Abstract
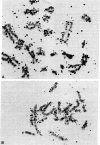
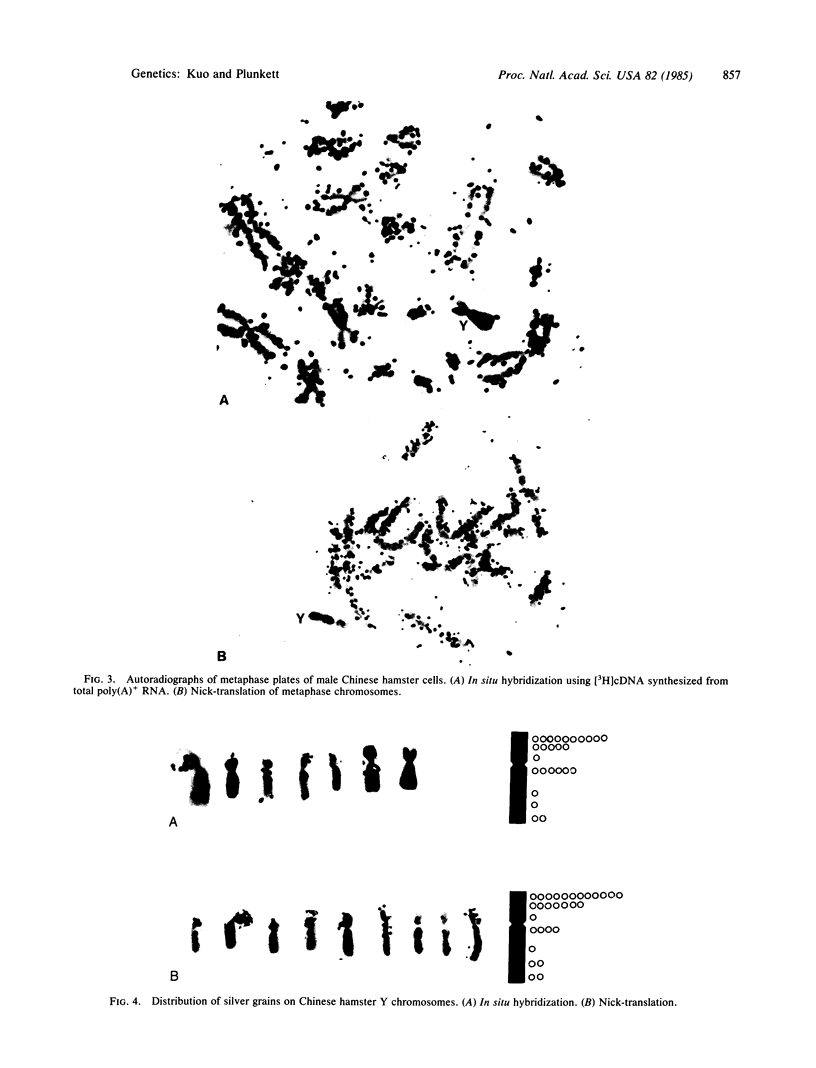
Chinese hamster metaphase chromosomes were labeled by nick-translation, which involved pretreatment of metaphase chromosomes with low levels of DNase I followed by incubation with DNA polymerase I and radioactively labeled nucleotides. The labeled DNA was located on nuclease-hypersensitive regions of the chromosomes, as suggested by the following observations. (i) The labeled DNA was hypersensitive to the subsequent DNase I digestion. (ii) The labeled DNA contained no nucleosomes. DNA reassociation kinetic analysis suggested that the labeled DNA was enriched in repetitive DNA sequences. Base composition analyses showed that the labeled DNA was highly enriched in guanine and adenine residues, suggesting that the nick-translation reaction was asymmetrical and the strand enriched in purine was preferentially translated. Autoradiographic analysis revealed that the label was distributed on every chromosome, but there was a lower grain density on the Y chromosome, which is heterochromatic and exhibits a relatively low level of gene activity. The locations of silver grains on the Y chromosomes were generally consistent with that revealed by the in situ hybridization using [3H]cDNA synthesized from the total Chinese hamster messenger RNA. These observations suggest that a specific subset of genomic DNA on active chromatin is the preferred site of the nick-translation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avramis V. I., Plunkett W. 2-fluoro-ATP: a toxic metabolite of 9-beta-D-arabinosyl-2-fluoroadenine. Biochem Biophys Res Commun. 1983 May 31;113(1):35–43. doi: 10.1016/0006-291x(83)90428-x. [DOI] [PubMed] [Google Scholar]
- Blumenthal A. B., Dieden J. D., Kapp L. N., Sedat J. W. Rapid isolation of metaphase chromosomes containing high molecular weight DNA. J Cell Biol. 1979 Apr;81(1):255–259. doi: 10.1083/jcb.81.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Castellot J. J., Jr, Miller M. R., Pardee A. B. Animal cells reversibly permeable to small molecules. Proc Natl Acad Sci U S A. 1978 Jan;75(1):351–355. doi: 10.1073/pnas.75.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. Y., Folsom V., Wooley J. DNase I-hypersensitive sites in the chromatin of immunoglobulin kappa light chain genes. Proc Natl Acad Sci U S A. 1983 May;80(9):2427–2431. doi: 10.1073/pnas.80.9.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Finer M. H., Fodor E. J., Boedtker H., Doty P. Endonuclease S1-sensitive site in chicken pro-alpha 2(I) collagen 5' flanking gene region. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1659–1663. doi: 10.1073/pnas.81.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., McCune R. A., Gerhardt K. O., Agris P. F. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J Chromatogr. 1982 Jul 9;230(2):297–308. [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Hen R., Borrelli E., Sassone-Corsi P., Chambon P. An enhancer element is located 340 base pairs upstream from the adenovirus-2 E1A capsite. Nucleic Acids Res. 1983 Dec 20;11(24):8747–8760. doi: 10.1093/nar/11.24.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian K., Fasman G. D. Nick translation of HeLa cell nuclei as a probe for locating DNase I-sensitive nucleosomes. J Biol Chem. 1984 Mar 10;259(5):3343–3349. [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Kaye J. S., Bellard M., Dretzen G., Bellard F., Chambon P. A close association between sites of DNase I hypersensitivity and sites of enhanced cleavage by micrococcal nuclease in the 5'-flanking region of the actively transcribed ovalbumin gene. EMBO J. 1984 May;3(5):1137–1144. doi: 10.1002/j.1460-2075.1984.tb01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Micrococcal nuclease as a probe of DNA sequence organization and chromatin structure. Cell. 1981 Nov;27(1 Pt 2):57–64. doi: 10.1016/0092-8674(81)90360-3. [DOI] [PubMed] [Google Scholar]
- Kerem B. S., Goitein R., Richler C., Marcus M., Cedar H. In situ nick-translation distinguishes between active and inactive X chromosomes. Nature. 1983 Jul 7;304(5921):88–90. doi: 10.1038/304088a0. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Gelinas R., Weintraub H. Detection of an altered DNA conformation at specific sites in chromatin and supercoiled DNA. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4389–4393. doi: 10.1073/pnas.80.14.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T. Analysis of DNA attached to the chromosome scaffold. J Cell Biol. 1982 May;93(2):278–284. doi: 10.1083/jcb.93.2.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T. Comparison of chromosomal structures isolated under different conditions. Exp Cell Res. 1982 Mar;138(1):221–229. doi: 10.1016/0014-4827(82)90109-4. [DOI] [PubMed] [Google Scholar]
- Kuo M. T., Iyer B., Schwarz R. J. Condensation of chromatin into chromosomes preserves an open configuration but alters the DNase I hypersensitive cleavage sites of the transcribed gene. Nucleic Acids Res. 1982 Aug 11;10(15):4565–4579. doi: 10.1093/nar/10.15.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T., Saunders G. F. Location of messenger specifying sequences in mammalian chromosomes. Chromosoma. 1977 Sep 27;63(3):241–252. doi: 10.1007/BF00327452. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Levitt A., Axel R., Cedar H. Nick translation of active genes in intact nuclei. Dev Biol. 1979 Apr;69(2):496–505. doi: 10.1016/0012-1606(79)90307-5. [DOI] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Mills F. C., Fisher L. M., Kuroda R., Ford A. M., Gould H. J. DNase I hypersensitive sites in the chromatin of human mu immunoglobulin heavy-chain genes. Nature. 1983 Dec 22;306(5945):809–812. doi: 10.1038/306809a0. [DOI] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Röhme D., Fox H., Herrmann B., Frischauf A. M., Edström J. E., Mains P., Silver L. M., Lehrach H. Molecular clones of the mouse t complex derived from microdissected metaphase chromosomes. Cell. 1984 Mar;36(3):783–788. doi: 10.1016/0092-8674(84)90358-1. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Cereghini S., Yaniv M. Fine structure of the regulatory region of simian virus 40 minichromosomes revealed by DNAase I digestion. J Mol Biol. 1982 Sep 15;160(2):133–146. doi: 10.1016/0022-2836(82)90171-1. [DOI] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]