Almost fifty years ago, Julius Adler initiated a program of research to gain insights into the basic biochemistry of intelligent behavior by studying the molecular mechanisms that underlie the chemotactic responses of Escherichia coli. All living organisms share elements of a common biochemistry for metabolism, growth and heredity — why not intelligence? Neurobiologists have demonstrated that this is the case for nervous systems in animals ranging from worms to man. Motile unicellular organisms such as E. coli exhibit rudimentary behaviors that can be loosely described in terms of cognitive phenomena such as memory and learning. Adler’s initiative at least raised the prospect that, because of the numerous experimental advantages provided by E. coli, it would be the first organism whose behavior could be understood at molecular resolution.
Adler was soon joined in this project by a number of competing laboratories, all focused on elucidating the biochemical mechanisms that underlie E. coli sensory–motor regulation. Analogues of cognitive behaviors such as memory and learning were elucidated, the genes that encode the E. coli sensory–motor system were identified and sequenced, the component proteins were purified, and their structure–function relationships determined — the nanobrain was revealed. E. coli sensory–motor regulation is clearly very different from sensory–motor regulation in animals. It was originally assumed that E. coli would be quite rudimentary and primitive, but this turned out to be a misconception. Bacteria are highly refined organisms — they have been living on earth for billions of years, continuously evolving at very high rates with huge populations and short generation times. Bacteria are extremely small, but size is certainly not a useful guide when it comes to assessing cognitive abilities. If our experience with the evolution of computers is any guide, reductions in the size of components are associated with increases in computational power.
E. coli swim by rotating long helical flagellar filaments that extend from molecular rotary motors embedded in their cell envelopes. The sense of rotation of these nanomotors controls the motion of each cell. When the flagella rotate synchronously, they act cooperatively to propel the cell forward. When one or more motors reverse their sense of rotation, the flagella become uncoordinated and the bacterium tumbles briefly until flagellar coordination is restored and the cell swims off in a new direction. Motor reversals are controlled by the so-called nanobrain, a sensory–motor regulatory organelle located at one or both poles of the cell that functions as a molecular brain to control motor function. As a cell swims, if its nanobrain senses that conditions are improving, tumbling is suppressed and the cell tends to continue on course. If, on the other hand, the nanobrain senses that conditions are not improving or are getting worse, it increases the activity of a protein kinase that generates a phosphorylated response regulator that binds to flagellar motors and promotes reversals so that the cell tends to tumble and swim off in a new and potentially more favorable direction.
Bacterial memory
E. coli respond to changes in stimulatory ligand concentrations rather than absolute levels. As they swim they constantly compare past and present to determine whether conditions are getting better or worse — whether to continue swimming on course, or to tumble and change direction. This chemotactic mechanism was first demonstrated experimentally by rapidly adding an attractant amino acid or sugar to cells swimming in minimal media. In the absence of attractant, E. coli swim and tumble intermittently, affecting a random walk. On addition of attractant, all cells sense they are swimming in a direction of increasing attractant concentration, and uniformly suppress their tendency to tumble and change direction. After a brief period of this smooth swimming behavior, the cells adapt; and despite the continued presence of attractant chemicals, they resume their prestimulus random walk of runs punctuated with tumbles. Once the adaptation process is complete, dilution back into minimal media causes all the cells to simultaneously tumble as if they all sensed they were headed in the wrong direction. After a few seconds, however, they adapt and resume their characteristic swim/tumble random walk despite the continued presence of lowered attractant concentrations.
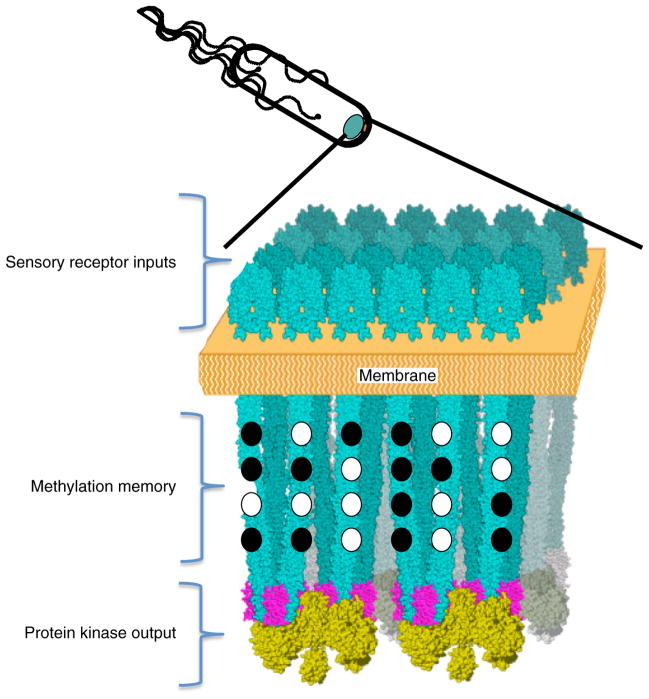
This simple go, no-go navigation strategy requires some sort of memory to allow temporal comparisons of past and present. In bacteria, this memory function is an intrinsic, highly conserved and essential part of the sensory–motor regulation mechanism. Each E. coli nanobrain consists of an array of several thousand alpha-helical coiled-coil protein fibers that interact laterally with one another to form a tightly packed cortical layer about 250 nm in diameter just below the cytoplasmic membrane (Figure 1). The fiber ends that pass through the cytoplasmic membrane into the periplasm interact with a complex layer of sensory receptor domains whose conformations depend on the binding of various stimulatory ligands, including several different amino acids and sugars. The intracellular aspect of the fibrous cortical layer interacts with and controls a protein kinase and phosphatase that release a small phosphoprotein that binds to flagellar motors to induce reversals in rotation, leading to tumbles. Each of the thousands of coiled-coil fibers that constitute the nanobrain consists of a four-helix bundle with at least eight potentially anionic glutamate side chains that can be either exposed as a minus charge or capped with a methyl group. The chemistry of capping and uncapping involves the methyl esterification and methyl ester hydrolysis of these eight glutamyl carboxylates. A specific methyl transferase enzyme catalyzes capping, and a specific methyl esterase catalyzes the uncapping reaction. Thus, each fiber can potentially be in one of 28 different states of modification. The ensemble of carboxylate methylations among the thousands of fibers controls the relationship between sensory inputs and protein kinase outputs. As a bacterium swims, any change in sensory-receptor input generates a change in kinase-mediated response regulator output that acts within milliseconds to modulate the probability of a tumble. Within seconds, the altered sensory input induces a positive or negative change in fiber methylation that leads to behavioral adaptation. If the environment is improved, and the levels of attractive sensory inputs have increased, the bacterium tends to continue on course. If conditions appear to be worse, the bacterium tends to change direction. The altered fiber interactions and resultant changes in kinase activity that engender behavioral responses feed back to alter the methylating and demethylating activities so as to change the state of receptor modification to restore a constant steady-state swimming behavior and at the same time refresh the memory function that provides a standard of comparison to evaluate future environments. Thus, analysis of E. coli sensory–motor regulation, where the molecular details are well defined, indicates that memory operates by a negative feedback mechanism that is directly linked to the signal transduction processes that translate sensory information into appropriate motor responses.
Figure 1. The E. coli nanobrain.
Sensory-motor regulation in motile bacteria such as E. coli involves a so-called nanobrain consisting of an array of several thousand alpha-helical transmembrane protein fibers clustered together at one pole. Receptor protein domains that specifically bind attractants such as serine, aspartate, ribose, etc. are associated with the extracytoplasmic sensory receptor input ends of these fibers, and a protein kinase that serves to control flagellar function is associated with the opposite, protein kinase output end of each fiber. The binding of attractant ligands induces changes in fiber conformation that act to control protein kinase activity and thereby modulate swimming behavior. Each of the thousands of fibers that constitute the nanobrain consists of a four-helix bundle with at least eight potentially anionic glutamate side chains that can be either exposed as a minus charge or neurtralized by methylation. Attractants induce increases in methylation (filled ovals) that counteract the effects of attractant binding and restore behavior to a preset, adapted steady state value. The new steady state level of methylation provides a memory function that acts as a reference to control further behavioral modifications. Subsequent increases (or decreases) in attractant concentration produce attractant (or repellent) behavioral responses that lead, in turn, to increases (or decreases) in methylation that act to restore steady state behavior, and provide a new baseline ‘memory’ for the assessment of future attractant or repellent stimuli.
The E. coli memory mechanism has several additional features that might be relevant for understanding the general role of memory in cognitive processes. Firstly, five different homologous genes encode the coiled-coil fibers that come together to form the core of the nanobrain. Whereas the methyl accepting memory storage and kinase output portions of each of these different fibers have very similar sequences and functions, the portions that interact with sensory receptor domains are quite divergent. Each fiber type represents a different sensory modality, channeling a different spectrum of sensory information into the nanobrain core. Information concerning the temporal coincidence of various different types of sensory inputs can therefore act to generate distinct memories, and a single bacterial cell could begin to formulate a rough olfactory map of its surroundings by the methylation changes induced by sensory inputs that come together in time (e.g. a specific mix of amino acids and a higher temperature might come from one source and therefore be coupled in time, while certain sugars and a relatively acidic pH might be associated with another source, etc.).
Secondly, the kinetics of methylation and demethylation are robustly regulated to ensure that within a few minutes in a constant environment the overall extent of nanobrain methylation will be just sufficient to, on average, reduce kinase activity to a low steady state value just sufficient to, on average, generate a random walk of runs punctuated by tumbles. This adaptive principle allows sensory responsiveness to be maintained over several orders of magnitude of stimulus concentrations.
Thirdly, the memory and behavioral responses of each individual bacterial cell are unique to that cell. Methylation and demethylation occur with different probabilities at different positions in different fibers in response to different stimuli so there is enormous variability in the particular pattern of modifications in the nanobrain of a particular cell, and it is safe to assume that no two cells will ever have the same pattern of glutamyl modification despite having identical genetic backgrounds and growing in the same culture media. Individual behavior is therefore unpredictable. There are always some individuals that swim away from attractants or toward repellents. One imagines that such deviant behaviors might be beneficial on occasion (e.g., an attractant odorant might mask a toxin).
Fourthly, in prokaryotes, the methylation memory system seems to be specifically used for the regulation of sensory–motor function. The nanobrain mechanism with its coiled-coil fibers and methylating and demethylating enzymes is conserved in virtually all motile bacteria, but is not found in immotile bacteria. There is also no evidence for homologs of the nanobrain methyl-accepting fibers in eukaryotic signal transduction systems. In contrast, the sensory receptors, sensory–motor kinase, and phosphorylated response regulator have numerous homologous relatives that are involved in a number of different regulatory roles in both prokaryotic and eukaryotic systems. This highlights the importance of sensory–motor regulation in cognitive processes such as memory. For metazoans that do not have nervous systems, like plants, intelligence, learning and memory are more difficult to define and the relevant chemistry is largely absent. Animals are distinct from plants in their highly developed sensory–motor abilities: the essential neuronal function is, after all, sensory–motor in nature.
Memory and learning
Considerations of vertebrate memory mechanisms generally assume that learning and memory are part of the same phenomenon viewed from a different perspective — memory being a means to achieve a learned response. Measures of memory generally involve behavioral outputs that depend on prior learning. When a rat learns where to swim to find a safe platform, he must remember where the platform is located. Intrinsic, procedural memories such as muscle memory can be viewed as a product of learning. After one learns to ride a bike, one always remembers how to ride a bike. Explicit, declarative memories, like remembered strings of numbers, could be viewed as learned responses. Few would argue with the idea that one can learn to remember or that one remembers what is learned.
In searching for the molecular substrates of memory in vertebrates, research has largely focused on mechanisms responsible for neuronal plasticity. The idea is that memory, like learning, derives in large part from altered synaptic connections between neurons that result from prior neuronal activities. Stronger and/or more synaptic connections lead to long-term potentiation (LTP) and weaker and/or fewer synaptic connections lead to long-term depression (LTD). These changes cause altered neuronal network circuitries that affect subsequent responses, i.e. learning. The behavioral paradigms and underlying molecular mechanisms employed to investigate synaptic strength may be more easily related to learning than memory per se, but this hardly matters if memory and learning are simply different ways of looking at the same cognitive phenomena.
There are numerous molecular mechanisms that serve to modulate synaptic connections in response to prior neuronal activities. Some of these are autonomous consequences of synaptic activation that involve mechanisms more or less contained within and intrinsic to a given activated synapse. Chemical synapses are organized around a synaptic cleft formed from the closely apposed, highly differentiated pre-synaptic axonal and post-synaptic dendritic membranes of two neurons. The pre-synaptic membrane is studded with synaptic vesicles filled with various neurotransmitters ready to be released into the cleft in response to the arrival of an action potential with its associated wave of elevated calcium. Glutamate is the principal excitatory neurotransmitter in the brain, and the generation of post-synaptic action potentials depends on the membrane depolarization produced by glutamate-induced opening of ionotropic glutamate receptors. Glutamate-induced opening of one of the most abundant glutamate receptors, the NMDA receptor, requires coordinate depolarization of the post-synaptic membrane. Action potentials that release sufficient glutamate to activate alternative post-synaptic glutamate channels such as AMPA-type receptors can depolarize post-synaptic membranes so as to subsequently allow NMDA-mediated glutamate signaling. This provides a mechanism for LTP at glutamatergic synapses and helps explain why NMDA receptors play a central role in learning. NMDA receptor activation is only one of many intrinsic control mechanisms associated with synaptic function, however. For instance, elevations in calcium associated with neurotransmission activate protein kinases and phosphatases that regulate a number of target activities associated with LTP and LTD.
In addition to ligand-gated ion channels, neurotransmitters generally interact with metabotropic G-protein coupled receptors, or GPCRs. Numerous different GPCRs clustered within synaptic membranes provide a sort of sensory cortex attuned to a wide range of neurotransmitter inputs. Different receptors modulate different signaling networks in response to different sets of agonists and antagonists. Clearly, metabotropic receptor signaling plays an important and pervasive role in the modulation of synaptic strength in response to previous neuronal activity. There are, for instance, at least three different metabotropic dopamine receptors that interact with heterotrimeric G-proteins that generally promote increased synapse strength, and there are at least two other dopamine receptors that activate other G-proteins that generally promote decreased synaptic strength. The ensemble of GPCRs, G-proteins and cellular signal transduction components at a particular synapse varies with genetic background, epigenetic status, age and history. GPCR regulatory outputs primarily act to control the network of interacting protein kinases that catalyze the transfer of phosphoryl groups from ATP to specific serine and threonine residues in regulatory targets. It has been estimated that roughly a third of all cytoplasmic proteins are subject to kinase regulation.
Receptor-mediated signaling mechanisms that act locally to modulate synaptic strength are thought to account for short-term memories. Stronger and more persistent receptor activation can result in the activation of signal transduction networks that extend beyond individual synapses to effect changes in nuclear kinase activities that lead to changes in epigenetic modifications and alterations in gene expression. Processes that lead to changes in gene expression are thought to be responsible for long-term memories. There are numerous different protein kinases in a given neuron, each responsible for the phosphorylation of a specific set of cellular targets. Moreover, protein kinases are themselves targets of kinase-mediated phosphorylation. This extensive regulatory network of receptors, G-proteins, kinases and their regulatory targets ensures that the dynamic functional connectivity between neurons is continuously evolving so that LTP and LTD would be expected to be the rule rather than the exception, with every train of action potentials potentially triggering both short and long-term changes in neuronal function. Neurologic processes ranging from associative learning to addiction clearly depend on metabotropic signaling mechanisms. In a sense, all of these cognitive phenomena have an important memory component.
Arguments have been made that changes in E. coli gene expression that lead to alterations in a cell’s repertoire of sensory receptors are analogous to neuronal processes associated with learning. For instance, E. coli that grow without ever being exposed to ribose are relatively unresponsive to ribose, whereas E. coli grown in the presence of ribose ‘learn’ to sense ribose, and exhibit a strong favorable motor response to ribose later in life. This learned behavior is conditional; if E. coli are grown in a culture where they are exposed to glucose in addition to ribose, their subsequent responses to ribose tend to be considerably reduced due, at least in part, to glucose-induced reductions in levels of cAMP. In E. coli, cAMP serves to activate a transcription factor that functions as a co-activator of the gene that encodes the ribose receptor, so that the lowered levels of cAMP caused by glucose tend to attenuate ribose-induced expression of the ribose receptor, and thereby reduce subsequent responses to ribose. This type of learning seems to imply a bacterial memory component analogous to the long-term procedural memory mediated by neuronal information processing systems. All living organisms, including plants, animals and both motile and immotile bacteria share essentially the same mechanisms of signal transduction and transcriptional regulation as those used by E. coli to control the expression of their sensory receptors. Convincing arguments have been advanced that all cells ‘learn’, since all cells can effectively alter their ‘behaviors’ in response to sensory inputs. Nevertheless, nanobrains in bacteria and nervous systems in animals clearly have unique features that make them particularly well suited to solve the cognitive problems associated with motility and motor control.
Molecular logic of memory and learning
There appears to be a molecular logic to the chemistries that underlie biological processes. Similar types of biochemical mechanisms are applied to solve analogous problems in a wide range of different organisms. Thermodynamic considerations provide useful systemic parameters to understand the utility of a particular biochemical mechanism. Metabolic energy, for instance, is generally used to drive cellular processes by coupling the free energy of nucleoside triphosphate hydrolysis to the generation of conformational changes in proteins that lead to the performance of various types of useful work. ATP and GTP hydrolysis play essential roles in a wide range of different learning paradigms in both neuronal and non-neuronal systems. Protein kinase regulatory networks, the so-called kinomes, in association with phosphoprotein phosphatases, are essentially regulatory ATPases. And the GPCR-mediated G-protein signaling systems that directly or indirectly control protein kinase activities are regulatory GTPases. The use of nucleoside triphosphates as a universal bioenergetic currency makes sense. ATP-coupled reactions are readily reversible, and electrochemical gradients can be used to regenerate ATP from ADP + Pi. Both ATP and the products of ATP hydrolysis are highly polar, water-soluble metabolites, easily retained within membrane-bound cytosolic compartments. Moreover, the free energies of ATP or GTP hydrolysis are well suited to their roles in signal transduction. Under physiological conditions the free energy of GTP hydrolysis or of phosphotransfer from ATP to serine or threonine hydroxyl side chains is highly favored as is the hydrolysis of protein phosphoserine or phosphothreonine groups to produce Pi. Thus, both G-protein-mediated signaling and kinase/phosphatase-mediated protein phosphorylation and dephosphorylation are under kinetic control, so that tightly regulated balances between kinase and phosphatase activities determine the level of phosphorylation of a given target protein.
Nanobrain memory in bacteria depends on a different biochemistry: the addition and removal of protein methyl groups rather than phosphoryl groups. In the case of methylation, the central metabolite is S-adenosylmethionine (SAM) rather than ATP. The logic of methylation is not readily understood in terms of the underlying bioenergetics. Methyl group transfer from SAM to a wide range of different nucleophiles is thermodynamically favored, but in proteins and nucleic acids the products are generally methylamines or methyl ethers that are not readily demethylated. Protein carboxyl methylation, such as occurs in the nanobrain, is an exception. In this case, however, the product of demethylation, methanol, tends to be toxic and cannot be effectively recycled. Moreover, the bioenergetics of carboxyl methylation do not appear to be optimal. The ‘cost’ per methylation event is equivalent to that expended in the hydrolysis of over 12 ATPs. Why not employ phosphorylation chemistries that just use 1 ATP per transaction?
The chemical changes elicited by methylation are relatively subtle compared to phosphorylation. In the bacterial nanobrain, glutamates are converted to methylglutamates, which are chemically very similar to glutamines. The structural similarity between glutamines and methylglutamates may be one reason for the use of methylation rather than phosphorylation. Glutamine to glutamate is a conservative substitution. In fact, about half of the eight or more potential methyl accepting glutamates per nanobrain fiber are synthesized de novo as glutamines, which can later be converted to glutamates by the same enzyme that hydrolyzes methylglutamates. Thus, the genetically encoded sequences of glutamines and glutamates act as a preprogramed memory that is subject to modification in response to sensory cues. Besides protein carboxyl methylation, irreversible N-methylations at arginine, lysine and histidine side chains also involve relatively modest alterations in structure and function. Whereas phosphorylations tend to act as switches to directly turn specific target activities on or off, methylation generally functions to modulate preexisting activities with the effects being most evident at a systemic level.
In both animals and bacteria defects in methylation metabolism are associated with defects in cognitive functions associated with memory. Adler initially discovered the methylation memory mechanism in E. coli when he observed that methionine auxotrophs were uniquely defective in their ability to adapt to attractant stimuli. In vertebrates, deficiencies in folate- and B12-dependent SAM synthesis are associated with neurodegenerative diseases such as Alzheimer’s, and supplementation with these vitamins has recently been shown to reduce the rate of brain atrophy in demented individuals with elevated plasma homocysteine, which is a marker for defects in methylation metabolism. Unlike phosphorylation, which is a very common post-translational modification, protein carboxyl methylation is extremely rare. The bacterial nanobrain provides the only known instance of regulatory carboxyl methylation in prokaryotes. In eukaryotes there are only two examples. One involves a membrane-associated SAM-dependent isoprenyl cysteine methyl transferase (ICMT) that reversibly methylates the α-carboxyl of a highly conserved, isoprenylated cysteine residue at the carboxyl terminus of a number of different membrane-associated signal transduction components, including all RAS-related and heterotrimeric G-proteins. ICMT is highly expressed in the cerebellum, which is thought to play a central role in procedural and motor memory.
The only other known instance of regulatory protein carboxyl methylation in eukaryotes involves methylation and demethylation of the α-carboxyl of the carboxy-terminal leucine in the catalytic subunit of phosphoprotein phosphatase 2A, PP2A. A specific cytosolic enzyme, PPMT, catalyzes the transfer of a methyl group from SAM to PP2A, and the resulting methyl ester is hydrolyzed by a specific methyl esterase, PME. PP2A accounts for the majority of the Phospho-Ser/Thr phosphatase activity in nervous tissue, and chronic PP2A demethylation is associated with Alzheimer’s disease. Methylated forms of PP2A are largely responsible for dephosphorylation of the microtubule regulatory protein Tau. The Tau protein contains over 40 serine and threonine residues that are subject to phosphorylation by numerous different protein kinases. Hyperphosphorylated Tau destabilizes axonal microtubules and forms aggregates termed neurofibrillary tangles that are a hallmark of Alzheimer’s dementia. It seems likely that the carboxyl methylation-dependent PP2A activity that regulates Tau and controls microtubule function plays an important role in regulating neural plasticity under healthy physiological conditions.
Conclusions
Is there a eukaryotic analogue of the bacterial nanobrain memory mechanism — a mechanism to essentially provide a continuous trace of the past that can be used to monitor and evaluate changes in outcome and effect appropriate responses? One imagines that without altered environmental conditions, memory input of the bacterial type would be minimal. A significant change that was out of line with the memory reference would induce a response. Associated with this response would be a change in memory that would match the altered conditions and lead to a return of steady state behavioral outputs. One can imagine such a mechanism functioning at a systemic level — the mental state that comes to mind is ‘attention’. But whereas in nanobrains mechanisms that underlie memory can be understood in terms of molecular structure and function, understanding memory in nervous systems requires an appreciation of cellular interactions. In addition to the molecular biochemistry, one must consider the entorhinal cortex, hippocampus, cerebral cortex, cerebellum, etc. The dimensions of an entire bacterial cell are only about the size of a single chemical synapse, and there are over 1014 synapses in the human brain. Nevertheless, because the constraints of time and space that apply to bacterial behavior are so alien to our own, bacteria provide an intriguing alternative perspective on fundamental cognitive processes such as memory and learning that have hitherto been examined almost exclusively within the context of human and animal behavioral paradigms.
Further reading
- Adler J. My life with nature. Annu Rev Biochem. 2011;80:42–70. doi: 10.1146/annurev-biochem-121609-100316. [DOI] [PubMed] [Google Scholar]
- Barco A, Bailey CH, Kandel ER. Common molecular mechanisms in explicit and implicit memory. J Neurochem. 2006;97:1520–1533. doi: 10.1111/j.1471-4159.2006.03870.x. [DOI] [PubMed] [Google Scholar]
- Braithwaite SP, Stock JB, Lombroso PJ, Nairn AC. Protein phosphatases and Alzheimer’s disease. Prog Mol Biol Transl Sci. 2012;106:343–379. doi: 10.1016/B978-0-12-396456-4.00012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Hall DD, Hell JW. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol Rev. 2009;89:411–452. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaud G, Refsum H, de Jager CA, Jacoby R, Nichols TE, Smith SM, Smith AD. Preventing Alzheimer’s disease-related gray matter atrophy by B-vitamin treatment. Proc Natl Acad Sci USA. 2013;110:9523–9528. doi: 10.1073/pnas.1301816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialog between genes and synapses. Biosci Rep. 2004;24:475–522. doi: 10.1007/s10540-005-2742-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Stock JB. Protein carboxyl methylation and the biochemistry of memory. Biol Chem. 2009;390:1087–1096. doi: 10.1515/BC.2009.133. [DOI] [PubMed] [Google Scholar]
- Muzzio IA, Kentros C, Kandel E. What is remembered? Role of attention on the encoding and retrieval of hippocampal representations. J Physiol. 2009;587:2837–2854. doi: 10.1113/jphysiol.2009.172445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivka PF, Falke JJ. Isolated bacterial chemosensory array possesses quasi- and ultrastable components: functional links between array stability, cooperativity, and order. Biochemistry. 2012;51:10218–10228. doi: 10.1021/bi301287h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol. 2012;24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]