Abstract
In culture, human pluripotent stem cells (PSCs) are phenotypically (for instance, SSEA3 expression level) and functionally (capacity to survive after single-cell dissociation) heterogeneous. We report here that the side scatter (SSC) signal measured by flow cytometry, a variable correlated with membrane irregularity and cell granularity, is very high in PSCs, even higher than in blood polymorphonuclear cells, and markedly heterogeneous. Moreover, SSC intensity rapidly and strongly decreases upon PSC differentiation into any of the three germ layers. PSCs with high SSC (HSSC cells) or low SSC (LSSC cells) values both express pluripotency markers, but HSSC cells are characterized by more frequent simultaneous expression of the membrane pluripotency factors SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24 and by higher mitochondrial content. Functionally, HSSC cells are more likely to generate colonies upon single cell passage than LSSC cells. SSC monitoring might provide a simple, but robust and rapid method to estimate pluripotency variations in culture and unveils a new phenotypic and functional heterogeneity in PSCs.
Keywords: Animals; Antigens, CD24; genetics; metabolism; Antigens, Surface; genetics; metabolism; Biological Markers; metabolism; Cell Differentiation; Cell Line; Clone Cells; Embryonic Stem Cells; cytology; metabolism; Flow Cytometry; Gene Expression; Genetic Heterogeneity; Germ Layers; cytology; metabolism; Humans; Mice; Mice, SCID; Pluripotent Stem Cells; cytology; metabolism; Proteoglycans; genetics; metabolism; Stage-Specific Embryonic Antigens; genetics; metabolism; Teratoma; metabolism; pathology
Introduction
Pluripotent stem cells (PSCs), either human embryonic stem cells (hESCs) or human induced pluripotent stem cells (hiPSCs), retain many features of the inner cell mass (ICM), particularly the expression of pluripotency transcription factors (such as POU5F1 and NANOG) [1]. In addition, expression fluctuations [2–4], not only at the mRNA but also at the protein level [5–7], have been reported for pluripotency transcription factors, surface markers [5–12], epigenetic marks [10,13,14] and lineage-specific genes [15]. PSCs might thus permanently shift between cell compartments characterized by different levels of pluripotency [9,12]. The apparently stochastic fluctuation in the expression of pluripotency genes, cell surface glycoproteins or epigenetic marks might reflect the unstable pluripotency equilibrium of cultured PSCs and a better control of culture conditions could help minimizing these variations [2]. Alternatively, this heterogeneity may be inherent to cell fate decisions as it is also observed in state transitions during neurogenesis or hematopoiesis [16] and may even be a common biological feature [17].
Flow cytometry is a powerful, quantitative method to study PSC diversity at the single cell level [18] and has significantly contributed to the characterization of the fluctuations of pluripotency cell surface markers in PSCs [9,11,12]. In addition to the quantification of fluorescence emission (biomarker detection), other types of information can be derived from the analysis of flow cytometry data. Specifically, as the laser beam passes through the cell, light is deflected and refracted. This scattered light is collected by the forward scatter (FSC) and by the side scatter (SSC) photodiodes. FSC intensity mainly correlates with the cell size, whereas SSC is a measure of the cell refractive index that depends on the cell granularity or internal complexity (for instance, cytoplasmic membrane wrinkling, number and shape of vesicles and mitochondria, development of the endoplasmic reticulum and nucleus structure).
We recently reported that polychromatic flow cytometry can provide information on the expression kinetics of the cell surface pluripotency markers SSEA3, SSEA4, TRA-1-81 and TRA-1-60 [11]. During that work, we noted that human PSCs had very heterogeneous SSC values. Here, we thoroughly investigated SSC intensity variations in different PSC lines and show that PSCs fluctuate between extreme SSC states. Moreover, PSCs with high SSC (HSSC cells) express more often many cell surface pluripotency markers simultaneously than cells with low SSC (LSSC). HSSC and LSSC cells show differences in cell cycle distribution. HSSC cells are more prone to apoptosis and display higher mitochondrial content. However, despite their higher apoptosis rate, HSSC cells have a much higher clonogenic self-renewal capacity than LSSC cells.
Material and Methods
Cell culture
The following hES cell lines were used: HUES1, from Douglas Melton’s laboratory [19] and HD83 and HD291 cells that we derived in our laboratory [11,20]; The hiPSC cell line M4C7 is one of a series of clones obtained by reprogramming the human fibroblast foreskin hFF1 cell line (ATCC) by using lentiviral vectors to express human OCT4/POU5F1, SOX2, NANOG and LIN28 [21]. M4C7 cells grow as typical PSCs and display all the features of PSCs: OCT4 and ABCG2 expression, phosphatase alkaline activity, a typical PSC expression signature by microarray analysis and can differentiate into cells of the three germ layers. All cell lines were routinely cultured in 35 mm wells in KO-DMEM medium supplemented with 20% Knockout serum replacement (KO-SR) (Invitrogen, Carlsbad, CA), 0.1mM non-essential amino acids, 2 mM L-Glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μM β-mercaptoethanol and 10 ng/ml bFGF (PSC culture medium) and were passaged mechanically (HD83 and HD291) or semi-mechanically using Collagenase IV (HUES1) or mechanically and then enzymatically using TrypLE Select (Invitrogen) (M4C7). Mouse embryonic fibroblasts (MEFs) or human foreskin fibroblasts (hFF1) were irradiated and used as feeders at 34 000 cells/cm2. HFF1 cells and MEFs were maintained in DMEM medium supplemented with 15% heat-inactivated fetal bovine serum (FBS) (Invitrogen). For low-serum condition, PSCs were grown in PSC culture medium with 0.1% instead of 20% KO-SR.
For in vitro differentiation, PSCs were pre-treated with 10 μM Y27632, a p160 Rho-associated kinase (ROCK) inhibitor (Sigma-Aldrich, St. Louis, MO), for 1 hour. Cells were then dissociated with 1X TrypLE Select (Invitrogen) at 37°C for 10 min, seeded at 5 000 cells/cm2 in 96-well plates pre-coated with 1mg/ml Matrigel (endoderm and ectoderm differentiation) or with feeder cells (mesoderm differentiation). PSCs were again incubated with 10 μM Y27632 for 24 hours and then PSC differentiation into endodermal (HUES1 cells), mesodermal (HD291 cells) or ectodermal cells (M4C7 cells) was induced as previously described [11]. Briefly, endodermal differentiation was induced by addition of activin A to low-serum medium [22]; ectodermal differentiation was induced by adding Noggin and SB431542, two specific inhibitors of SMAD signaling [23]; and mesoderm was obtained by co-culture with OP9 cells and VEGF [24]. Differentiation was accompanied by the sharp down-regulation of the pluripotency specific transcription factor OCT4 and by the induction of lineage-specific markers, such as CD45 (mesoderm), FOXA2 and CXCR4 (endoderm), Nestin and CD56 (ectoderm). In addition, hematopoietic differentiation (mesoderm model) was documented by the appearance of white blood cells, as indicated by May-Grünwald-Giemsa staining [11].
Quantification of stem cell markers, mitochondrial content, apoptosis rate and cell cycle distribution by flow cytometry
For flow cytometry analyses, cells were dissociated with TrypLE at 37°C for 10 minutes. Cell surface pluripotency markers were quantified using a cocktail of five antibodies against CD24-PB (EXBIO), SSEA3-PE (BD), SSEA4-PERCP (R&D), TRA-1-60-FITC, (BD) and TRA-1-81-APC (BD) and the Sytox Blue Dead Cell Stain (Invitrogen).
Mitochondrial content was determined using MitoTRacker Red CMXRos probes (Invitrogen). PSC culture medium was removed from the 35 mm dishes and cells were incubated with 100 nM MitoTRacker probes in pre-warmed (37°C) PSC medium without serum at 37°C for 30 minutes. Cells were then washed with PBS, dissociated with TrypLE, suspended in 500 μl PBS and incubated with 6 μl anti-CD44-APC antibody (BD) to exclude feeder cells.
To evaluate the cell cycle distribution, dissociated cells were fixed in cooled 70% EtOH at 4°C for 15 min, permeabilized using 0.1% Triton X-100 (v/v) and stained with 10 μg/ml propidium iodide plus 250 μg/ml RNase A for 30 min. To quantify the apoptosis rate, TrypLE-dissociated cells were incubated with the anti-CD44 antibody for 30 min and then stained with Annexin-V-FITC (BD Biosciences) in Annexin-V binding buffer (BD) for 20 min, as recommended by the manufacturer. Sytox Blue stain was added to exclude dead cells.
Flow cytometry data acquisition was carried using a CyAn instrument (Beckman Coulter, Fullerton, CA) and data analyzed using the KALUZA software (Beckman Coulter).
FACS-sorting of HSSC and LSCC cells
PSCs were pretreated with 10 μM Y27632 for 1 h, stained with Sytox Blue (Invitrogen) and incubated with an anti-CD44 antibody (Pharmingen) to label feeder cells. LSSC and HSSC cells were then sorted using an Astrios instrument (debris and aggregated cells were eliminated by specific gating), plated in 96-well plates pre-coated with irradiated hFF1 cells and cultured in the presence of 10 μM Y27632 in PSC culture medium for 24 hours.
Teratoma formation
Animals were handled in strict accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the EU directives (Council Directive 86/6009/EEC). NOD/SCID/IL2RG−/− mice (Charles River, France) were maintained in standard conditions (12 h 90 lux light and 12 h dark) and fed ad libitum with a standard rodent diet. 300 000 sorted HSSC and LSSC cells were centrifuged and pellets resuspended in 30% Matrigel diluted in KO-SR in a final volume of 200 μl per injection. Before subcutaneous injection of the cell mixture, mice were lightly anesthetized with 35 mg/kg ketamine (Merial, France) and 14 mg/kg xylasine (Bayer Healthcare, Germany), shaved on the left flank and disinfected with 70% ethanol. Teratoma appearance and growth were followed weekly. Approximately eight weeks after the graft, teratomas were dissected, fixed in 3.7% formaldehyde, dehydrated and embedded in paraffin; 4-μm sections were processed for Hematoxylin-Eosin-Safran staining.
Transmission Electron Microscopy (TEM)
FACS-sorted HSSC and LSSC cells were immersed in a solution of 2.5% glutaraldehyde in 0.1M Sorensen’s buffer (pH 7.4) at 4°C overnight. They were then rinsed in Sorensen’s buffer and post-fixed in 0.5% osmic acid in the dark, at room temperature for 2 h. After two rinses in Sorensen’s buffer, cells were dehydrated in a graded series of ethanol solutions (30–100%) and embedded in EmBed 812 using an Automated Microwave Tissue Processor for Electron Microscopy (Leica EM AMW). Thin sections (70 nm; Leica-Reichert Ultracut E) were collected at different levels of each block, counterstained with uranyl acetate and observed using a Hitachi 7100 transmission electron microscope at the Centre de Ressources en Imagerie Cellulaire de Montpellier (France)
Statistical analysis
Statistical comparisons were made using the Student’s t-test and p-values <0.05 were considered to be significant. The SSC distribution of the different cell subpopulations was analyzed with the R software (R.2.13, http://www.r-project.org/) and Bioconductor, version 2.8 [25]. The flowCore package [26] was used to import the flow cytometry data in R and the SSC distribution of each cell subpopulation is shown as dot plots.
Results
SSC intensity values in PSCs are very heterogeneous
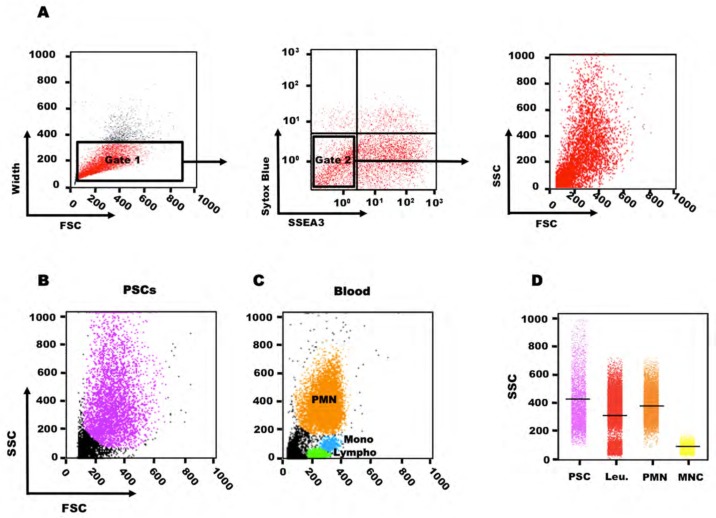
To thoroughly evaluate the variations in SSC intensity in human PSCs, we assessed SSC intensity distribution in hES (Figure 1 and Figures 2B–D) and hiPS cell lines (Figure 2A) by flow cytometry. In this study, to quantify the PSCs, we excluded dead cells, aggregated cells and debris (Figure 1A). All PSC lines showed a wide SSC distribution, including a sub-population of cells with very high SSC intensity. Comparison with the SSC values of blood cells (assessed using the same settings) (Figure 1C) indicated that SSC intensity in PSCs was 6.5-fold higher than in blood mononuclear cells and even higher than in granulocytes, which are known to have very high SSC values (Figure 1D). In addition, the SSC values of PSCs were markedly heterogeneous (from very low to very high SSC intensity), suggesting that each PSC population includes cells with variable morphological features (internal and/or membrane complexity). To understand why PSCs display such a huge SSC variability, we investigated several features (morphology, expression of pluripotency markers, clonogenic self-renewal, apoptosis and cell cycle distribution) in PSCs with either high SSC (HSSC) or low SSC values (LSSC).
Figure 1. Analysis of the side scatter parameter in pluripotent stem cells (PSCs) and human blood cells.
Scatter plot of SSC versus FSC values in HD83 human ES cells. (A) Gating strategy. To quantify the PSCs, we eliminated aggregated cells (gate 1) and dead cells using Sytox blue stain (gate 2). Debris cells were evaluated based on FSC and SSC. PSCs are in pink. (C) Scatter plot of SSC versus FSC values in human blood cells. Based on the SSC and FSC distribution, granulocytes (orange) can be differentiated from monocytes (blue) and lymphocytes (green). (D) Comparison of the SSC distributions of PSCs, human blood leucocytes, granulocytes and mononuclear cells. Flow cytometry data were imported in the R statistical software using the flowCore package. The SSC distribution of each cell subpopulation was extracted and represented as a dot plot.
SSC: side scatter; FSC: forward scatter; PMN: granulocytes; Mono: mononuclear cells; Lympho: lymphocytes; Leu: leukocytes; MNC: mononuclear cells.
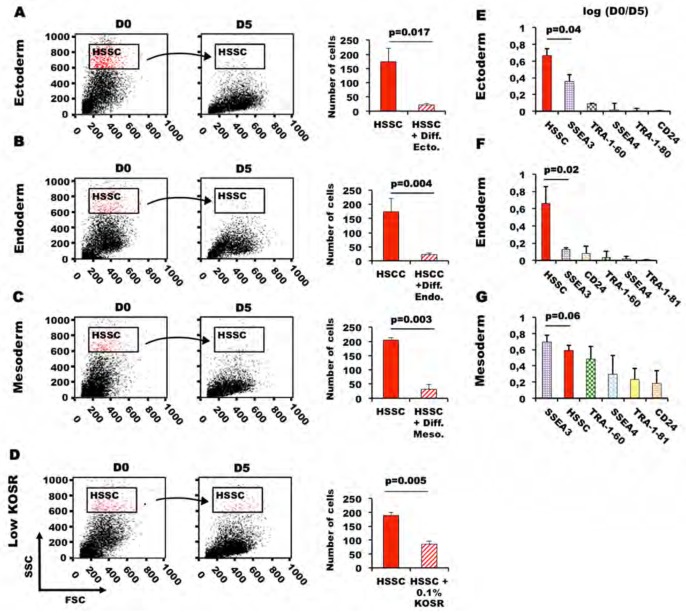
Figure 2. SSC values are very heterogeneous within PSCs.
HiPSCs (M4C7) (A) and ES cells (HUES1 (B) and HD291 (C)) were directly differentiated into ectoderm, endoderm and mesoderm, respectively; whereas the ES line HD83 (D) was cultured in low-serum (0.1 % KOSR) medium. Samples were collected at day 5 and SSC analyzed by flow cytometry. The bar plots show the number of cells with high SSC values (HSSC) at day 0 and 5. (E, F and G) Loss of expression of specific pluripotency markers and reduction of the HSSC population between day 0 and 5 after induction of differentiation. We used 5-color flow cytometry to simultaneously identify cells that were positive for the surface pluripotency markers SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24. The percentage of positive cells was quantified after exclusion of cell aggregates, debris and dead cells. Data are presented as the log of the ratio of the mean ± SD at day 0 and at day 5 of three independent experiments using M4C7 (ectoderm differentiation) HUES1 (endoderm differentiation), and HD291 cells (mesoderm differentiation). * p-values are calculated using the Student’s t test.
The size of the HSSC population is highly correlated with the pluripotency state
First, we tested whether loss of pluripotency and cell differentiation could be associated with a decrease in SSC intensity by measuring the size of the HSSC population in the four PSC lines following induction of differentiation [11], or culture in low-serum conditions (0.1% KO-SR). The size of the HSSC populations was drastically decreased after only five days of differentiation into ectodermal (M4C7 hiPSC; 6-fold reduction), endodermal (HUES1 ES cells; 7-fold decrease) or mesodermal cells (HD291 ES cells; 7.8-fold reduction) (Figures 2A, B and C), and also when HD83 cells were induced to differentiate in low-serum medium (Figure 2D). As the decrease of the HSSC populations was a very early event during PSC differentiation, we investigated whether HSCC reduction was temporally associated with changes in the expression levels of the cell surface pluripotency markers SSEA3, SSEA4, TRA-1-60, TRA-1-81 and CD24 (Figures 2E, F and G). The decrease of the HSSC cell subset was the most remarkable change between day 0 and day 5 of differentiation in the ectoderm and endoderm differentiation models, whereas reduction in SSEA3 expression, one of the most sensitive pluripotency markers [9,11], came first only during mesoderm differentiation. Hence, in two of the three differentiation pathways tested, the decrease of the HSSC population significantly exceeded in magnitude loss of SSEA3 expression. Conversely, SSEA4, TRA-1-81 and CD24 expression were not changed or only slightly decreased in all the differentiation models at day 5 of differentiation. These results indicate that SSC could represent a new parameter for monitoring loss of pluripotency during PSC differentiation.
HSSC cells express simultaneously different pluripotent markers
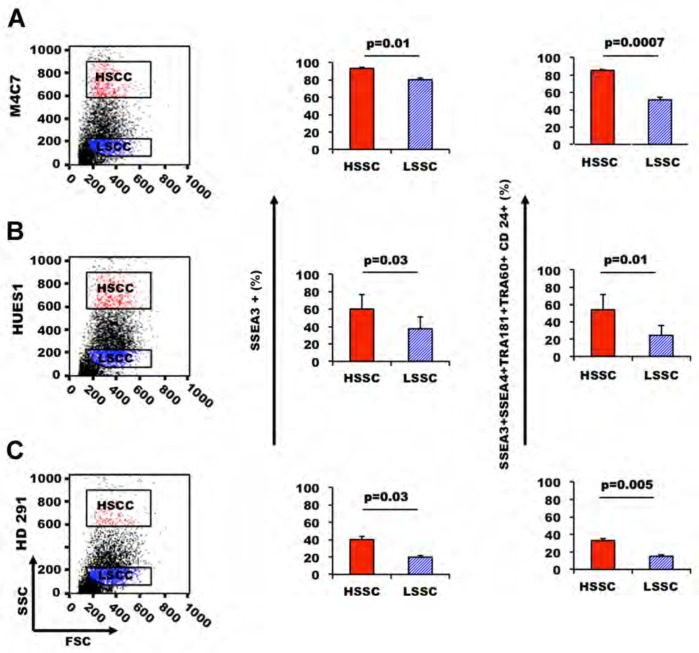
We previously showed that expression of five common cell surface pluripotency markers (SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24) is highly variable within a PSC population and that only a portion of these cells expresses simultaneously all five markers [11,27]. We thus asked whether HSSC and LSSC cells had distinct expression profiles of pluripotent markers. HSSC cells from the hiPS cell line M4C7 contained a significantly higher percentage of SSEA3-positive cells than LSSC cells. In addition, 85–87% of HSSC cells expressed simultaneously SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24, whereas only 51–54% of LSSC cells did (Figure 3A). Similar results were obtained with the hES cell lines HUES1 and HD291 (Figures 3B and 3C), indicating that this finding was not limited to a specific PSC line and suggesting that high SSC may be correlated with pluripotency.
Figure 3. HSSC cells express high level of pluripotent markers.
We compared the expression of cell surface pluripotency markers in the HSSC and LSSC populations of pluripotent stem cells from three PSC lines (the hiPS cell line M4C7 (A) and of the ES cell lines HUES1 (B) and HD291 (C)). The bar plots show the percentage of cells that express either SSEA3 alone or the five markers SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24 simultaneously. Data are representative of three independent experiments.
The clonogenic self-renewal capacity of PSCs is correlated with high SSC values
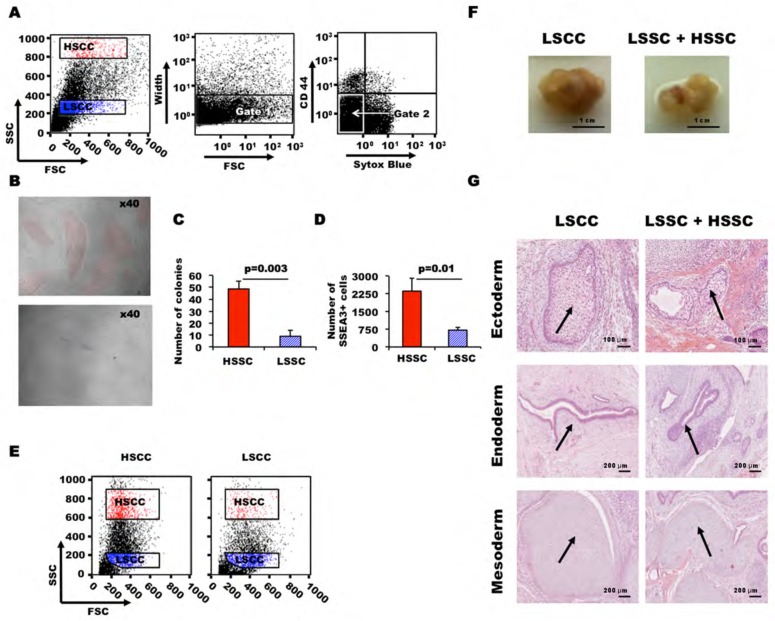
To examine more closely the link between SSC intensity and pluripotency, we compared the developmental potential of HSSC and LSSC cells. To quantitate their clonogenic self-renewal capacity, PSCs were FACS-sorted into HSSC and LSSC cells (Figure 4A) and cultured at low density in 96-well plates pre-coated with irradiated hFFs for four days. Microscopy analysis revealed that HSSC cells had higher self-renewal capacity and generated larger colonies than LSSC cells (49 +/− 6 and 9 +/− 5 colonies per 500 cells, respectively) (Figures 4B and C). Moreover, the number of SSEA3-positive cells per well was higher in colonies generated from HSSC than from LSSC (Figure 4D). Despite their lower clonogenic self-renewal capacity, LSSC cells could form bona fide PSC colonies that contained both HSSC and LSSC cells, as indicated by flow cytometry analysis of the colonies after 5 days of culture (Figure 4E). This finding demonstrates that PSCs fluctuate between these extreme SSC states. Finally, to confirm the pluripotency status of the LSSC population, we grafted immuno-compromised mice with FACS-sorted LSSC cells or with a mixture of LSSC and HSSC cells [28,29]. Both cell preparations produced teratomas of similar size that included patches of endoderm, mesoderm and ectoderm (Figures 4G and H), indicating that LSSC cells are nevertheless pluripotent. Taken together, these results demonstrate that HSSC cells have a higher clonogenic self-renewal capacity, but that each SSC subset contains pluripotent cells that can generate both HSSC and LSSC cells.
Figure 4. Clonogenic self-renewal efficiency of HSSC and LSSC PSCs.
(A–E) FACS-sorted HSSC and LSSC cells were plated in 96-well plates (500 cells/well) and cultured for 5 days. (A) During cell sorting, debris, aggregates, dead cells and feeder cells were excluded using the FSC-SSC and FSC-Width parameters and labelling with the anti-CD44 antibody and Sytox Blue stain, respectively. (B) Representative phase contrast images of PSC colonies from HSSC (upper panel) and LSSC cells (lower panel) at day 5 after sorting, highlighted in red and blue respectively. (C and D) Bar plots display the number of colonies and of SSEA3-positive cells per well from FACS-sorted HSCC and LSSC cells measured by quantitative flow cytometry using Flow Check beads. (E) FSC and SSC analysis of HSCC and LSSC cells after 5 days of culture show that each population regenerates both cell types. (F, G) Teratomas obtained by subcutaneous injection of LSSC and LSSC + HSSC cells in NOD/SCID/IL2RG−/− mice: tumors (F) and Hematoxylin-Eosin-Safran staining of tumor sections (G) arrows showing glycogenated squamous epithelium (ectoderm), epithelium with caliciform cells (endoderm) and cartilage (mesoderm).
HSSC and LSSC cells have distinct apoptosis rates and cell cycle profiles
To further characterize the biological features of HSSC and LSSC PSCs, we evaluated their apoptosis rate and cell cycle distribution. Indeed, during FACS sorting of the SSC populations, we noticed that HSSC cells were less viable than LSSC cells (data not shown). To quantify apoptosis and cell death, M4C7 cells were stained with Annexin-V and Sytox Blue, respectively, and then the percentages of early and late apoptotic cells, live cells and dead cells were quantified in the HSSC and LSSC populations (Figure 5A). Early and late apoptosis as well as cell death were significantly higher (2.4-, 4- and 6.7-fold, respectively) in HSSC cells than in LSSC cells (Figure 5B).
Figure 5. Cell cycle distribution and apoptosis rate in HSSC and LSSC cells.
Apoptosis rates and cell cycle distribution were analyzed in M4C7 cells (hiPSCs). (A) Representative dot plot of the Annexin-V- and Sytox Blue-stained PSCs. Fibroblast feeder cells were excluded by labeling with anti-CD44 antibodies. (B) Quantification of live, dead, early and late apoptotic cells in the HSSC and LSSC subpopulations indicates that HSSC cells have higher apoptosis rates than LSSC cells. (C and D) M4C7 cells were stained with propidium iodide and the anti-CD44 antibody to exclude fibroblast feeder cells and the cell cycle distribution quantified in HSCC and LSSC cells. Data are the mean ± SEM of three independent experiments.
Next, we investigated cell cycle progression in HSSC and LSSC cells. The percentage of cells in the G2/M phase was 2.1-fold higher, whereas the percentage of cells in G0/G1 and S phase was reduced by 1.5- and 2-fold, respectively, in HSSC in comparison to LSSC cells (Figures 5C and D).
HSSC and LSSC cells show distinct ultrastructural characteristics and mitochondrial content
As differences in SSC intensity are related to irregularities in the cell membrane, cytoplasm or nucleus, we compared the ultrastructure of HSSC and LSSC PSCs by transmission electron microscopy (TEM). The most notable differences between the two cell populations were that HSSC cells were characterized by many rounded mitochondria that were distributed around the nucleus, whereas LSSC cells had many lipid vesicles in the cytoplasm (Figures 6A and B). The higher mitochondrial content of HSSC cells was confirmed also by quantification of the mitochondrial-specific dye MitoTracker by flow cytometry (see Material and Methods). Indeed, MitoTracker accumulation was significantly higher in HSSC cells than in LSSC cells (Figure 6C).
Figure 6. Ultrastructural analysis and mitochondrial content of HSSC and LSSC cells.
(A and B) Analysis of the ultrastructure of both HSSC and LSSC cells by transmission electron microscopy reveals specific characteristics for each subset: high level of rounded mitochondria in HSSC cells (asterisks) and many lipid vesicles in the cytoplasm of LSSC cells (arrows). Cells were FACS-sorted and aggregates, debris, dead cells and feeder cells were excluded as detailed in Figure 4. (C) Flow cytometry using MitoTracker probes indicates that HSSC cells have a significantly higher mitochondrial content than LSCC cells. Data are the mean ± SEM of three independent experiments.
Plural pluripotent stem cell compartments
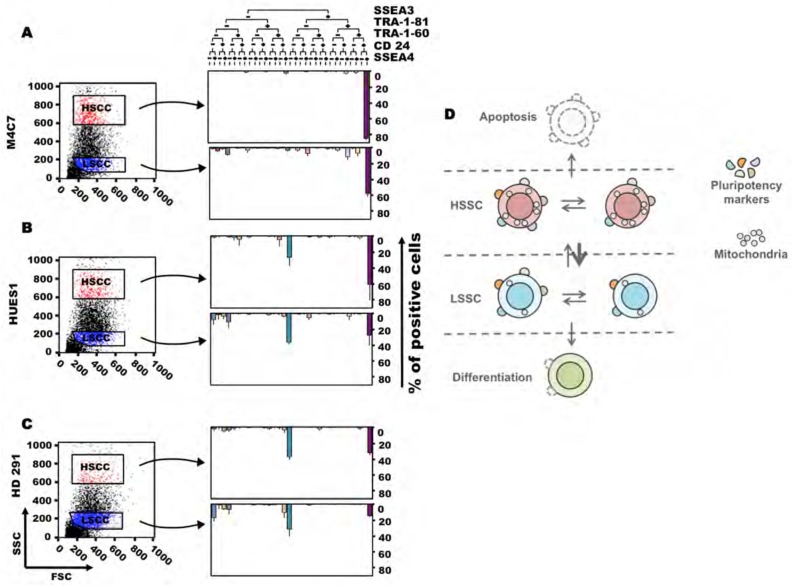
The SSC parameter can thus delineate different PSC cell populations that do not overlap with the differential expression of the cell surface pluripotency marker SSEA3 (Figure 3). Indeed, both HSSC and LSSC populations contained SSEA3-positive and -negative cells. As differential expression of SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24 can delineate several PSC subsets that co-exist in variable percentages in the different PSC lines 8, we assessed the association of these different markers and the SSC parameter (Figures 7A, B and C). Like for SSEA3 alone, there was no overlap of the SSEA3, SSEA4, TRA-1-81, TRA-1-60 and CD24 compartments with the HSSC and LSSC populations, suggesting that SSC might represent another level of PSC heterogeneity. Moreover, many sub-populations expressed few pluripotency markers.
Figure 7. Plural pluripotent stem cell compartments.
The cell subsets identified based on the expression of the cell surface pluripotency markers TRA-1-60, TRA-1-81, SEEA3, SEEA4 and CD24 in the in HSSC and LSSC subpopulations of M4C7 (hiPSCs) (A) and HUES1 (B) and HD291 (ES) (C) cells are detailed as bar plots. Results were plotted as tree diagrams using the KALUSA software. Each subset is represented by a different color. This analysis shows that there is no superimposition of the compartments defined by the expression of cell surface markers and SSC intensity. Data are the mean ± SEM of three independent experiments. (D) Schematic representation of the stem cell compartments defined by cell surface pluripotency markers and SSC, the permanent exchange between these compartments and the preferential exit toward apoptosis for HSSC cells and toward differentiation for LSSC cells.
Discussion
The molecular definition of stem cells is an old goal that has been only partially met. Single adult stem cells cannot be isolated and they can only be enriched based on specific factors the expression of which has been reported to be heterogeneous [5–7,16]. This is also true for PSCs. For instance, SSEA3 is a cell surface marker strongly associated with pluripotency, but a PSC colony will contain both SSEA3-positive and -negative cells. Although SSEA3-negative cells are less prone to self-renewal and appear primed for differentiation, they can still generate SSEA3-positive cells in vitro and are pluripotent [9,12]. Our work reveals that also SSC, a basic parameter that is routinely monitored when cells are analyzed by flow cytometry, is very heterogeneous in human PSCs and many cells have very high or very low SSC values. SSC heterogeneity was comparable in different human PSC lines (three ES and one hiPS cell line), although hiPSCs are often thought to be more heterogeneous than ES cells and thus less stable [30]. As a matter of fact, expression of cell surface pluripotency markers in the hiPS cell line M4C7 appeared to be less variable than in the three ES cell lines analyzed.
The wide SSC distribution could not be explained by major morphological or phenotypically differences, as cycling and non-cycling cells, apoptotic and non-apoptotic cells, SSEA3-positive and -negative cells were detected in both HSSC and LSSC populations, although some variations were observed. Neither it was explained by culture biases, such as the use of the Rock inhibitor Y27632. Indeed, cell lines that were cultured using mechanical passaging (HD83 cells) or semi-enzymatic passaging (HUES1cells) and that thus were not exposed to this inhibitor were also highly heterogeneous (see Figure 2D and 2B). The only consistent difference was the higher number of round-shaped mitochondria near the nucleus and the 2.1-fold increase in mitochondrial content in HSSC in comparison to LSSC cells, suggesting that HSSC cells have a higher mitochondrial energy production than LSSC cells. This morphological feature could be related to random segregation of mitochondria at cell division, as previously suggested for the cell-to-cell variability in mitochondrial mass observed in a the Hela cell line[31].
We then show that the size of the HSSC population diminishes dramatically and rapidly (day 5) following directed PSC differentiation or serum deprivation. SSC could thus be used as a simple, rapid and inexpensive tool to follow loss of pluripotency during cell culture or differentiation of PSCs, although it cannot substitute the analysis of pluripotency marker expression or functional assays, such as teratoma formation.
Although LSSC cells express lower amounts of pluripotency markers (suggesting that differentiation takes place essentially in this compartment), both HSSC and LSSC populations can form new PSC colonies, which contain HSSC and LSSC cells, and teratomas. In the model proposed by Enver et al, the SSEA3-positive ES compartment gives rise also to SSEA3-negative cells that can revert to the SSEA3-positive compartment or retain their multi-lineage differentiation potential and then differentiate [9,12]. Here we show that both HSSC and LSSC compartments contain SSEA3-positive and -negative cells (Figure 7C) and that LSSC cells can generate HSSC cells, albeit at a lower rate than HSSC cells. PSCs might thus fluctuate between compartments, as suggested by our findings (see Figure 4) and similar experiments based on sorting for SSEA3 [9]. Overall, there might be a permanent shuffling between compartments, with a preferential exit toward apoptosis for cells with the highest SSC and toward differentiation for cells with the lowest SSC values (Figure 7D).
Given the well-known PSC heterogeneity for some specific markers, it is surprising that SSC heterogeneity was never previously reported. As PSC heterogeneity may be a fundamental feature of undifferentiated cells [32] and especially of pluripotent stem cells [16], their high and variable SSC values may have important biological implications, as suggested by the higher mitochondrial content and clonogenic self-renewal capacity of the HSSC population.
Acknowledgments
Supported by grants from the Association Française contre les Myopathies (AFM), the Région Languedoc-Roussillon (Chercheur d’Avenir 09-13198 01) and the Agence Nationale de la Recherche (ANR-07-BLAN-0076-01, ANR-08-BIOT-012-02). We thank E. Andermarcher for critical reading of this manuscript. We thank Montpellier RIO Imaging (MRI) for access to the flow cytometry department, the Centre de Ressources en Imagerie Cellulaire (CRIC) for TEM and the Plateau Animalerie Inserm U583, Institut des Neurosciences de Montpellier (INM) for mice husbandry. The authors are grateful to Chantal Cazevieille and Cécile Sanchez for their technical assistance and interpreting TEM images. We thank the “Réseau d’Histologie Expérimentale de Montpellier (RHEM)”histology facility for processing our teratoma samples and Pascal Roger for the interpretation of the Hematoxylin-Eosin-Safran stained sections.
Abbreviations list
- hiPSCs
Human-induced pluripotent stem cells
- hFF
Human foreskin fibroblasts
- PSCs
Pluripotent stem cells
- hESCs
Human embryonic stem cells
- MEFs
Murine embryonic fibroblasts
- SSC
Side scatter
- FSC
Forward scatter
- HSSC
High side scatter
- LSSC
Low side scatter
Footnotes
Author Disclosure Statement
All authors disclose any potential conflicts of interest.
References
- 1.Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22:1987–97. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–4. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 3.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–85. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Halley JD, Smith-Miles K, Winkler DA, Kalkan T, Huang S, Smith A. Self-organizing circuitry and emergent computation in mouse embryonic stem cells. Stem Cell Res. 2012;8:324–33. doi: 10.1016/j.scr.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Gu B, Zhang J, Chen Q, Tao B, Wang W, Zhou Y, Chen L, Liu Y, Zhang M. Aire regulates the expression of differentiation-associated genes and self-renewal of embryonic stem cells. Biochem Biophys Res Commun. 2010;394:418–423. doi: 10.1016/j.bbrc.2010.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu B, Zhang J, Wang W, Mo L, Zhou Y, Chen L, Liu Y, Zhang M. Global expression of cell surface proteins in embryonic stem cells. PLoS ONE. 2010;5:e15795. doi: 10.1371/journal.pone.0015795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu B, Zhang J, Wu Y, Zhang X, Tan Z, Lin Y, Huang X, Chen L, Yao K, Zhang M. Proteomic analyses reveal common promiscuous patterns of cell surface proteins on human embryonic stem cells and sperms. PLoS ONE. 2011;6:e19386. doi: 10.1371/journal.pone.0019386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui L, Johkura K, Yue F, Ogiwara N, Okouchi Y, Asanuma K, Sasaki K. Spatial distribution and initial changes of SSEA-1 and other cell adhesion-related molecules on mouse embryonic stem cells before and during differentiation. J Histochem Cytochem. 2004;52:1447–57. doi: 10.1369/jhc.3A6241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Thykjaer T, Maltby E, Smith K, Abu Dawud R, Jones M, Matin M, Gokhale P, Draper J, Andrews PW. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–40. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 10.Hong SH, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS, Bhatia M. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell. 2011;9:24–36. doi: 10.1016/j.stem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez JM, Gerbal-Chaloin S, Milhavet O, Qiang B, Becker F, Assou S, Lemaitre JM, Hamamah S, De Vos J. Brief report: benchmarking human pluripotent stem cell markers during differentiation into the three germ layers unveils a striking heterogeneity: all markers are not equal. Stem Cells. 2011;29:1469–74. doi: 10.1002/stem.681. [DOI] [PubMed] [Google Scholar]
- 12.Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods. 2006;3:807–15. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanasijevic B, Dai B, Ezashi T, Livingston K, Roberts RM, Rasmussen TP. Progressive accumulation of epigenetic heterogeneity during human ES cell culture. Epigenetics. 2009;4:330–8. doi: 10.4161/epi.4.5.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS One. 2009;4:e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez Arias A, Brickman JM. Gene expression heterogeneities in embryonic stem cell populations: origin and function. Curr Opin Cell Biol. 2011;23:650–6. doi: 10.1016/j.ceb.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–73. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho MS, Fryga A, Laslett AL. Flow cytometric analysis of human pluripotent stem cells. Methods Mol Biol. 2011;767:221–30. doi: 10.1007/978-1-61779-201-4_16. [DOI] [PubMed] [Google Scholar]
- 19.Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, Melton DA. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–6. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 20.Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Vos J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 22.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–41. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 23.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klimchenko O, Mori M, Distefano A, Langlois T, Larbret F, Lecluse Y, Feraud O, Vainchenker W, Norol F, Debili N. A common bipotent progenitor generates the erythroid and megakaryocyte lineages in embryonic stem cell-derived primitive hematopoiesis. Blood. 2009;114:1506–17. doi: 10.1182/blood-2008-09-178863. [DOI] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, Spidlen J, Strain E, Gentleman R. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assou S, Le Carrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot JP, Gasca S, Hovatta O, Hamamah S, Klein B, De Vos J. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–73. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lensch MW, Schlaeger TM, Zon LI, Daley GQ. Teratoma formation assays with human embryonic stem cells: a rationale for one type of human-animal chimera. Cell Stem Cell. 2007;1:253–8. doi: 10.1016/j.stem.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Prokhorova TA, Harkness LM, Frandsen U, Ditzel N, Schroder HD, Burns JS, Kassem M. Teratoma formation by human embryonic stem cells is site dependent and enhanced by the presence of Matrigel. Stem Cells Dev. 2009;18:47–54. doi: 10.1089/scd.2007.0266. [DOI] [PubMed] [Google Scholar]
- 30.Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S, Jan T, Wilson KD, Leong D, Rosenberg J, Yao M, Robbins RC, Wu JC. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest. 2011;121:1217–21. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.das Neves RP, Jones NS, Andreu L, Gupta R, Enver T, Iborra FJ. Connecting variability in global transcription rate to mitochondrial variability. PLoS Biol. 2010;8:e1000560. doi: 10.1371/journal.pbio.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–7. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]