Abstract
The past decade has witnessed an impressive expansion of our knowledge of retinal photoreceptor signal transduction and the regulation of the visual cycle required for normal eyesight. Progress in human genetics and next generation sequencing technologies have revealed the complexity behind many inherited retinal diseases. Structural studies have markedly increased our understanding of the visual process. Moreover, technical innovations and improved methodologies in proteomics, macromolecular crystallization and high resolution imaging at different levels set the scene for even greater advances. Pharmacology combined with structural biology of membrane proteins holds great promise for developing innovative accessible therapies for millions robbed of their sight or progressing toward blindness.
Keywords: Rhodopsin, arrestin, transducin, RPE65, retinoid isomerase, ABCA4, RPE, photoreceptor
Introduction
Recent elucidation of the complete transcriptome of the retina and accurate analysis of the retinal proteome by mass spectrometry continue to reveal protein products encoded by retinal transcripts and their modifications [1]. Many of these proteins contribute to the formation and function of photoreceptors, which are unique highly-differentiated light-sensing cells that comprise more than 80% of cells in the retina and initiate the process of vision [2]. Each of the genes encoding these proteins is susceptible to potential blinding mutations. A detailed representation of these protein structures and their assemblies will lead to improved understanding of retinal processes, pathologies, and rational development of drugs to combat retinal diseases.
Photoreceptor cells and specifically the sites of phototransduction, namely rod and cone outer segments (ROS and COS), contain many proteins that are integrated into plasma or disc membranes [3, 4]. Fortunately, the challenge of crystallizing such proteins is rapidly being overcome by novel methods that are becoming routine. Vision research has produced outstanding contributions to the field of structural biology and physiology. Notable examples include determining the first structure of a membrane-bound G protein-coupled receptor (GPCR), vertebrate rhodopsin [5–8]. This was followed by structures of an invertebrate homolog [9, 10], rhodopsin mutants [11], many rhodopsin photoactivated intermediates and other derivatives [12–19], structures of G proteins alone and in complex with other proteins, and the structure of the RPE65 isomerase involved in visual chromophore regeneration [20, 21]. As the breadth of our structural knowledge of phototransduction/visual cycle proteins expands from an estimated 10%, we should be able to translate genetic changes into visual pathology and use network pharmacology to design and develop novel therapeutics to halt the progression of blinding retinal diseases.
Rhodopsin and GPCRs
Determination of the X-ray structure of rhodopsin from a native source offered the first atomic view of an archetypical GPCR [5], revealing arrangements of the transmembrane helices around the 11-cis-retinyldene ligand and the unanticipated feature of an extracellular domain. Different crystallization conditions have trapped different conformations of rhodopsin’s intracellular loop-III [7, 12], which comprises part of the heterotrimeric G protein binding site. New insights into rhodopsin activation have been provided by X-ray structures for the activated forms (Meta II [11, 14]) and chromophore-free forms, called opsins [13, 14]).
Because most GPCRs have unique extracellular regions accessible to pharmacological agents, these new advances will improve future structure-based drug design. But what is truly remarkable is that the overall folding of these receptors, including their transmembrane and cytoplasmic regions, is highly analogous (Fig 1). Thermal motions of atoms are reflected in their B-factors. Although there could be parameters other than thermal motion that potentially influence B-factor values, it appears that flexibility (quantified by their thermal motion) differs among these receptors. This suggests that these structures are fine-tuned for their rates of activation, amplification of the light signal, and timing of desensitization.
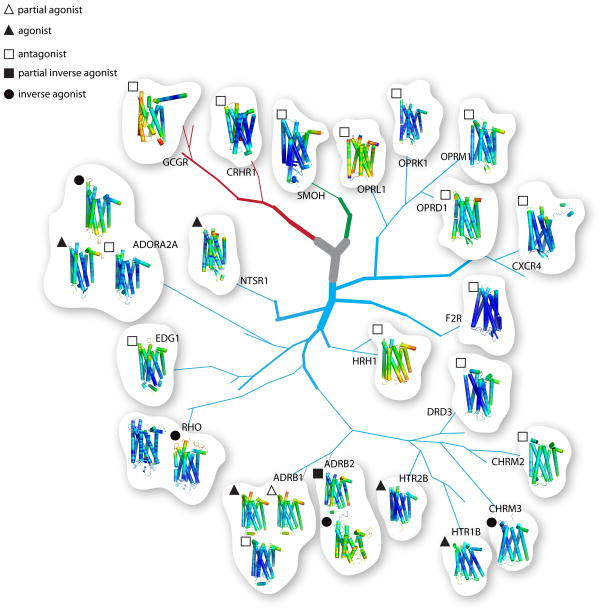
Fig. 1. Phylogenetic tree of GPCRs with known X-ray structures.
Branches and sub-branches (detailed methods are described elsewhere) of the GPCR phylogenetic tree are shown (adapted from Katritch et al. [25]). Branches of varying thickness represent different subgroups. Known structures from class A (rhodopsin subfamily, light blue branches), class B (secretin subfamily, red branches), and class F (frizzled, green branches) GPCRs are shown, but other groups such as the adhesion and glutamate GPCRs are not included. Structures of known GPCRs obtained in the presence of different ligands are clustered together. Types of ligands are identified by geometrical shapes placed at the top left corner of each individual GPCR structure. Structures with partial agonists are denoted with open triangles, agonists are highlighted with closed triangles, antagonists are identified by open squares, partial inverse agonists are marked by closed squares and inverse agonists are shown with closed circles. GPCRs bound to different inhibitors are identified by their gene names. B-factors for each individual GPCR structure were used to generate the GPCR rainbow color coding (blue through red, for minimum and maximum B-factor values, respectively). The gene names of the clustered structures and their corresponding PDB codes are: RHO (1U19 and 3CAP); ADRB1 (2VT4, 2Y02, and 2Y01); ADRB2 (2R4R and 2RH1); HTR2B (4IB4); HTR1B (4IAQ); CHRM3 (4DAJ); CHRM2 (3UON); DRD3 (3PBL); HRH1 (3RZE); F2R (3VW7); CXCR4 (3ODU); OPRD1 (4EJ4); OPRM1 (4DKL); OPRK1 (4DJH); OPRL1 (4EA3); NTSR1 (4GRV); ADORA2A (3QAK; 3VGA), and 3EML); EDG1 (3V2W); CRF1R (4K5Y); GCGR (4L6R); SMOH (4JKV).
Although structures derived from X-ray crystallography have provided a wealth of knowledge, complementary biophysical techniques have also offered valuable insights into the structure/function of rhodopsin. For example, the activation mechanism that could involve an intricate molecular network of internal water molecules was first suggested based on X-ray structural analysis [7], and later supported by evidence obtained through complementary biophysical studies such as radiolytic footprinting and hydrogen-deuterium exchange [22–24].
To date, structures have been solved for roughly two dozen GPCRs from three different classes (Fig. 1). Due to crystallization difficulties, several methodologies have been employed to obtain well-diffracting crystals, including co-crystallization with a Fab fragment of a monoclonal antibody and fusion with T4 lysozyme (T4L) or cytochrome b562RIL onto the N-terminus or into the third intracellular loop [25–28]. Other methods involved the use of a novel class of detergents, bicelles [29], lipid cubic phase [30–32] or mutagenic approaches to thermodynamically stabilize the target GPCRs [26]. This multitude of structures provides a valuable snapshot of GPCR conformations with a variety of ligands. Recently, members of class B (red branch [33, 34]) and class F (green branch [35]) have been added to the growing list of GPCRs with known structures from class A (light blue branch).
Photoactivation of rhodopsin
A number of excellent reviews devoted to various aspects of the photoactivation of rhodopsin have been written [36] based on high resolution structures determined by X-ray crystallography and probed by NMR or FTIR [5–10, 12, 13, 15–19, 37–40]. Major conclusions from these studies can be summarized as follows: 1) isomerization of the chromophore initially causes retention of all-trans-retinylidene in the active site within the space occupied by 11-cis-retinylidene; 2) relaxation of the protein and the photoisomerized chromophore lead to an inward movement of helix VI by about 6 Å, along with small adjustments of other helices; 3) photoactivation of rhodopsin produces fluctuations and relaxation of the overall GPCR structure; 4) internal embedded water molecules provide a key non-protein component of activation, transmitting activation-associated structural changes from the chromophore active site to the protein cytoplasmic surface [22, 41]. These changes were first probed with structures of rhodopsin and opsin (Fig. 2), but the complex of this GPCR with the G protein transducin (Gt) has not yet been determined at atomic resolution. Notably, multiple rhodopsin crystals represent a single conformation trapped under specific crystallization conditions, but in reality rhodopsin could switch between different conformations in native membranes [19, 39]. Ultimately, photoactivated rhodopsin relaxes prior to adopting a specific conformation as it forms a transient complex with Gt (Fig. 2) [24].
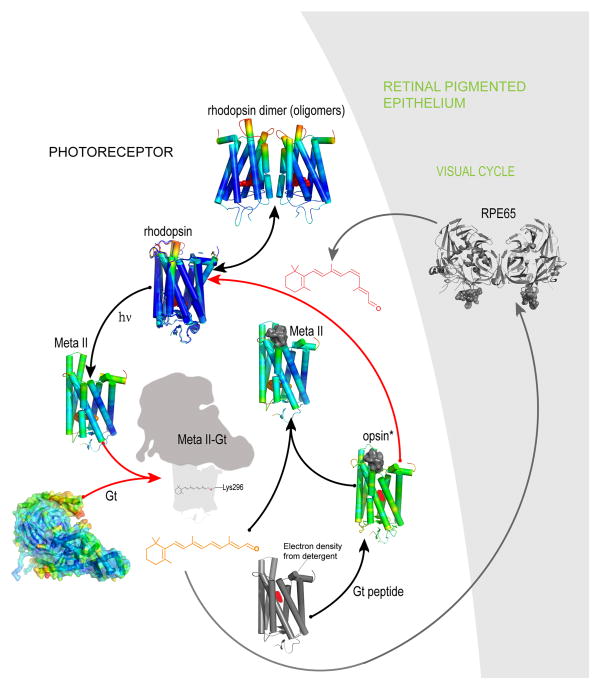
Fig. 2. Structures of rhodopsin, opsin and activated intermediates determined by X-ray crystallography.
Color coding of the shown structures was derived from B- factors of the αC atoms as noted in Fig. 1. Ground state rhodopsin (with bound 11-cis-retinal depicted as a red sphere) is activated by a photon of light. Prepared under different experimental conditions, presented GPCR structures were superimposed on ground state rhodopsin and generated using PDB codes: 1U19, 3C9l, 1GZM, 2I35, and 2I36. Photoisomerization of 11-cis-retinal to all-trans-retinal (shown as orange spheres) is accompanied by conformational changes in the transmembrane helices together with a variety of local environmental changes (such as rearrangement of the internal water molecular network). Some of these changes are reflected in B-factor values. The Meta II structure (PDB code 3PXO) contains the all-trans-retinal chromophore. A potential structure of the Meta II-Gt complex is shown in dark grey and light grey shades and indicated by red arrows. The structures of both opsin* and opsin* with bound Gt peptide lack a visual chromophore (PDB codes 3CAP (in gray cartoons because the B-factors for this structure are incorrect) and 3DQB, respectively) and both can be converted to a Meta II state [14]) in the presence of all-trans-retinal (orange sticks) (black arrows, PDB code 3PQR). Opsin structures are not apo-receptors, because they contained bound detergent molecules (depicted in red) [90]. The Gt peptide is represented as grey surface. However, addition of 11-cis-retinal (red sticks) to opsin* crystals cannot successfully regenerate ground state rhodopsin (red arrow). RPE65 (4F3D), a component of the visual cycle, is shown as a dark gray cartoon on a light gray background denoting the retinal pigmented epithelium (RPE). Recycling of all-trans-retinal to 11-cis-retinal through the visual cycle is indicated by gray arrows. Rhodopsin has propensity to oligomerize in both native and expression systems [91–93].
Molecular interactions stabilizing rhodopsin
The mechanism of rhodopsin folding is not well understood. However, a number of mutations in rhodopsin have been shown to associate with its structural destabilization and misfolding, thus leading to loss of visual function. Hence, understanding the molecular interactions critical for proper folding of this receptor in its native environment is of great interest but limited by available methodologies. In 2006, we first employed atomic force microscopy (AFM)-based single molecule force spectroscopy (SMFS) to map, for the first time, specific segments with highly conserved and functionally important residues in native bovine rhodopsin that stabilize its secondary structure [42]. Further studies comparing rhodopsin with G90D rhodopsin, a mutant associated with congenital stationary night blindness, detected decreased thermodynamic stability and increased mechanical rigidity of most structural segments in the mutant, providing insights into the nature of its pathology. Furthermore, SMFS was proven useful for detecting molecular interactions stabilizing rhodopsin in its different photo-states and various animal species [43, 44]. Further similar work followed with the β2-adrenergic receptor [45]. The differences and similarities between two receptors were noted [46].
Rhodopsin and its G protein, transducin
Protein conformational dynamics represent the quintessential mechanism of GPCR-mediated signal transduction [47]. Following absorption of a photon of light, a series of activated conformational and dynamic changes are initiated in rhodopsin that culminate in its signaling in the Meta II state. This activation translates into movements of helixes V and VI [14]. But more generally, activation results in a general relaxation of the rhodopsin molecule [24]. This relaxation, and perhaps formation of several coexisting states [19], expose binding sites on Meta II rhodopsin that are required for optimal interaction with the phototransduction-specific Gt. This interaction involves a conformational change in the α5 helix of Gt. From a dynamic perspective, a similar sequence of events was found to take place for rhodopsin and the β2-adrenergic receptor. For example, activation placed both receptors in a much more dynamic state, whereas binding to their corresponding G proteins reduced this flexibility [24]. The same trend was documented for the cognate G proteins; both free Gs and Gt exhibited increased dynamics upon binding to their activated receptors [24, 48]. Although a high resolution structure of the Meta II-Gt complex has not yet been determined, the available X-ray structure of the β2-adrenergic receptor in complex with its G protein (Gs) [49] provides a glimpse of the domain organization for such a complex. The modified β2-adrenergic receptor-bound α-helical domain of Gs exhibits a profound rigid body movement as compared with the X-ray structure of the free G protein [50], as well as with the structures of more than 40 heterotrimeric G protein structures determined to date. Notably, if this rigid structure is inserted into a simulated [51] phospholipid bilayer (Fig. 3), a conformational clash of the α-helical domain of Gs with the membrane becomes apparent. Therefore, the exact orientation of the α-domain of Gs relative to the lipid bilayer requires further evaluation in light of this apparent overlap.
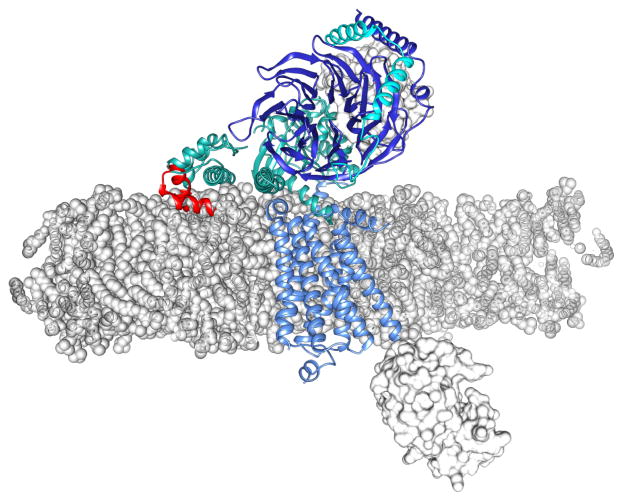
Fig. 3. The β2-adrenergic-Gs complex modeled into a phospholipid membrane bilayer.
The β2-adrenergic receptor in complex with Gs (PDB code: 3SN6) is inserted in a lipid bilayer. The phospholipid units (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) of the lipid bilayer are depicted as gray spheres and the model has been described in detail elsewhere [51]. The position of the β2-adrenergic transmembrane domain (light blue cartoon) along the z axis of the phospholipid bilayer was chosen by superposition with rhodopsin (not shown). The position of rhodopsin (PDB code: 1U19) in turn is presented such that the palmitoyl fatty acyl chains attached to residues Cys322 and Cys323 are positioned at the same depth as the phospholipid fatty acyl chains of the membrane leaflet. Both the Nb35 nanobody and T4 lysozyme [49] are manifested as light gray structures. The Gs domains are represented as follows: Gsα and Gsγ are shown as cyan cartoons whereas Gsβ is pictured as a blue cartoon. Amino acid residues (Asn121 to Leu153) of the Gsα domain that overlap with the phospholipid head group region of the bilayer model are shown as a red cartoon. The face-on view of the transmembrane region is presented as a z-plane section of the phospholipid bilayer.
Lower resolution structures obtained by using negative staining coupled to electron microscopy (EM) have also been solved for both complexes [52–54]. Although EM models of the two receptors appear similar in shape, interpretations of the data differ. In the case of the β2-adrenergic receptor, the extra electron density in the transmembrane region was attributed to detergent [54], whereas for rhodopsin, the extra space left after fitting in the monomeric receptor was attributed to the presence of a second rhodopsin molecule, in agreement with the previously determined 2:1 rhodopsin:Gt stoichiometry (Fig. 4) [52, 53, 55]. We attempted to resolve this inconsistency by using the same detergent as that in the β2-adrenergic receptor study, but the presence of the second receptor molecule remained the only satisfactory explanation for the observed additional density [53, 56]. In contrast, EM single particle analysis of the T4L-β2AR-Gs complex demonstrated only a single density corresponding to the T4L tag linked to the N-terminus of β2-adrenergic receptor. Nevertheless, extensive biochemical analyses strongly suggest that the rhodopsin dimer is in fact asymmetric in nature [53, 55, 56]. This asymmetry allows the interaction of Gt with the C-terminus of only one of the two rhodopsin molecules, consistent with the model obtained from the EM study.
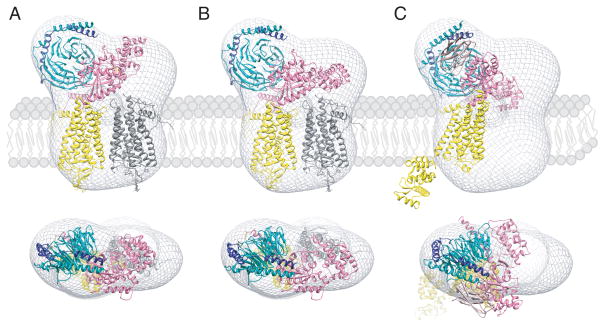
Fig. 4. Orientation of GPCR-G protein complexes in a phospholipid membrane.
Semi-empirical models of the complex formed between light-activated rhodopsin dimer and a Gt heterotrimer (A and B) [52] and the T4L-β2AR-Gs-nanobody complex (C) [48] were fitted into a 3D molecular envelope calculated from projections of negatively stained, bis[succinimidyl] 2,2,4,4-glutarate (DSG) crosslinked rhodopsin*-Gt complexes purified in lauryl maltose neopentyl glycol (LMNG). Fitting of the rhodopsin*-Gt model generated with the structure of inactive Gt (PDB code: 1GOT) leaves a significant unoccupied density above one of the rhodopsin molecules (A), which becomes occupied after a 30o hinge-like motion of the α-helical domain is applied (B). Though fitting the T4L-β2-adrenergic receptor-Gs-nanobody structure into our EM 3D map leaves sufficient space to accommodate a second molecule of this receptor, conformation of the Gsα helical domain is inconsistent with our EM-density (C). Thus, the favored structure of the rhodopsin-Gt complex appears to be that shown in (B). Photoactivated rhodopsin (Rho*) that binds the C-terminal peptide derived from Gtα and the T4L-β2-adrenergic receptor-nanobody molecule are both depicted in yellow. The second rhodopsin molecule in (A) and (B) is shown in gray. Gtα, Gtβ, Gtγ are colored pink, dark cyan and dark blue, respectively.
Rhodopsin, rhodopsin kinase (GRK1), the capping protein arrestin and its splice variant p44
Photoactivated rhodopsin is phosphorylated by rhodopsin kinase and binds a 48 kDa capping protein called arrestin. Lack of either GRK1 or arrestin causes slowly progressive Oguchi disease characterized by profound delayed dark adaptation [57]. This visual phenotype is largely mimicked by knockout mice [58, 59]. Detailed analysis of a GRK1 Oguchi patient revealed that light adaptation was normal in low light backgrounds but abnormal in higher light environments indicating GRK1 involvement in rhodopsin phosphorylation at high light intensity [60]. A high resolution structure of GRK1 has been elucidated [61] as well as bipartite structures of rod- [62, 63] and cone-specific arrestins [64]. Similar to the (rhodopsin)2-Gt complex, at least in some experimental conditions, one arrestin molecule binds to an asymmetric dimer of rhodopsin [65]. Palczewski and colleagues isolated a truncated form of arrestin called p44 from bovine retina wherein the 15-amino acid-long C-terminal region was replaced by a single amino acid. As shown in subsequent work, p44 was a novel splice form of the arrestin transcript. Significantly, a physiological assay revealed that p44 binds to photoactivated rhodopsin even without phosphorylation, indicating a mechanism for full-length arrestin binding that involves displacement of the C-terminal region of arrestin by the C-terminal region of phosphorylated photoactivated rhodopsin. The structure of p44 was determined under two different crystallization conditions that resulted in different conformations of this protein [66, 67]. A hysteresis model was proposed for the more recent of these two structures that assumed a stable “active” conformation because opsin was present during the crystallization [66, 67]. However, this hysteresis model would be extremely rare for a protein without chemical modification in any biological system. Neither rhodopsin kinase nor an arrestin/p44 structure combined with any form of rhodopsin is currently available. Moreover, the structure of β-arrestin-1 with a phospho-peptide derived from vasopressin receptor 2 in complex with a Fab fragment shows additional C-terminal region conformational differences in β-arrestin-1 as compared to its inactive conformation [68].
Enzymatic production of 11-cis-retinal: the key step of rhodopsin regeneration
To maintain vertebrate vision, the spent all-trans-retinal chromophore released from rhodopsin in photoreceptor cell outer segments must be converted back to 11-cis-retinal, a process largely accomplished in an adjacent layer of the retina called the retinal pigmented epithelium (RPE) [69]. The key enzyme responsible for this cis-trans isomerization is retinoid isomerase (RPE65), with its bovine orthologous structure determined by Kiser and colleagues [20, 70]. RPE65 is a membrane-associated Fe2+-containing metalloenzyme. Importantly, disabling mutations of RPE65 cause a childhood blinding conditions termed Leber congenital amaurosis (LCA) and early-onset retinitis pigmentosa [71]. The overall structure of RPE65 is that of a seven-bladed β-propeller with single-strand extensions on blades VI and VII and a two-strand extension on blade III that contributes to dimerization of this protein. The Fe2+- ion is coordinated by four His residues and three second sphere Glu residues with each blade of the propeller contributing a single residue to the metal ion coordination. There is also a hydrophobic tunnel that allows the retinyl ester substrate to diffuse to the iron-containing catalytic site [20]. More recently crystals were obtained from enzymatically active native protein embedded in a lipid-detergent sheet. Based on these structures and complementary studies, we proposed that the Lewis acidity of iron could be used to promote ester dissociation and generation of a carbocation intermediate required for retinoid isomerization [42].
Other proteins of the phototransduction and retinoid cycle
Research discoveries over the past several years have significantly enhanced our understanding of the structural regulation of visual processes in the eye. In ROS, 11-cis-retinal binds to the apo-protein opsin forming rhodopsin, a functional light receptor. Consequently, a number of proteins involved in signal transduction are activated. Light-induced structural changes in rhodopsin lead to activation of Gt, resulting in formation of the Gα-GTP complex which activates retinal cGMP phosphodiesterase (PDE6). It is also known that transducin translocates between photoreceptor compartments (from ROS to inner segments) during the dark-light cycle [72]. This movement could be supported in part by helper proteins. One of these proteins, UNC119, was co-crystalized with an acylated α-subunit of transducin N-terminal peptide. This structure revealed that the lipid chain is buried deeply in UNC119’s hydrophobic cavity [73]. The recently determined EM structure of PDE6γ in complex with prenyl-binding protein delta (PrBP/delta), revealed probable locations of its isoprenylation sites, protein subunits, and catalytic sites [74]. Activated PDE6 catalyzes the hydrolysis of cGMP followed by closure of the cyclic nucleotide gated (CNG) channels and photoreceptor hyperpolarization, in effect attenuating synaptic glutamate release. Native CNG channels from rod cells are composed of CNGA1 and CNGB1 subunits in a 3:1 stoichiometry. The stoichiometry is not known for cone CNG channels. However, recently solved crystal structures of the parallel 3-helix coiled-coil domains of CNGA1 and the cone-localized CNGA3 (domains that regulate subunit assembly) were similar, suggesting a shared mechanism controlling the stoichiometry of rod and cone CNG channels.
Absorption of a photon of light causes isomerization of 11-cis-retinal to its all-trans-configuration and subsequent release of all-trans-retinal into the cytoplasm. However, a small fraction of all-trans-retinal is released into the luminal space of ROS and must be transferred to the cytoplasm to prevent accumulation of toxic condensation products of retinal and phosphatidylethanolamine. The ABCA4 transporter is involved in this process and EM and hydrogen-deuterium exchange studies have provided the first direct structural information about the membrane topology of this transporter and mechanistic insights into its function [75]. Released all-trans-retinal is reduced to all-trans-retinol by NADPH-dependent all-trans-retinol dehydrogenases (primarily RDH12 and RDH8) in the ROS and then all-trans-retinol diffuses into RPE cells co-chaperoned by inter-photoreceptor retinoid-binding protein (IRBP). Upon entry into the RPE, all-trans-retinol is bound by cellular retinol-binding protein (CRBP1) and then esterified by lecithin-retinol acyltransferase (LRAT) [76]. Besides LRAT, two other subfamilies belong to the LRAT-like family of proteins: the largely uncharacterized neuronal sensitive proteins (NSE1-2) and H-Ras-like tumor suppressor proteins (HRASLS1-5). Although the structure of LRAT has yet to be determined, a recently solved structure of HRASLS3 sheds light on the catalytic properties of LRAT [77]. Formed retinyl esters can be either stored in specialized compartments of RPE cells called retinosomes [78, 79], or used as substrates for the retinoid isomerase RPE65, which catalyzes the formation of 11-cis-retinol [80]. The last retinoid is rapidly bound by cellular retinal-binding protein (CRALBP) and its subsequent oxidation to 11-cis-retinal is catalyzed by NAD(P)+-dependent 11-cis-retinol RDHs (RDH5, RDH11 and RDH12) [81]. Structures of human CRALBP and of its mutant R234W which causes Bothnia dystrophy, with the endogenous ligand 11-cis-retinal were determined at 3.0 and 1.7 Å resolution, respectively [82]. The high redundancy of RDHs highlights the importance of maintaining low cellular levels of retinoids due to their high toxicity. 11-cis-Retinal is released from CRALBP by membrane acidic lipids [83] into the interphotoreceptor matrix where IRBP shuttles and releases it to photoreceptors, allowing the chromophore to regenerate accumulated opsin. A novel hydrophobic cavity that binds 11-cis-retinal and all-trans-retinal and is distinct from the long-chain fatty acid-binding site has recently been identified in the IRBP structure [84].
Finally, structures of the globular domains of C1q and tumor necrosis factor related protein 5 (C1QTNF5), a membrane associated protein involved in adhesion of RPE cells, provide insights into the structural basis of autosomal-dominant late-onset retinal macular degeneration (L-ORMD) [85].
Supramolecular organization of visual signaling machinery
Photoreceptor rod and cone cells are highly differentiated neurons with clearly defined substructures. In particular, the ROS house all components of the phototransduction signaling machinery in disc membranes, well-defined compartments containing about 50% internal membranes by volume [3]. Thus, almost all chemical reactions occurring in discs are interfacial, including those involving enzymes and other proteins. Animal models of many human retinal diseases provide powerful tools for studying the mechanisms of vision and their impairment by genetic mutations. The mouse has several advantages as the species of choice, including a non-redundant set of genes involved in vision, relative ease of genetic manipulation, and a close mechanistic resemblance to human vision. Segregation of nearly all involved proteins in a single cellular compartment renders the visual signalling system very suited to modern structural methodologies developed for studying biologically relevant complexes in their native environments. Cryo-EM and tomography, in particular, yield a wealth of information about large protein complexes, subcellular substructures and internal segments of the cell. Nickel et. al. first used cryo-EM tomography to obtain three-dimensional morphological information about a vitrified ROS structure from murine retina [86]. As one of the largest structures imaged by this technique, this allowed distance measurements among the various membrane components of the ROS to define the space available for phototransduction and provide a glimpse into the unfixed three-dimensional architecture of this highly differentiated neuron. Wensel and colleagues extended these cryo-EM tomography studies to obtain 3D maps of the connecting cilium and adjacent cellular structures of the ROS, a modified primary cilium, from wild-type and genetically defective mice [4]. After finding that the ciliary rootlet is involved in cellular transport and stabilizes the axoneme, they proposed a model for disc morphogenesis in which basal discs are enveloped by the plasma membrane.
Imaging studies of the retina remain at the forefront of progress in reconstruction analyses to visualize multiple cells simultaneously at high resolution. Novel applications include scanning EM (SEM) coupled with serial ion ablation (SIA) technology [87], electron microscopic high resolution imaging reconstruction such as serial block-face electron microscopy (SBEM) [88], and two-photon microscopy [89]. These continuously evolving techniques provide indispensable information about the higher order organization of the retina at subcellular resolution.
Conclusions
Progress in the structural biology of individual photoreceptor proteins must be followed with studies to assign their position within the supramolecular assemblies that underlie visual function. Hybrid microscopy techniques such as cryo-EM at sub-nanometer resolution, hybrid serial sectioning combined with high resolution transmission EM and correlative microscopy already allow imaging of retinal layers in unprecedented detail. Moreover, serial sectioning-coupled scanning and transmission EM permit three dimensional reconstructions that expedite understanding the global pathological features associated with genotypic disorders. The final challenge is to understand at an atomic level what therapies can be employed to prevent devastating blinding diseases.
Highlights.
Recent structural advances related to vertebrate visual transduction proteins are described.
Progress pertaining to rhodopsin and its complex with rod G protein is summarized.
The RPE65 structure reveals a novel mechanism for retinoid isomerization.
Structural analyses of other proteins involved in visual processes are reviewed.
New approaches to study relevant cellular complexes are discussed.
Acknowledgments
We thank Drs. Leslie T. Webster Jr., David T. Lodowski and members of the Palczewski laboratory for helpful comments on this manuscript. The work of our laboratory is supported by funding from the National Eye Institute of the National Institutes of Health (grants EY008061 and EY019478 (KP)). K.P. is John H. Hord Professor of Pharmacology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Mustafi D, Maeda T, Kohno H, Nadeau JH, Palczewski K. Inflammatory priming predisposes mice to age-related retinal degeneration. J Clin Invest. 2012;122:2989–3001. doi: 10.1172/JCI64427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arshavsky VY, Burns ME. Photoreceptor signaling: supporting vision across a wide range of light intensities. J Biol Chem. 2012;287:1620–1626. doi: 10.1074/jbc.R111.305243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •3.Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. Visual perception in humans is initiated through absorption of photons by photoreceptors in the retina, a layered sensory organ containing all the necessary functional and structural proteins to support this process. How this remarkable tissue develops and operates over such an incredible dynamic range and how retinoids are recycled are some of the most fascinating questions in biology. With current methodology, it is now possible to identify all components of the retina and trace mutations to multiple blinding diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••4.Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, Pittler SJ, Chiu W, Wensel TG. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. Using cryo-electron tomography, Wensel and colleagues obtained for the first time the 3D structure of the connecting cilium and adjacent cellular structures of a modified primary cilium in the rod outer segment isolated from wild type and disease model mice. This work revealed new structural features involved in cellular transport that is impaired by the involved genetic defects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Okada T, Palczewski K. Crystal structure of rhodopsin: implications for vision and beyond. Curr Opin Struct Biol. 2001;11:420–426. doi: 10.1016/s0959-440x(00)00227-x. [DOI] [PubMed] [Google Scholar]
- 7.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 9.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 10.Shimamura T, Hiraki K, Takahashi N, Hori T, Ago H, Masuda K, Takio K, Ishiguro M, Miyano M. Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J Biol Chem. 2008;283:17753–17756. doi: 10.1074/jbc.C800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •11.Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci U S A. 2012;109:119–124. doi: 10.1073/pnas.1114089108. Constitutively active mutants are ubiquitously found among GPCRs and their increased basal activity often correlates with a pathological outcome. The structure of the M257Y constitutively active rhodopsin mutant in complex with the C-terminal fragment from the cognate G protein alpha subunit trapped in its active Meta II state revealed the molecular basis for this receptor’s constitutive activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 14.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- •15.Ye S, Huber T, Vogel R, Sakmar TP. FTIR analysis of GPCR activation using azido probes. Nat Chem Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. FTIR difference spectroscopy and novel amber codon suppression technology introducing a site-specific vibrational probe (azidoF) into protein structure, was used to understand conformational changes associated with GPCR activation in a lipid membrane. The azido label senses the polarity of its environment and thus, incorporated in rhodopsin, allows monitoring changes of the electrostatic environment of selected side chains during the conformational transitions associated with receptor activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goncalves JA, South K, Ahuja S, Zaitseva E, Opefi CA, Eilers M, Vogel R, Reeves PJ, Smith SO. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc Natl Acad Sci U S A. 2010;107:19861–19866. doi: 10.1073/pnas.1009405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye S, Zaitseva E, Caltabiano G, Schertler GF, Sakmar TP, Deupi X, Vogel R. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 18.Ahuja S, Eilers M, Hirshfeld A, Yan EC, Ziliox M, Sakmar TP, Sheves M, Smith SO. 6-s-cis Conformation and polar binding pocket of the retinal chromophore in the photoactivated state of rhodopsin. J Am Chem Soc. 2009;131:15160–15169. doi: 10.1021/ja9034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••19.Struts AV, Salgado GF, Martinez-Mayorga K, Brown MF. Retinal dynamics underlie its switch from inverse agonist to agonist during rhodopsin activation. Nat Struct Mol Biol. 2011;18:392–394. doi: 10.1038/nsmb.1982. Light activation of rhodopsin induces isomerization of its inverse agonist 11-cis-retinal to all-trans-retinal and transition of dark state rhodopsin to Meta I and the following Meta II state. In the membrane environment these changes in retinal structure and dynamics trigger activating fluctuations of transmembrane helices H5 and H6 in the Meta I-Meta II equilibrium of rhodopsin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc Natl Acad Sci U S A. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••21.Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR, Palczewski K. Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc Natl Acad Sci U S A. 2012;109:E2747–2756. doi: 10.1073/pnas.1212025109. The crystal structure of RPE65 in a membrane-like environment displayed an unusual packing arrangement wherein RPE65 was embedded in a lipid-detergent sheet. This structure also uncovered key residues involved in substrate uptake and processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •22.Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc Natl Acad Sci U S A. 2009;106:14367–14372. doi: 10.1073/pnas.0901074106. Radiolytic protein footprinting and rapid labeling with H2O18 revealed local structural changes and reorganization of internal waters in the rhodopsin molecule upon its photoactivation. Because multiple structures of the GPCR superfamily possess conserved embedded water molecules, they are likely important to GPCR function in general. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodowski DT, Palczewski K, Miyagi M. Conformational changes in the g protein-coupled receptor rhodopsin revealed by histidine hydrogen-deuterium exchange. Biochemistry. 2010;49:9425–9427. doi: 10.1021/bi101502v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, Chance MR, Palczewski K. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20:826–840. doi: 10.1016/j.str.2012.03.017. Hydroxyl radical labeling and deuterium uptake revealed a conformational difference between light-activated rhodopsin alone and upon binding to its cognate G protein, transducin. While photoactivation caused a structural relaxation of rhodopsin, coupling to Gt led to tightening and rigidifying this receptor. In contrast, nucleotide-free transducin in complex with rhodopsin was significantly more accessible to deuterium uptake allowing it to accept GTP and mediate complex dissociation, a critical step for signal transmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. Recently the number of GPCR crystal structures has rapidly expanded providing a solid framework for experimental and molecular modeling studies as well as an understanding of GPCR biological and therapeutic mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci U S A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol. 2011;21:567–572. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day PW, Rasmussen SG, Parnot C, Fung JJ, Masood A, Kobilka TS, Yao XJ, Choi HJ, Weis WI, Rohrer DK, Kobilka BK. A monoclonal antibody for G protein-coupled receptor crystallography. Nat Methods. 2007;4:927–929. doi: 10.1038/nmeth1112. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 30.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Xu F, Cherezov V, Hanson MA, Stevens RC. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 33.Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–449. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36.Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. Structural changes that occur in rhodopsin due to its photoactivation include a rearrangement of TM5-TM6 helices that opens a crevice at the cytoplasmic side of the receptor into which the C terminus of the Gα subunit can bind. The mechanism relies on dynamic interactions between conserved residues and could therefore be common to other GPCRs. [DOI] [PubMed] [Google Scholar]
- 37.Sekharan S, Sugihara M, Weingart O, Okada T, Buss V. Protein assistance in the photoisomerization of rhodopsin and 9-cis-rhodopsin--insights from experiment and theory. J Am Chem Soc. 2007;129:1052–1054. doi: 10.1021/ja066970p. [DOI] [PubMed] [Google Scholar]
- •38.Hornak V, Ahuja S, Eilers M, Goncalves JA, Sheves M, Reeves PJ, Smith SO. Light activation of rhodopsin: insights from molecular dynamics simulations guided by solid-state NMR distance restraints. J Mol Biol. 2010;396:510–527. doi: 10.1016/j.jmb.2009.12.003. Measurements of structural restraints in Meta II state of rhodopsin provided by solid-state NMR in combination with molecular dynamics simulations enhanced our understanding of the mechanism of rhodopsin activation. These studies revealed that upon light illumination multiple switches on the extracellular side of rhodopsin trigger structural changes that converge to disrupt the ionic lock between helices H3 and H6 on the intracellular side of this receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struts AV, Salgado GF, Brown MF. Solid-state 2H NMR relaxation illuminates functional dynamics of retinal cofactor in membrane activation of rhodopsin. Proc Natl Acad Sci U S A. 2011;108:8263–8268. doi: 10.1073/pnas.1014692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahuja S, Crocker E, Eilers M, Hornak V, Hirshfeld A, Ziliox M, Syrett N, Reeves PJ, Khorana HG, Sheves M, Smith SO. Location of the retinal chromophore in the activated state of rhodopsin*. J Biol Chem. 2009;284:10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci U S A. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanuj Sapra K, Park PS, Filipek S, Engel A, Muller DJ, Palczewski K. Detecting molecular interactions that stabilize native bovine rhodopsin. J Mol Biol. 2006;358:255–269. doi: 10.1016/j.jmb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Kawamura S, Gerstung M, Colozo AT, Helenius J, Maeda A, Beerenwinkel N, Park PS, Muller DJ. Kinetic, energetic, and mechanical differences between dark-state rhodopsin and opsin. Structure. 2013;21:426–437. doi: 10.1016/j.str.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawamura S, Colozo AT, Muller DJ, Park PS. Conservation of molecular interactions stabilizing bovine and mouse rhodopsin. Biochemistry. 2010;49:10412–10420. doi: 10.1021/bi101345x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zocher M, Fung JJ, Kobilka BK, Muller DJ. Ligand-Specific Interactions Modulate Kinetic, Energetic, and Mechanical Properties of the Human beta(2) Adrenergic Receptor. Structure. 2012;20:1391–1402. doi: 10.1016/j.str.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zocher M, Bippes CA, Zhang C, Muller DJ. Single-molecule force spectroscopy of G-protein-coupled receptors. Chem Soc Rev. 2013;42:7801–7815. doi: 10.1039/c3cs60085h. [DOI] [PubMed] [Google Scholar]
- 47.Preininger AM, Meiler J, Hamm HE. Conformational flexibility and structural dynamics in GPCR-mediated G protein activation: a perspective. J Mol Biol. 2013;425:2288–2298. doi: 10.1016/j.jmb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••49.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. This paper presents the first crystal structure of the active state ternary complex composed of agonist-occupied monomeric β2AR and nucleotide-free Gs heterotrimer. Within this complex the largest conformational changes in the β2AR include a 14 Å outward movement at the cytoplasmic end of transmembrane segment 6 (TM6) and an α-helical extension of the cytoplasmic end of TM5. Surprisingly, in Gs a major displacement of the α-helical domain of Gαs relative to the Ras-like GTPase domain was observed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 51.Tieleman DP, Berendsen HJ, Sansom MS. An alamethicin channel in a lipid bilayer: molecular dynamics simulations. Biophys J. 1999;76:1757–1769. doi: 10.1016/s0006-3495(99)77337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jastrzebska B, Ringler P, Lodowski DT, Moiseenkova-Bell V, Golczak M, Muller SA, Palczewski K, Engel A. Rhodopsin-transducin heteropentamer: three-dimensional structure and biochemical characterization. J Struct Biol. 2011;176:387–394. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •53.Jastrzebska B, Ringler P, Palczewski K, Engel A. The rhodopsin-transducin complex houses two distinct rhodopsin molecules. J Struct Biol. 2013;182:164–72. doi: 10.1016/j.jsb.2013.02.014. Reconstructions of negatively-stained nucleotide-free rhodopsin-transducin complex purified in lauryl-maltose-neopentyl-glycol produced a 3D map that accommodated two rhodopsin molecules, one Gt heterotrimer and a detergent belt. Visualization of triple succinylated Concanavalin A-rhodopsin-Gt complexes unequivocally demonstrated a pentameric assembly of the rhodopsin-Gt complex in which the photoactivated rhodopsin dimer serves as a platform for binding the Gt heterotrimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••55.Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013;27:1572–1584. doi: 10.1096/fj.12-225383. The retinoid chromophores 11-cis-retinal and 9-cis-retinal were used to monitor each monomer of the dimeric model GPCR, rhodopsin within a stable complex with nucleotide-free Gt. Interestingly only 50% of activated rhodopsin in the rhodopsin-Gt complex was trapped in a Meta II conformation, while 50% evolved toward an opsin conformation and could be regenerated with 9-cis-retinal. Thus, each of the monomers contributes unequally to the pentameric complex of the rhodopsin dimer and Gt heterotrimer, validating the oligomeric structure of the complex and the asymmetry of the GPCR dimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vahedi-Faridi A, Jastrzebska B, Palczewski K, Engel A. 3D imaging and quantitative analysis of small solubilized membrane proteins and their complexes by transmission electron microscopy. J Electron Microsc (Tokyo) 2013;62:95–107. doi: 10.1093/jmicro/dfs091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dryja TP. Molecular genetics of Oguchi disease, fundus albipunctatus, and other forms of stationary night blindness: LVII Edward Jackson Memorial Lecture. Am J Ophthalmol. 2000;130:547–563. doi: 10.1016/s0002-9394(00)00737-6. [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Simon MI, Matthes MT, Yasumura D, LaVail MM. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness) Invest Ophthalmol Vis Sci. 1999;40:2978–2982. [PubMed] [Google Scholar]
- 59.Fan J, Sakurai K, Chen CK, Rohrer B, Wu BX, Yau KW, Kefalov V, Crouch RK. Deletion of GRK1 causes retina degeneration through a transducin-independent mechanism. Journal of Neuroscience. 2010;30:2496–2503. doi: 10.1523/JNEUROSCI.6254-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cideciyan AV, Zhao X, Nielsen L, Khani SC, Jacobson SG, Palczewski K. Null mutation in the rhodopsin kinase gene slows recovery kinetics of rod and cone phototransduction in man. Proc Natl Acad Sci U S A. 1998;95:328–333. doi: 10.1073/pnas.95.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 63.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 64.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 2.3A: evolution of receptor specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- •65.Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. Rod arrestin induces uptake of the agonist all-trans-retinal in only half the population of phosphorylated opsin in the native membrane. The entry of the agonist induces conformational changes only in arrestin’s N-domain, while C-domain engages the apo-receptor even before agonist is added suggesting that each domain of arrestin binds its own receptor molecule indicating a dimeric nature of the receptor bound to arrestin and its asymmetric properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Granzin J, Cousin A, Weirauch M, Schlesinger R, Buldt G, Batra-Safferling R. Crystal structure of p44, a constitutively active splice variant of visual arrestin. J Mol Biol. 2012;416:611–618. doi: 10.1016/j.jmb.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 67.Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, Sommer ME. Crystal structure of pre-activated arrestin p44. Nature. 2013;497:142–146. doi: 10.1038/nature12133. [DOI] [PubMed] [Google Scholar]
- 68.Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, Paduch M, Tripathi-Shukla P, Koide A, Koide S, Weis WI, Kossiakoff AA, Kobilka BK, Lefkowitz RJ. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Lintig J, Kiser PD, Golczak M, Palczewski K. The biochemical and structural basis for trans-to-cis isomerization of retinoids in the chemistry of vision. Trends Biochem Sci. 2010;35:400–410. doi: 10.1016/j.tibs.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiser PD, Palczewski K. Membrane-binding and enzymatic properties of RPE65. Prog Retin Eye Res. 2010;29:428–442. doi: 10.1016/j.preteyeres.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cideciyan AV. Leber congenital amaurosis due to RPE65 mutations and its treatment with gene therapy. Prog Retin Eye Res. 2010;29:398–427. doi: 10.1016/j.preteyeres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr, Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 73.Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, Xiao R, Montelione GT, Gerstner CD, Davis MW, Inana G, Whitby FG, Jorgensen EM, Hill CP, Tong L, Baehr W. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci. 2011;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •74.Goc A, Chami M, Lodowski DT, Bosshart P, Moiseenkova-Bell V, Baehr W, Engel A, Palczewski K. Structural Characterization of the Rod cGMP Phosphodiesterase 6. J Mol Biol. 2010;401:363–373. doi: 10.1016/j.jmb.2010.06.044. Visualization of rod cGMP phosphodiesterase 6 (PDE6) in complex with prenyl-binding protein delta (PrBP/delta) after transmission electron microscopy of negatively stained particles allowed identification of the probable location of isoprenylation, PDE6γ, and catalytic sites in the 3D structure of the PDE6αβγ complex which was determined up to 18 Å resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsybovsky Y, Orban T, Molday RS, Taylor D, Palczewski K. Molecular Organization and ATP-Induced Conformational Changes of ABCA4, the Photoreceptor-Specific ABC Transporter. Structure. 2013;21:854–860. doi: 10.1016/j.str.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Golczak M, Kiser PD, Sears AE, Lodowski DT, Blaner WS, Palczewski K. Structural basis for the acyltransferase activity of lecithin:retinol acyltransferase-like proteins. J Biol Chem. 2012;287:23790–23807. doi: 10.1074/jbc.M112.361550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imanishi Y, Gerke V, Palczewski K. Retinosomes: new insights into intracellular managing of hydrophobic substances in lipid bodies. J Cell Biol. 2004;166:447–453. doi: 10.1083/jcb.200405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orban T, Palczewska G, Palczewski K. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J Biol Chem. 2011;286:17248–17258. doi: 10.1074/jbc.M110.195198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chander P, Gentleman S, Poliakov E, Redmond TM. Aromatic residues in the substrate cleft of RPE65 protein govern retinol isomerization and modulate its progression. J Biol Chem. 2012;287:30552–30559. doi: 10.1074/jbc.M112.364596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kiser PD, Golczak M, Palczewski K. Chemistry of the Retinoid (Visual) Cycle. Chem Rev. 2013 doi: 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He X, Lobsiger J, Stocker A. Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W. Proc Natl Acad Sci U S A. 2009;106:18545–18550. doi: 10.1073/pnas.0907454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saari JC, Nawrot M, Stenkamp RE, Teller DC, Garwin GG. Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids. Mol Vis. 2009;15:844–854. [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez-Fernandez F, Bevilacqua T, Lee KI, Chandrashekar R, Hsu L, Garlipp MA, Griswold JB, Crouch RK, Ghosh D. Retinol-binding site in interphotoreceptor retinoid-binding protein (IRBP): a novel hydrophobic cavity. Invest Ophthalmol Vis Sci. 2009;50:5577–5586. doi: 10.1167/iovs.08-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tu X, Palczewski K. Crystal structure of the globular domain of C1QTNF5: Implications for late-onset retinal macular degeneration. J Struct Biol. 2012;180:439–446. doi: 10.1016/j.jsb.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••86.Nickell S, Park PS, Baumeister W, Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J Cell Biol. 2007;177:917–925. doi: 10.1083/jcb.200612010. Imaging of a vitrified rod outer segment (ROS) of photoreceptor cells by cryoelectron tomography provided a glimpse into the three-dimensional architecture of this highly differentiated neuron. Image reconstructions revealed spacers that likely maintain the proper distance between adjacent discs and between discs and the plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mustafi D, Avishai A, Avishai N, Engel A, Heuer A, Palczewski K. Serial sectioning for examination of photoreceptor cell architecture by focused ion beam technology. J Neurosci Methods. 2011;198:70–76. doi: 10.1016/j.jneumeth.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mustafi D, Kevany BM, Genoud C, Okano K, Cideciyan AV, Sumaroka A, Roman AJ, Jacobson SG, Engel A, Adams MD, Palczewski K. Defective photoreceptor phagocytosis in a mouse model of enhanced Scone syndrome causes progressive retinal degeneration. FASEB J. 2011;25:3157–3176. doi: 10.1096/fj.11-186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palczewska G, Maeda T, Imanishi Y, Sun W, Chen Y, Williams DR, Piston DW, Maeda A, Palczewski K. Noninvasive multiphoton fluorescence microscopy resolves retinol and retinal condensation products in mouse eyes. Nat Med. 2010;16:1444–1449. doi: 10.1038/nm.2260. Dear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park JH, Morizumi T, Li Y, Hong JE, Pai EF, Hofmann KP, Choe HW, Ernst OP. Opsin, a Structural Model for Olfactory Receptors? Angew Chem Int Ed Engl. 2013;52:11021–11024. doi: 10.1002/anie.201302374. [DOI] [PubMed] [Google Scholar]
- 91.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 92.Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mansoor SE, Palczewski K, Farrens DL. Rhodopsin self-associates in asolectin liposomes. Proc Natl Acad Sci U S A. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]