote activity of distinct complex intracellular signaling networks and ultimately lead to divergent physiological responses.
Additionally, distinct populations of phosphorylation sites on the β2AR have been identified for GRK2 compared to GRK6 [3] suggesting bias occurring at the most proximal level of GPCR signal transduction. Interestingly, carvedilol, a weak β-arrestin biased agonist at the β2AR [10], stimulated receptor phosphorylation only at GRK6-specific sites, whereas the full agonist isoproterenol stimulated β2AR phosphorylation at both GRK2- and GRK6-specific sites. These findings demonstrate that ligands may possess the ability to stimulate unique phosphorylation “barcodes” on the receptor, which may in turn result in activation of distinct β-arrestin-mediated cellular functions.
Signaling bias may refer to preferential activation of β-arrestin-dependent signaling compared to G protein-dependent signaling, or vice versa. However a recent study by Blättermann et al. serves as the first example of a biased ligand that discriminates between Gα and Gβγ subunits adding a new dimension to the understanding of biased agonism. In this study of the oxoeicosanoid receptor (OXE-R), they identified an allosteric OXE-R-specific ligand, Gue1654, which selectively inhibits Gβγ but not Gαi signaling induced by the OXE-R agonist, 5-oxo-ETE [11]. Indeed the authors speculated that Gue1654 exerts its effects on the OXE-R as a biased allosteric modulator that causes a change in the unbiased Gβγ and Gαi signaling profile of 5-oxo-ETE to a pattern of exclusive Gαi activation.
These results are somewhat difficult to reconcile with the current understanding of G protein activation where heterotrimeric G protein interacts with active receptor through the Gα subunit causing exchange of Gα-bound GDP with GTP and activation of Gα with release of Gβγ subunits enabling them to initiate signaling on their own [12]. However, it has been indicated that Gi protein activation, specifically, may involve rearrangement of the Gβγ and Gα subunits rather than dissociation [13, 14]. Thus, it is plausible that Gue1654-bound activated OXE-R induces a Gi protein active conformation where solely Gα activity is initiated. Using a bioluminescence resonance energy transfer (BRET)-based assay, it was shown that Gue1654 inhibited agonist stimulated recruitment of Gγ subunit to OXE-R while having no effect on Gα or Gβ recruitment to the receptor [11]. These observations suggest that Gue1654 may promote a spatial separation and/or rearrangement of the Gβγ to the agonist bound OXE-R.
Quantification of Ligand Bias
Quantifying ligand bias is important not only to pharmacologically characterize a compound but also in the design of biased drugs, e.g., for lead optimization and selection of candidate compounds. While qualitative approaches, such as a comparison of efficacies or an assessment of relative rank order of potencies [15–17], can identify extremely biased compounds, they usually cannot identify weakly biased compounds or compare different levels of bias between compounds. Thus, different approaches have been developed to calculate “bias factors” that quantify the level of bias towards G protein- or β-arrestin-mediated signaling [15, 18]. These approaches are based on using simple fits of concentration-response data alone [18], fitting such data combined with dissociation constants from independent binding experiments [18] or fitting concentration-response data with more complicated routines [16]. There is some controversy with regards to the best way to calculate bias factors [15, 16, 19], although it appears that bias factors calculated using these different approaches are, in general, similar to one another.
Methodological Advances
A common means of determining agonist efficacy for specific transducer pathways is by measurement of second messenger production or downstream signaling events in cell-based assays, such as by monitoring production of cAMP or phosphorylation of ERK1/2. However, as noted above, downstream signals are often amplified to different extents depending on the assay being used [18, 20, 21]. Thus, conclusions about transducer efficacy drawn solely based on downstream signaling events may be imprecise and misleading. In this context, novel approaches to determine transducer efficacy directly are of significant interest. One such approach has recently been applied to study direct G protein coupling and activation using BRET-based assays on two receptor systems; angiotensin II type 1a receptor (AT1AR) and oxytocin receptor (OTR) [22, 23]. Apart from characterizing direct interaction between receptor and transducer, an advantage of this approach is that a panel of transducer subtypes can be investigated providing detailed understanding of receptor-transducer pharmacology that is hard to obtain otherwise.
Using this experimental setup it was demonstrated that angiotensin II stimulation of AT1AR caused activation of multiple G proteins, including Gi1, Gi2, Gi3, GoA, GoB, Gq, G11, G13, and Gs. In addition, when stimulated by the β-arrestin-biased agonist [1Ser4Ile8Ile]-angiotensin II (SII), a very small partial activation of several of the same G proteins, including Gi/o and Gq/11, was observed. These results are in conflict with previous findings that SII is a complete β-arrestin-biased agonist with trivial to no effects on Gq activity [24]. In the new study it was shown that SII-stimulated intracellular calcium mobilization and ERK1/2 phosphorylation was blocked by the Gq/11 specific inhibitor YM-254890. In addition, it was shown that SII inhibited cAMP production and that this effect together with ERK1/2 phosphorylation was sensitive to pre-treatment with PTX.
Using the same methodology as Sauliere et al. [22], Busnelli et al. [23] showed that oxytocin stimulation recruits and activates several G proteins including Gi1, Gi2, Gi3, GoA, GoB, and Gq to the OTR. A panel of oxytocin analogs was tested and showed a general inability to stimulate Gi1, GoA, and GoB, indicating Gq, Gi2, and Gi3 subtype bias [23]. Most interesting was a previously identified Gi/o biased ligand, atosiban [25], and a new oxytocin analogue, DNaLOVT, which showed subtype-specific bias towards Gi3 and Gi1, respectively. In contrast to oxytocin, neither of these peptides induced β-arrestin recruitment or β-arrestin-dependent receptor endocytosis demonstrating these analogs are Gi subtype-specific biased ligands [23, 25].
These studies highlight the difficulties in exploring biased agonism in vitro. Cellular signaling profiles are highly sensitive to the systems, cell types, receptor/transducer expression levels, and readouts used to assess them. In their study, Sauliere et. al note differences in the relative contributions of various G protein subtypes as well as β-arrestins 1 and 2 to angiotensin or SII-induced ERK 1/2 activation between overexpressed and endogenous cell types [22]. Whether differences reported in the activity of the biased ligand SII to promote levels of G protein activation are due to differences in cell types, expression levels of receptors, transducers or signaling endpoints such as ERK, or the even varying assays used remains to be determined.
Conformational plasticity and multiple ligand-specific conformations of receptor enable biased GPCR signaling
The “two-state” receptor model (inactive and active states) was previously widely accepted to explain GPCR conformation and function. In this model, all ligands with similar functional capabilities stabilize a common receptor conformation. However there is a growing body of evidence supporting “multi-state” models in which GPCRs are dynamic proteins with a high level of plasticity, manifesting in thermally accessible multiple distinct ligand-specific conformations [26–39]. Recently a novel chemical labeling/quantitative mass spectrometry based proteomics approach was applied to monitor the conformational changes of the β2AR in the presence of nine functionally distinct ligands [27]. Unexpectedly, two patterns of conformational rearrangements of the receptor were observed: one consistent with classic agonism and the other with ligand-specific conformational changes, even amongst functionally similar ligands. These results provide direct evidence for multiple conformational states of the β2AR. Similar evidence supporting a “multi-state” model has been reported for other GPCRs including rhodopsin [40], V2R [34], ghrelin receptor [41], M2 muscarinic acetylcholine receptor (mAChR) [36], cholecystokinin-2 receptor (CCK2R) [37], thromboxane A2 receptor (TP) [42], chemokine receptor CCR5 [38], and glucagon-like peptide-1 receptor (GLP-1R) [35].
Conceptually, GPCRs can be visualized as oscillating amongst a series of conformational intermediates associated with a complex energy landscape [28, 33, 39, 40, 43–45]. This energy landscape is influenced by both ligands and effector proteins bound to the receptor [39, 41]. Pharmacologically distinct ligands regulate receptor activity by shifting the conformational equilibrium and the shape of this landscape. A biased ligand, therefore, should shift the receptor’s conformational equilibrium to a specific state or states that preferentially activates a specific signaling network inside the cell without activating others. Accumulating experimental data has demonstrated that the conformations of a GPCR stabilized by unbiased ligands, G protein-biased ligands, or β-arrestin-biased ligands are distinct from one another [12, 27, 34, 41, 46, 47]. Characterization and comparison of these distinct conformations by a variety of biophysical techniques will provide new insights into the structural basis of biased GPCR signaling.
Molecular and structural mechanisms underlying biased agonism
Recent structural determination efforts as well as biochemical and biophysical studies are beginning to shed light on the mechanisms by which biased ligands regulate receptor activity. The data suggest that conformational changes in transmembrane domain (TM) 7-helix (H) 8 and extracellular loop (ECL) 2 may all play a role in β-arrestin-mediated signaling, whereas conformational changes in TM3, 5 and 6 as well as intracellular loop (ICL) 3 have been associated with G protein-mediated signaling [12, 31, 34, 36, 38, 42, 48, 49]. A site-specific (19)F-NMR study of the β2AR in complex with various ligands revealed that binding of an unbiased agonist induced a G protein-specific active conformation of TM6, whereas binding of putatively β-arrestin-biased ligands primarily impacted the conformational states of TM7 and H8 [31]. Similarly, fluorescence spectroscopy of the V2R showed that conformational changes of TM6-ICL3 are associated with G protein-dependent signaling whereas changes of TM7-H8 domains are associated with β-arrestin-mediated signaling [34].
In addition, it has been recently observed that insertion of a mutation in the interface between TM6 and TM7 that sterically hinders their movement could induce G protein-biased signaling for the CC-chemokine receptor CCR5 [38]. In contrast, G protein-mediated signaling was found to be suppressed in the thromboxane receptor by disruption of a GxxGxxxL helical interaction motif in TM5 [42]. In the muscarinic acetylcholine receptor, mutation of residues Tyr2.61 and Trp3.28 in an allosteric binding pocket introduces biased signaling properties to previously unbiased ligands suggesting these residues may act as molecular switches or gatekeeper residues for functional selectivity [36].
In addition to the biophysical assays described above, several X-ray crystal structures of GPCRs in putatively biased agonist-bound states have been reported in recent years [48, 49]. Comparison of these structures with those of receptors occupied by unbiased ligands has provided further insight into the structural basis of biased agonism [48–51]. For example, for the β1AR, putativelyβ-arrestin-biased ligands interact with additional residues in a “minor” binding pocket near TM7 and ECL2, but have weaker contact with TM5 compared to unbiased ligands [49, 50]. For serotonin receptors, ergotamine (ERG) exhibits strong functional selectivity for β-arrestin-mediated signaling at the 5-HT2B subtype, but is relatively unbiased at the 5-HT1B receptor. Comparison between the crystal structures of these two serotonin receptors occupied by ERG demonstrated the “P5.50-I3.40-F6.44” trigger motif (previously shown to be an interface between TM5, 3 and 6 near the base of the ligand binding pocket in the β2AR) and the D(E)RY motif (TM3) adopt active-like conformation in the 5-HT1B/ERG structure. In contrast, an intermediate active conformation or inactive state is observed in the 5-HT2B/ERG structure [48, 51]. In addition, another key receptor activation microswitch, the NPxxY motif in TM7, as well as the overall conformation of TM7 exhibit more pronounced activation features in the 5-HT2B/ERG structure [48, 51]. For class B GPCRs, recent X-ray crystal structures of the transmembrane domains of the glucagon-like peptide-1 receptor (GLP-1R) and corticotrophin-releasing factor receptor 1 have also provided a structural framework to explain receptor activation and biased agonism [52, 53]. Mutagenesis studies of the human GLP-1R suggested that ECL2 and distinct clusters of polar transmembrane residues of the GLP-1R may serve important roles in receptor activation and biased signaling [35, 54].
The co-crystal structure of the β2AR/Gs complex has also provided structural information about the interface between the receptor and G protein. The receptor interacts with the G protein via TM5, 6, ICL2 and 3. There is no substantial contact with TM7 and H8 [12], domains previously identified to be important in β-arrestin-dependent signaling [34]. However these are still very early days in terms of understanding the structural basis of biased signaling.
Clinical application
The recognition that biased ligands can activate distinct subsets of downstream signaling cascades relative to unbiased ligands has led to a paradigm shift in terms of how ligand efficacy is defined and characterized. In addition, it has stimulated significant interest in the potential clinical implications of these agents. Indeed recent evidence supports the hypothesis that biased ligands may possess unique pharmacologic properties compared to traditional unbiased ligands.
Over the past few years, several biased agonists have begun to advance in clinical development. One example is TRV120027, a β-arrestin-biased ligand at the AT1AR currently in clinical development for the treatment of acute heart failure. Previous work has shown that β-arrestin-biased agonists at the AT1AR stimulate cardiac contractility in isolated cardiac myocytes [55] as well as in in vivo animal studies [56, 57]. This in vivo effect appears to be secondary to a β-arrestin-dependent mechanism promoting myofilament response to calcium via altered protein phosphorylation [58].
In a canine model of heart failure, infusion of TRV120027 resulted in significant increases in cardiac output and renal blood flow as well as decreases in mean arterial pressure (MAP), right atrial pressure, and pulmonary capillary wedge pressure (PCWP) suggesting a unique profile of pharmacologic activity for this biased agonist [57]. When co-administered with the loop diuretic furosemide, furosemide-mediated diuresis and naturesis was maintained, as was glomerular filtration rate, while PCWP was significantly decreased compared to furosemide alone [59]. In first-in-human studies, TRV120027 was found to be safe and tolerable when administered via intravenous infusion and resulted in significant reductions in MAP in those patients with elevated levels of plasma renin activity, a common feature in patients with acute heart failure [60].
In addition to the development of a biased agonist at the AT1AR, recent work has evaluated the use of G protein-biased agonists at the μ-opioid receptor (MOR) as a means to reduce the side effects of unbiased MOR agonists such as morphine. In β-arrestin2 knock out mice, early evidence suggested that morphine analgesia was enhanced and prolonged with reduced desensitization [61], whereas morphine-induced constipation and respiratory depression were reduced compared to wild type animals [62]. It was therefore speculated that a G protein-biased ligand might maintain the analgesic effects of opioids such as morphine while simultaneously reducing unwanted side effects such as respiratory depression and gastrointestinal dysfunction commonly observed with clinically used MOR agonists. Indeed, TRV130, a novel MOR G protein-biased agonist was observed to be potently analgesic while causing less respiratory depression and gastrointestinal dysfunction compared to equianalgesic doses of morphine when administered in mice and rats [63]. Additionally, in first-in-human studies, TRV130 exhibited favorable pharmacokinetic and pharmacodynamics profiles as well as excellent tolerability [64]. These results suggest this G protein-biased agonist could represent a step forward in the clinical development of biased ligands by displaying a profile of maintained desired analgesic effects while simultaneously reducing unwanted side effects mediated via the same receptor.
Conclusions
The classical paradigm of ligand efficacy has undergone major revisions over the past several years with the introduction of concepts such as biased agonism. The recognition that ligands can induce specific receptor activation profiles has stimulated significant interest in obtaining a better understanding of the physiologic, pharmacologic, structural and biophysical mechanisms underlying this phenomenology. Recent research has broadened the understanding of ligand bias and demonstrated that bias may originate at the most proximal sites of cellular signaling such as, for example, receptor phosphorylation patterns. In addition, ligand bias has also been identified for activation of specific subsets of a single transducer e.g. distinct G protein activation patterns. The proliferation of atomic-level information about receptors and receptor/transducer interactions has further enhanced understanding of how specific receptor conformational changes may engender biased patterns of cellular function. Finally, the recent ongoing clinical development of several biased agonists for a variety of indications suggests that future drug design may increasingly consider incorporation of an assessment of ligand bias as a potential means to develop safer and perhaps more effective medications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.May LT, et al. Allosteric modulation of G protein-coupled receptors. Annual review of pharmacology and toxicology. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 2.Zidar DA, et al. Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc Natl Acad Sci U S A. 2009;106(24):9649–54. doi: 10.1073/pnas.0904361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Nobles KN, et al. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci Signal. 2011;4(185):ra51. doi: 10.1126/scisignal.2001707. Using the beta2 adrenergic receptor as a protypical receptor, the authors demonstrate that ligands can induce unique phosphorylation patterns or “bar codes” on the receptor, which in turn leads to different functional profiles within the cell. This paper provides the first evidence for existence of such a “bar code” and demonstrates support for ligand bias at the most proximal levels of GPCR function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones BW, Hinkle PM. Arrestin binds to different phosphorylated regions of the thyrotropin-releasing hormone receptor with distinct functional consequences. Mol Pharmacol. 2008;74(1):195–202. doi: 10.1124/mol.108.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones BW, et al. Phosphorylation of the endogenous thyrotropin-releasing hormone receptor in pituitary GH3 cells and pituitary tissue revealed by phosphosite-specific antibodies. J Biol Chem. 2007;282(17):12893–906. doi: 10.1074/jbc.M610854200. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, et al. Functional antagonism of different G protein-coupled receptor kinases for beta-arrestin-mediated angiotensin II receptor signaling. Proc Natl Acad Sci U S A. 2005;102(5):1442–7. doi: 10.1073/pnas.0409532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren XR, et al. Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc Natl Acad Sci U S A. 2005;102(5):1448–53. doi: 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenoy SK, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281(2):1261–73. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 9.Kara E, et al. A phosphorylation cluster of five serine and threonine residues in the C-terminus of the follicle-stimulating hormone receptor is important for desensitization but not for beta-arrestin-mediated ERK activation. Mol Endocrinol. 2006;20(11):3014–26. doi: 10.1210/me.2006-0098. [DOI] [PubMed] [Google Scholar]
- 10.Wisler JW, et al. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proc Natl Acad Sci U S A. 2007;104(42):16657–62. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11• •.Blattermann S, et al. A biased ligand for OXE-R uncouples Galpha and Gbetagamma signaling within a heterotrimer. Nat Chem Biol. 2012;8(7):631–8. doi: 10.1038/nchembio.962. The authors demonstrate that Gue1654 acts as a biased allosteric modulator at the oxoeicosanoid receptor shifting the signaling profile of the unbiased endogenous ligand to biased Gαi-mediated signaling only. This work represents the first example of a biased ligand discriminating between different G protein subunits. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunemann M, Frank M, Lohse MJ. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci U S A. 2003;100(26):16077–82. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank M, et al. G Protein activation without subunit dissociation depends on a G{alpha}(i)-specific region. J Biol Chem. 2005;280(26):24584–90. doi: 10.1074/jbc.M414630200. [DOI] [PubMed] [Google Scholar]
- 15.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12(3):205–16. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 16.Kenakin T, Christopoulos A. Measurements of ligand bias and functional affinity. Nat Rev Drug Discov. 2013;12(6):483. doi: 10.1038/nrd3954-c2. [DOI] [PubMed] [Google Scholar]
- 17.Kenakin T, et al. A simple method for quantifying functional selectivity and agonist bias. ACS chemical neuroscience. 2012;3(3):193–203. doi: 10.1021/cn200111m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Rajagopal S, et al. Quantifying ligand bias at seven-transmembrane receptors. Mol Pharmacol. 2011;80(3):367–77. doi: 10.1124/mol.111.072801. In this work, a method for quantification of signaling bias is proposed incorporating equimolar and equiactive responses as well as estimates of coupling efficiency derived from the operational model using experimentally determined dissociation constants. This method was validated on two different receptors and serves as a general method for quantification of ligand bias. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajagopal S. Quantifying biased agonism: understanding the links between affinity and efficacy. Nat Rev Drug Discov. 2013;12(6):483. doi: 10.1038/nrd3954-c1. [DOI] [PubMed] [Google Scholar]
- 20.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9(5):373–86. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onaran HO, Costa T. Where have all the active receptor states gone? Nat Chem Biol. 2012;8(8):674–7. doi: 10.1038/nchembio.1024. [DOI] [PubMed] [Google Scholar]
- 22•.Sauliere A, et al. Deciphering biased-agonism complexity reveals a new active AT1 receptor entity. Nat Chem Biol. 2012;8(7):622–30. doi: 10.1038/nchembio.961. Utilizing a novel bioluminescence resonance energy transfer assay, the authors assessed ligand bias at the level of G protein subunits. This serves as the first example of the recognition of contributions of distinct G protein subunits to the signaling profiles of different ligands. [DOI] [PubMed] [Google Scholar]
- 23.Busnelli M, et al. Functional selective oxytocin-derived agonists discriminate between individual G protein family subtypes. J Biol Chem. 2012;287(6):3617–29. doi: 10.1074/jbc.M111.277178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei H, et al. Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc Natl Acad Sci U S A. 2003;100(19):10782–7. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reversi A, et al. The oxytocin receptor antagonist atosiban inhibits cell growth via a “biased agonist” mechanism. J Biol Chem. 2005;280(16):16311–8. doi: 10.1074/jbc.M409945200. [DOI] [PubMed] [Google Scholar]
- 26.Kobilka B. The structural basis of G-protein-coupled receptor signaling (Nobel Lecture) Angew Chem Int Ed Engl. 2013;52(25):6380–8. doi: 10.1002/anie.201302116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Kahsai AW, et al. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat Chem Biol. 2011;7(10):692–700. doi: 10.1038/nchembio.634. The authors developed a quantitative mass spectrometric strategy to investigate dynamic receptor conformational changes induced by different lignads. Contrary to the classical two-state model of receptor activation, multiple, ligand-specific conformations were observed supporting the presence of multiple active receptor conformations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deupi X, Kobilka BK. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25(5):293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106(23):9501–6. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao X, et al. Coupling ligand structure to specific conformational switches in the beta2-adrenoceptor. Nat Chem Biol. 2006;2(8):417–22. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- 31• •.Liu JJ, et al. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335(6072):1106–10. doi: 10.1126/science.1215802. Using site-specific FNMR labels in the beta2 adrenergic receptor in complexes with various ligands, including both putatively biased and unbiased ligands, the authors observed selective effects of different ligands on the conformational equilibria of the receptor. This work further demonstrates the heterogeniety of “active” receptor conformations induced by biased and unbiased ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altenbach C, et al. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci U S A. 2008;105(21):7439–44. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West GM, et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR beta2 adrenergic receptor revealed by HDX. Structure. 2011;19(10):1424–32. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Rahmeh R, et al. Structural insights into biased G protein-coupled receptor signaling revealed by fluorescence spectroscopy. Proc Natl Acad Sci U S A. 2012;109(17):6733–8. doi: 10.1073/pnas.1201093109. The authors utilized fluorescence spectroscopy on the arginine-vasopressin type 2 receptor to demonstrate that G protein-biased ligands stablize distinct conformations from beta-arrestin-biased ligands. This work again demonstrates the presence of multiple ligand-specific “active” receptor states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koole C, et al. Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. J Biol Chem. 2012;287(6):3642–58. doi: 10.1074/jbc.M111.309328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory KJ, et al. Stimulus bias provides evidence for conformational constraints in the structure of a G protein-coupled receptor. J Biol Chem. 2012;287(44):37066–77. doi: 10.1074/jbc.M112.408534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magnan R, et al. Distinct CCK-2 receptor conformations associated with beta-arrestin-2 recruitment or phospholipase-C activation revealed by a biased antagonist. J Am Chem Soc. 2013;135(7):2560–73. doi: 10.1021/ja308784w. [DOI] [PubMed] [Google Scholar]
- 38.Steen A, et al. Biased and constitutive signaling in the CC-chemokine receptor CCR5 by manipulating the interface between transmembrane helices 6 and 7. J Biol Chem. 2013;288(18):12511–21. doi: 10.1074/jbc.M112.449587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39• •.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152(3):532–42. doi: 10.1016/j.cell.2013.01.008. The authors utilized NMR spectrosocopy on the beta2 adrenergic receptor to demonstrate the presence of distinct conformational states not observed in previously published crystal strucutres as well as significant conformational heterogeniety in agonist- and inverse agonist-bound receptors. This observed heterogeniety provides evidence for the distinct conformational changes necessary to engage specific transducers and regulatory proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orban T, et al. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20(5):826–40. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mary S, et al. Ligands and signaling proteins govern the conformational landscape explored by a G protein-coupled receptor. Proc Natl Acad Sci U S A. 2012;109(21):8304–9. doi: 10.1073/pnas.1119881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey AJ, et al. Biased suppression of TP homodimerization and signaling through disruption of a TM GxxxGxxxL helical interaction motif. J Lipid Res. 2013;54(6):1678–90. doi: 10.1194/jlr.M036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abrol R, et al. Conformational ensemble view of G protein-coupled receptors and the effect of mutations and ligand binding. Methods Enzymol. 2013;520:31–48. doi: 10.1016/B978-0-12-391861-1.00002-2. [DOI] [PubMed] [Google Scholar]
- 44.Abrol R, et al. Characterizing and predicting the functional and conformational diversity of seven-transmembrane proteins. Methods. 2011;55(4):405–14. doi: 10.1016/j.ymeth.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kofuku Y, et al. Efficacy of the beta(2)-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat Commun. 2012;3:1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474(7352):521–5. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332(6027):322–7. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340(6132):615–9. doi: 10.1126/science.1232808. Taking advantage of the fact that ergotamine functions as an unbiased ligand at the 5HT1B receptor and a beta arrestin-biased agonist at the 5HT2B receptor, the authors presented X-ray crystal structures of both ligand-bound receptor complexes. Comparisons between these two structures provided the first X-ray crystallographic-level structural information regarding biased vs. unbiased active receptor states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warne T, et al. Crystal structures of a stabilized beta1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure. 2012;20(5):841–9. doi: 10.1016/j.str.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warne T, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469(7329):241–4. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C, et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340(6132):610–4. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hollenstein K, et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499(7459):438–43. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 53.Siu FY, et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499(7459):444–9. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wootten D, et al. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc Natl Acad Sci U S A. 2013;110(13):5211–6. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajagopal K, et al. Beta-arrestin2-mediated inotropic effects of the angiotensin II type 1A receptor in isolated cardiac myocytes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(44):16284–9. doi: 10.1073/pnas.0607583103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Violin JD, et al. Selectively engaging beta-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. The Journal of pharmacology and experimental therapeutics. 2010;335(3):572–9. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- 57.Boerrigter G, et al. Cardiorenal actions of TRV120027, a novel beta-arrestin-biased ligand at the angiotensin II type I receptor, in healthy and heart failure canines: a novel therapeutic strategy for acute heart failure. Circulation Heart failure. 2011;4(6):770–8. doi: 10.1161/CIRCHEARTFAILURE.111.962571. [DOI] [PubMed] [Google Scholar]
- 58.Monasky MM, et al. The beta-arrestin-Biased Ligand TRV120023 Inhibits Angiotensin II-Induced Cardiac Hypertrophy While Preserving Enhanced Myofilament Response to Calcium. American journal of physiology. Heart and circulatory physiology. 2013 doi: 10.1152/ajpheart.00327.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boerrigter G, et al. TRV120027, a novel beta-arrestin biased ligand at the angiotensin II type I receptor, unloads the heart and maintains renal function when added to furosemide in experimental heart failure. Circulation Heart failure. 2012;5(5):627–34. doi: 10.1161/CIRCHEARTFAILURE.112.969220. [DOI] [PubMed] [Google Scholar]
- 60.Soergel DG, et al. First Clinical Experience with TRV027: Pharmacokinetics and Pharmacodynamics in Healthy Volunteers. Journal of clinical pharmacology. 2013;53(9):892–9. doi: 10.1002/jcph.111. [DOI] [PubMed] [Google Scholar]
- 61.Bohn LM, et al. Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science. 1999;286(5449):2495–8. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- 62.Raehal KM, Walker JK, Bohn LM. Morphine side effects in beta-arrestin 2 knockout mice. The Journal of pharmacology and experimental therapeutics. 2005;314(3):1195–201. doi: 10.1124/jpet.105.087254. [DOI] [PubMed] [Google Scholar]
- 63.DeWire SM, et al. A G protein-biased ligand at the mu-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. The Journal of pharmacology and experimental therapeutics. 2013;344(3):708–17. doi: 10.1124/jpet.112.201616. [DOI] [PubMed] [Google Scholar]
- 64.Soergel DG, et al. First Human Dosing Experience with TRV130, a Biased Ligand at the Mu Opioid Receptor (P02.013) Neurology. 2013;80(Meeting Abstracts 1):P02.013. [Google Scholar]