Abstract
While many vertebrate transcription factor (TF) families are conserved, the C2H2 zinc finger (ZNF) family stands out as a notable exception. In particular, novel ZNF gene types have arisen, duplicated, and diverged independently throughout evolution to yield many lineage-specific TF genes. This evolutionary dynamic not only raises many intriguing questions but also severely complicates identification of those ZNF genes that remain functionally conserved. To address this problem, we searched for vertebrate “DNA binding orthologs” by mining ZNF loci from eight sequenced genomes and then aligning the patterns of DNA-binding amino acids, or “fingerprints,” extracted from the encoded ZNF motifs. Using this approach, we found hundreds of lineage-specific genes in each species and also hundreds of orthologous groups. Most groups of orthologs displayed some degree of fingerprint divergence between species, but 174 groups showed fingerprint patterns that have been very rigidly conserved. Focusing on the dynamic KRAB-ZNF subfamily—including nearly 400 human genes thought to possess potent KRAB-mediated epigenetic silencing activities—we found only three genes conserved between mammals and nonmammalian groups. These three genes, members of an ancient familial cluster, encode an unusual KRAB domain that functions as a transcriptional activator. Evolutionary analysis confirms the ancient provenance of this activating KRAB and reveals the independent expansion of KRAB-ZNFs in every vertebrate lineage. Most human ZNF genes, from the most deeply conserved to the primate-specific genes, are highly expressed in immune and reproductive tissues, indicating that they have been enlisted to regulate evolutionarily divergent biological traits.
Keywords: zinc finger genes, transcription factor evolution, vertebrate gene families
Introduction
Most eukaryotic transcription factors (TFs) are members of ancient protein families, and many are conserved across divergent evolutionary lineages. However, this latter generalization does not hold universally true, and the C2H2 zinc finger (ZNF) family stands out as a particularly significant exception. At several points in evolutionary history, novel gene types have arisen to encode proteins in which DNA-binding ZNF motifs are tethered to different types of chromatin-interacting or “effector” domains. Some of these innovations have subsequently been expanded by duplication into large cohorts of lineage-specific genes (Collins et al. 2001; Stubbs et al. 2011).
ZNF proteins that function as TFs typically contain an array of two or more tandemly arranged C2H2 motifs; each ZNF in such polydactyl fingered or “polydactyl” proteins can bind three adjacent nucleotides at target sites with amino acids in positions −1, 2, 3, and 6 in the alpha helical region of each motif playing the most critical DNA-recognition roles (Pavletich and Pabo 1991; Kim and Berg 1996). Adjacent motifs influence each other’s DNA binding, creating a complex “code” that links the pattern of DNA-binding amino acids in a protein to target-site preferences in DNA (Isalan et al. 1997; Wolfe et al. 2000). In the following discussion, we will refer to the pattern of DNA-binding amino acids within a polydactyl ZNF array as a protein’s “fingerprint.” It stands to reason that ZNF proteins with similar fingerprints should recognize similar DNA sequences, while even closely related proteins with divergent fingerprints should preferentially interact with different recognition sites in DNA. An extreme example of this type of fingerprint divergence is provided by PRDM9, an ancient protein that binds hotspots of meiotic recombination. PRDM9 orthologs encode proteins that are similar in overall sequence, but that nevertheless define hotspots uniquely in every species using ZNF arrays that have been positively selected for fingerprint divergence (Oliver et al. 2009; Baudat et al. 2010; Berg et al. 2010; Myers et al. 2010; Parvanov et al. 2010).
Interestingly, although PRDM9 is unique in invertebrate genomes, this single gene’s descendants have expanded to form the largest ZNF subfamily in mammalian genomes (Birtle and Ponting 2006). The human genome encodes hundreds of these so-called KRAB-ZNF genes, encoding proteins in which arrays of tandem ZNF motifs are tethered to an N-terminal effector domain called the Krüppel-associated box or KRAB (Constantinou-Deltas et al. 1992; Bellefroid et al. 1993; Huntley et al. 2006). The canonically structured mammalian “KRAB A” domain interacts with a universal cofactor, KAP1, which recruits histone deacetylase complexes to the ZNF-binding sites, and KRAB-ZNF proteins are thus thought to act as potent transcriptional repressors (Margolin et al. 1994; Pengue et al. 1994; Witzgall et al. 1994; Vissing et al. 1995). However, the vertebrate roots of the KRAB-ZNF family in particular, and of polydactyl ZNF genes in general, remain somewhat mysterious. For example, it is not known which human polydactyl proteins are conserved in structure and function in other vertebrate species or which among the otherwise conserved proteins, like PRDM9, might have been selected especially for DNA-binding diversity.
To address these questions, we used methods that we applied previously to identify mouse, dog, and primate genes (Huntley et al. 2006; Nowick et al. 2011) to collect consistent sets of polydactyl ZNF gene models from the opossum, chicken, zebra finch, lizard, frog, and updated mouse genomes. From these models, we extracted and aligned fingerprint patterns to identify proteins with similar or divergent DNA binding capacities. We identified hundreds of polydactyl ZNF loci in every genome including more than 100 predicted novel mouse genes, but surprisingly few encoding proteins with fingerprint patterns that are conserved between eutherians and other evolutionary groups. Notably, the subset that is deeply conserved includes only three KRAB-ZNF genes, all of which map to a single familial cluster. These ancient genes share certain features that are unusual in mammalian genomes, including a noncanonical KRAB domain sequence that does not bind KAP1 and functions as a transcriptional activator (Okumura et al. 1997; Conroy et al. 2002). These and other findings suggest a history in which the KRAB-ZNF proteins expanded and diverged independently in every vertebrate lineage including amphibians, where they expanded without KAP1-interacting capabilities, very possibly as activating TFs.
The rigid preservation of DNA-binding domains suggests that the conserved polydactyl ZNF genes have been stably integrated into essential regulatory relationships. Strikingly, however, the most deeply conserved genes are expressed at highest levels in human tissues that are the least conserved in structure and function, including placenta. Our results identify hundreds of novel polydactyl ZNF genes of both deeply conserved and lineage-specific types, providing new clues to the history and root functions of this dynamic TF family.
Materials and Methods
Genome Searches and Initial Data Analysis
Human KRAB-A, KRAB-B, KRAB-b, KRAB-C, KRAB-L, BTB/POZ, SCAN, and FINGER HMM matrices were retrieved from previous analysis (Huntley et al. 2006). Chicken KRAB-A-containing protein sequences from NCBI (sequences were trimmed according to HMMER result to retrieve KRAB-A sequences) and PFAM KRAB, BTB/POZ, and SCAN sequences were also retrieved. Sequence alignments for each motif type were generated using CLUSTALW 2.0.10 (Larkin et al. 2007) and submitted to the HMMER (hmmer-2.3.2, http://hmmer.janelia.org/, last accessed October 2013) profile HMM matrix building tool “hmmbuild” to generate matrices (and processed by “hmmcalibrate”). These matrices were used by the HMMER search program to identify all putative motif matches in a full six-frame translation of sequences from mouse (Mus musculus, mm9 genome build), opossum (Monodelphis domestica, monDom5), chicken (Gallus gallus, galGal3), zebra finch (Taeniopygia guttata, taeGut1), lizard (Anolis carolinensis, AnoCar2.0), and frog (Xenopus tropicalis, xenTro3) genomes from the UCSC genome browser (Meyer et al. 2013). An e-value cutoff of 0.001 was used for these analyses. For overlapping matches, the match with lowest e-value was retained.
Gene Model Construction
Gene model structures were constructed by the following procedure:
-
1.
Motifs were grouped if no genome gap was found between them (bridged gaps were ignored because the order and orientation of either side of the gap is known in this case). We included same-strand HMMER motifs closer than 30 kb from each other into gene models. For ZNF motifs, this threshold was more stringent, requiring 1 kb separation or less. Effector motifs (BTB/POZ, SCAN, or KRAB) were not included in gene models if they were near, but 3′ of, clusters of ZNFs.
For each ZNF cluster, we considered all possible combinations of nearby upstream effectors, including the six main KRAB subtypes (Looman et al. 2002a), as well as SCAN and BTB/POZ, to generate transcript models.
-
2.
We extended exon boundaries to maintain ORFs and include canonical intron splice sites (GT-AG, AT-AG, and GC-AG) identified the nearest start and stop codons in each model. We made sure no stop codon was identified in frame.
-
3.
Models were compared with existing ENSEMBL gene models and refined. Specifically, fragmented models were fused together if they were included in the same ENSEMBL gene model.
Finally, gene models encoding at least two intact tandem ZNF motifs were retained. For each gene, only the longest transcript model was retained. The requirement for an ORF encoding at least two ZNF motifs may possibly excluded some bona fide polydactyl ZNF genes in the poorly assembled draft genome sequences. Genes with very short ZNF arrays (encoding 2–3 fingers) would be particularly likely to be missed. In addition, assembly and sequence accuracy issues may have contributed to creation of artificially truncated ZNF arrays in some cases. For this reason, we cannot definitively argue for the absence of any gene, or motifs within specific genes, in a particular species, and both the models and the counts of predicted genes can only be considered as conservative estimates.
ZNF Fingerprint Extraction
For mouse, opossum, chicken, zebra finch, lizard, and frog ZNF motif sequences were retrieved based on HMMER search results. Next, they were aligned with a canonical ZNF sequence (“YECSECGKSFSRSSHLIVHQRIHTGERP,” a Finch C2H2 ZNF HMMER hit with e-value 5.8e−21). Amino acids immediately preceding the alpha-helix (position -1) and the second, third, sixth amino acids within the helix (before the first Histidine) residues were retrieved as the fingerprint (e.g., the fingerprint is “RSHV” for the standard finger above) (Elrod-Erickson et al. 1998). Previously established human and Dog ZNF gene models (Huntley et al. 2006; Nowick et al. 2011) were used to extract fingerprint data for comparison.
Fingerprint Alignments
Pairwise alignment of the 4-aa fingerprint sequences from genes from the eight species (frog, lizard, zebra finch, chicken, opossum, dog, mouse, and human) was carried out using the Global Alignment Algorithm with gap penalty = 1, mismatch penalty = 1, match penalty = −2, similar penalty = −0.5 (2 out of 3 or 3 out of 4 positions in the Fingerprints are the same), closematch penalty = −1 (only the second residue is different). The scores were first normalized by sequence length, then were shifted as Score (x,y) = Score (x,y) − Score (x,x) so that the score is always nonnegative and equal to zero if and only if two fingerprint sequences are identical. The normalized scores were used as a distance matrix and served as input for an agglomerative hierarchical clustering. The clustering was done in R using average linkage criteria (http://stat.ethz.ch/R-manual/R-patched/library/stats/html/hclust.html, last accessed February 27, 2014). The genes were grouped by cutting the clustering tree at a height of 2 (height of 1–5 are tried and 2 is chosen by considering both the discrimination power and stringency). Then, for each group, a multiple alignment using UPGMA guide tree was generated. After an initial alignment, we identified many conserved groups without a human ortholog included, even though human orthologs are well known to exist. These human genes, which were missed in our previous study due to the stringent requirement for at least three tandem ZNF-encoding motifs in the sequence in those genome scans, were retrieved from NCBI for a second round of fingerprint alignments. We included all ZNF-containing isoforms recorded for those proteins, so that many are represented by multiple fingerprint patterns in supplementary table S2, Supplementary Material online. For final counts of gene and ortholog numbers, we included only one isoform, encoding the longest protein, for each gene.
Tree Construction and Display
The tree of KRAB A motifs was generated using PhyML by the NNI search method, with SH-like branch support (Guindon et al. 2010) which is of the range 0–1.0: the larger score is more significant. Tree Graphs were generated using Python ete2 package (Huerta-Cepas et al. 2010). To obtain information regarding the history of all gene-linked human KRAB A domains, we used all human KRAB A sequences identified in previous studies regardless of C2H2 motif association (Huntley et al. 2006). For all other species, we used only KRAB A domains included in ZNF gene models that are described here.
RNA-seq and Cluster Analysis
RNA-seq expression data, including data from the public BodyMap project (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513/, last accessed February 27, 2014) and supplemented with published expression data from amnion, chorion, and decidua of human term placenta, were kindly provided in the form of processed uniquely mapped log2 fragment per million reads (FPKM) values by Dr Yi Xing (University of California, Los Angeles). The processing steps and sources of raw data are cited in the Xing laboratory’s recent article (Kim et al. 2012). We removed genes with FPKM values that were not at least 1 in any tissue and used Cluster 3.0 Software (de Hoon et al. 2004) to generate the heat maps from data centered to the median of each gene’s expression levels for Heirarchical clustering with Average Linkage.
RNA Preparation and Quantitative PCR
Animal work described in this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Illinois (Animal Assurance Number: A3118-01; approved IACUC protocol number 11030). RNA was prepared from snap-frozen dissected mouse embryos collected from timed matings at various stages of development and purified using Trizol (Invitrogen) extraction. cDNA was generated using Superscript III Reverse Transcriptase (Invitrogen, CA) according to manufacturer’s instructions. Interexonic qRT-PCR primers for Zfp282 (forward: 5′-TGACTGCAGACACAGGAACAG-3′, reverse: 5′-CTCTGCCAAATCCTGCTGGT-3′) and Zfp777 (forward: 5′-TTCCCAAGGTTCCTGTCACATTC-3′, reverse: 5′-CGTCTCACCCTCCTCAGAATC-3′) were synthesized from IDT (Coralville, Iowa). Reaction was carried out using Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) on the ABI7900HT system. Expression levels were calculated relative to the average expression of two housekeeping genes, Succinate dehydrogenase complex, sub unit A (Sdha: accession number BC011301) and Tyrosine 3-monooxygenase/tryptophan 5′monooxygenase activation protein, zeta polypeptide (Ywhaz, NM011740), as described (Livak and Schmittgen 2001; Veazey and Golding 2011).
In Situ Hybridization and Immunohistochemistry
Mouse embryos, placenta, and yolk sac were isolated at embryonic day 12.5 (E12.5) or E16.5 after timed matings. Chicken embryos were collected after incubation of freshly fertilized eggs at 37 °C. Dissected embryos were fixed in fresh 4% paraformaldehyde (PFA) and embedded in paraffin. Human normal term placenta was obtained from an anonymous donor at the Carle Foundation Hospital (Urbana, IL) and provided as PFA-fixed, dissected maternal and fetal tissue segments that were subsequently embedded in paraffin. Tissues were cut into 5-µm sections using a Leica RM2155 microtome and Super Plus charged slides.
For mouse in situ hybridization (ISH), probe sequences were generated from sequenced cDNA clones from the IMAGE consortium: mouse LIBEST_005352 clone 9530039E11 (accession number BY722098) and chicken clone LIBEST_011205 CSEQRBN13ChEST197h24 (accession number BU448580). Mouse primers for PCR were as follows: forward, 5′-AGGACAGACCAGAATGCATC-3′ and reverse, 5′-CGAAGCTACTGACAAGGTGT-3′; the chicken probe was generated using primers: forward, 5′-ACAAGACAACGCACAATGCC-3′ and reverse, 5′-TATCTGGAAGACCGTGTTGC-3′. Probe sequences were labeled and hybridized essentially as described (Elso et al. 2013). Immunohistochemistry (IHC) was also carried out essentially as described (Elso et al. 2013). We used primary antibodies: ZNF282 (AVIVA, Rb38361) and ZNF777 (AVIVA, Rb32569) diluted 1:200 (5 μg/ml). The sections were incubated with primary antibodies overnight at 4 °C, then washed and incubated with secondary antibody Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen), 1:200 diluted (1 μg/ml). The results were imaged with a Nanozoomer Scanner (Hamamatsu), Zeiss Apotome Fluorescence microscope, and Zeiss Confocal Microscope LSM 710.
Results
Identification of Polydactyl ZNF Genes in Sequenced Genomes
We used methods based on those described previously (Huntley et al. 2006; Nowick et al. 2011) to identify potential ZNF coding genes in the M. musculus (mouse, mm9 genome build), Mo. domestica (opossum, monDom5), G. gallus (chicken, galGal3), T. guttata (zebra finch, taeGut1), A. carolinensis (lizard, AnoCar2.0), and X. tropicalis (frog, xenTro3) genomes. Of these assemblies, only the mouse genome is finished sequence. With the expectation that many genes could be fragmented in the unfinished genomes, we built gene models requiring only two closely spaced ZNF HMMER matches, rather than three tandem ZNFs as we had in the previous human, dog, and primate genome analysis. We also scanned each genome for HMMER models corresponding to the BTB/POZ, SCAN, and KRAB effector domains and included exons encoding those domains into ZNF gene models where possible as previously described (Huntley et al. 2006; Nowick et al. 2011).
We gathered substantial numbers of polydactyl ZNF-encoding ORFs from every species including members of all subfamilies defined by association with the common known effector domains (table 1). These gene model sets are very likely to include recent pseudogenes; we examined overlap with the other annotated gene sets for additional model support. For the 1,194 mouse polydactyl ZNF models, we identified 799 overlapping with known genes and/or ENSEMBL gene models; the counts of previously annotated mouse and human gene 70 are roughly comparable (table 1).
Table 1.
ZNF Gene Models in Each Subfamily with and without ENSEMBL Model Overlap
| Species | ZNF-Only |
BTB/POZ |
KRAB |
SCAN |
SCAN-KRAB |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| All | ENSEMBL | All | ENSEMBL | All | ENSEMBL | All | ENSEMBL | All | ENSEMBL | |
| Humana | 212 | 212 | 42 | 42 | 366 | 366 | 29 | 29 | 28 | 28 |
| Mouse | 590 | 353 | 40 | 39 | 523 | 370 | 27 | 23 | 14 | 14 |
| Opossum | 868 | 548 | 23 | 23 | 709 | 513 | 0 | 0 | 20 | 19 |
| Chicken | 290 | 219 | 23 | 23 | 47 | 39 | 0 | 0 | 0 | 0 |
| Zebra finch | 1026 | 749 | 23 | 22 | 3 | 2 | 0 | 0 | 0 | 0 |
| Lizard | 723 | 484 | 30 | 28 | 122 | 67 | 89 | 54 | 240 | 126 |
| Frog | 473 | 293 | 34 | 33 | 158 | 112 | 0b | 0 | 0 | 0 |
aManually curated protein-coding loci from Huntley et al. (2006) and including human orthologs of mouse or other species genes from GenBank, as described in the text.
bExcluding the single model also detected by Emerson and Thomas (2011) but considered a false prediction, as discussed in the text.
However, we also found 210 mouse loci with ORFs encoding five or more contiguous ZNF motifs but without known gene assignment or ENSEMBL models; nine predicted genes share fingerprints with annotated human genes and models in other species and are likely orthologs (table 2). Furthermore, 131 of these novel mouse models overlap mouse EST sequences, most of which are derived from oocytes, preimplantation embryos, or dissected tissues from midgestation embryonic stages (supplementary table S1, Supplementary Material online; examples in tables 2 and 3). Many of the unknown genes are found in genomic clusters; some but not all of these clusters also include known genes. As EST collections from such tissues and cell types are relatively rare, the fact that EST overlaps with many of the mouse models were found only for these tissues is even more notable. Excluding these unannotated mouse genes, the counts of mouse genes in each ZNF subfamily are roughly similar to those in the human genome; if the novel models are taken into account, the number of KRAB-ZNF genes would be substantially higher in mouse than in the human genome (table 1).
Table 2.
Predicted Novel Mouse Genes with Excellent Fingerprint Match in Other Species
| Model | Conserved ina | Human Match | Type | Example EST/mRNA (accession number)b | Example EST Source(s) |
|---|---|---|---|---|---|
| ZF01023_1 | Md, Hs | GLI4 | Zx5 | AK084954 | Whole embryo; E14.5 haematopoietic |
| ZF04524_2 | Cl, Hs | ZNF471 | ABZx14 | M36516 | Oocyte; embryo eye |
| ZF02332_2 | Cl, Hs | ZNF582 | ABZx9 | BB619218; BU054342 | E8 whole; E12.5 brain |
| ZF02433_2 | Cl, Hs | ZNF570 | ABZx9 | AK138949; CJ048012 | Aorta; 11d pregnant uterus |
| ZF02869_1 | Cl, Hs | ZNF660 | Zx10 | BB193415; CB524555 | Spinal cord; E12.5 brain |
| ZF04379_1 | Gg, Md, Cl, Hs | ZNF853 | Zx5 | BG277278 | Maxillary process |
| ZF02438_1 | Cl, Hs | ZNF567 | AZx13 | None | No EST |
| ZF02320_1 | Cl, Hs | ZNF331 | Zx8 | None | No EST |
| ZF05506_1 | Md, Cl, Hs | ZNF16 | Zx9 | None | No EST |
aGg, chicken; Md, opossum; Cl, dog; Hs, human.
bOverlapping ESTs; only example ESTs are listed, other ESTs overlap with most models.
Table 3.
Predicted Novel Mouse Genes That Are Not Conserved in Other Species (Selected Examples)
| Example Model | Cluster Location | No. Clustered Genes; Knowna | Type | Example EST | EST Sources |
|---|---|---|---|---|---|
| ZF00123_1 | Chr1:119-121 Mb | 18 genes; none known | Zx17 | AK139669 | 2-cell embryo |
| ZF00134_1 | Zx16 | DV654250 | Oocyte | ||
| ZF00304_2 | Chr10:81 Mb | 20 genes; Zfp873 known | ABZx17 | CK635639 | E9.5-10.5 upper head |
| ZF00313_2 | ABZx16 | CF725361 | Midgestation embryo eye | ||
| ZF00529_2 | Chr12:18-25 Mb | 51 genes; none known | ABZx12 | BU519096; CJ049410 | Undifferentiated limb; E13 testis |
| ZF00537_2 | ABZx14 | BB452393 | E12 spinal ganglion | ||
| ZF00548_2 | ABZx18 | AV579126 | ES cells | ||
| ZF00551_2 | ABZx15 | BQ551390 | Mixed adult tissue | ||
| ZF06013_2 | ABZx13 | BM201758; BF714015 | E7.5 whole embryo; E10.5 branchial arches | ||
| ZF04218_1 | Chr6:130.4–131.2 Mb | 14 genes; none known | Zx17 | AU017585; DV645475 | 2-cell embryo, occyte |
| ZF04215_1 | Zx11 | AK136154; DV649857 | In vitro fertilized egg, oocyte | ||
| ZF04205_1 | Zx17 | BX513671 | 2-cell embryo | ||
| ZF04202_1 | Zx13 | DV65065 | Oocyte | ||
| ZF04199_1 | Zx14 | CA559522 | Unfertilized egg | ||
| ZF04196_1 | Zx13 | BG080473 | Mixed tissue |
aOne example of a Refseq ZNF gene is shown here for each cluster although several cluster members may be known.
Identifying “DNA-Binding Orthologs” for Human ZNF Genes
To identify ZNF genes encoding proteins with conserved DNA binding preferences, we extracted the ZNF DNA-contacting residues from translated gene models, including the dog and curated human gene models from our previous study (Huntley et al. 2006). We then carried out a global alignment of these fingerprint sequences from all species (see Materials and Methods). After an initial alignment, we found a number of deeply conserved protein groups (e.g., conserved fingerprints in mouse and nonmammalian species) that did not include a human protein member. Most of these cases involved known human genes encoding only one or two ZNF tandem domains in any single exon; these genes would have been missed with our previous approach. To include the missing human proteins in this analysis, we collected the human protein sequences from GenBank, extracted fingerprint patterns, and repeated the global alignments for a final set (supplementary table S2, Supplementary Material online).
In addition to fingerprint alignments, we used reciprocal best Blast, a standard method for ortholog identification used in most published studies (Huntley et al. 2006; Thomas and Schneider 2011; Corsinotti et al. 2013). Reciprocal Blast was the only way to positively identify orthologs in many large groups, like the SP1 and KLF families, which include large numbers of proteins with identical fingerprints (supplementary table S2, Supplementary Material online). Fingerprint alignments also clustered together groups of paralogs with similar fingers including lineage-specific duplicates; fingerprint alignments could not always resolve these groups.
We consolidated and manually curated the results from Blast and fingerprint matches to identify groups of orthologs to the human protein set (supplementary table S3, Supplementary Material online). Using these combined data, each human protein was classified as 1) primate-specific (detected in human only); 2) shared by eutherians (human and dog and/or mouse); 3) shared by mammals (at least human and opossum); 4) shared by amniotes (at least human and one bird or lizard); and 5) shared by tetrapods (at least human and frog) (table 4). Of special note, nine of the unannotated mouse models we discovered (discussed earlier) encode predicted proteins with fingerprint patterns that match annotated genes in human and other species extremely well. For example, four of the novel mouse models are clearly conserved in dog and human, and one model, matching human gene ZNF853, detects clear orthologs in dog, opossum, and both species of birds (tables 2 and 3). We counted genes (including PRDM9) with excellent, unique best-Blast matches in other species but no fingerprint match, as conserved genes with divergent ZNFs.
Table 4.
Numbers of Human ZNF Genes Conserved to Each Lineagea
| Gene Type | Te | Amn | Ma | Eu | P | Total |
|---|---|---|---|---|---|---|
| All | 175 | 18 | 63 | 213 | 208 | 677 |
| ZNF-only | 136 | 11 | 19 | 28 | 18 | 212 |
| BTB/POZ | 38 | 4 | 0 | 0 | 0 | 42 |
| SCAN | 0 | 0 | 10 | 15 | 4 | 29 |
| SCANKRAB | 0 | 0 | 17 | 10 | 1 | 28 |
| KRAB | 1 | 3 | 17 | 160 | 185 | 366 |
aTe, ortholog found in tetrapods; Amn, amniotes; Ma, mammals; Eu, Eutheria; P, primates only.
Here, we should note that these classifications should be considered as a minimal depth of conservation, as orthologs might be found by scanning additional species, different evolutionary groups, or finished genomes as they become available. As an example, several of the human genes conserved in frog, such as ZBTB16 (aka PLZF1), also recognize orthologs in Drosophila. Nevertheless, the classifications provide a solid overall view of family and subfamily history in vertebrate lineages.
As summarized in table 4 and consistent with previous estimates (Huntley et al. 2006), the KRAB-ZNF family contributes the vast majority of ZNF genes that are exclusive to eutherians or to primate lineages. In contrast, nearly all the human genes that are functionally conserved across amniotes or tetrapods are members of the ZNF-only and BTB/POZ-ZNF subfamilies. We also found SCAN-ZNF and KRAB-ZNF genes in most species, although no SCAN-ZNF and very few KRAB-ZNF proteins were conserved across vertebrate groups. Findings from each subfamily are highlighted further in following sections.
A Small Number of Deep Vertebrate Roots for the Human KRAB-ZNF Family
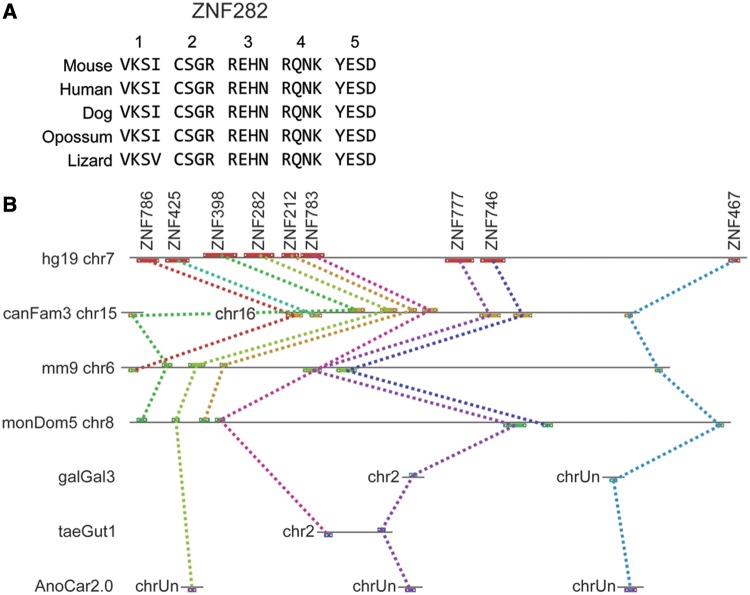
Of the 366 human protein-coding KRAB-ZNF (not including the SCAN-KRAB-ZNF genes, which are discussed later), only 181 genes (49.5%) found a convincing and unique (1:1) fingerprint match in one or both of the other eutherians; 185 genes were classified as primate-specific (table 4). Only 17 human KRAB-ZNF genes were found with fingerprint sequence conserved between eutherians and opossum. Looking for orthologs in nonmammalian species, we found only three human KRAB-ZNF proteins, ZNF282, ZNF777, and ZNF783, that have orthologous proteins in nonmammalian amniote groups; orthology is recognized both by overall protein sequence and fingerprint pattern similarities. In particular, the fingerprints of human, bird, and lizard ZNF777 and ZNF282 proteins are strikingly similar, as illustrated by the alignment of ZNF282 orthologs (fig. 1A). In contrast, while the lizard ortholog of ZNF783 is clearly similar in overall protein sequence and was identified as the best match to mammalian ZNF783 in fingerprint alignments as well, two ZNF motifs are deleted in our lizard gene model compared with the mammalian orthologs (supplementary table S2, Supplementary Material online).
Fig. 1.—
ZNF282 and neighboring genes are members of a deeply conserved gene cluster. (A) Fingerprint alignment of ZNF282 orthologs in mammalian and nonmammalian vertebrate species shows the rigid conservation of DNA-binding amino acids in the ZNFs of this gene. Each column contains the DNA-binding amino acids (positions 6, 3, 2, and −1 relative to the alpha helix) and rows correspond to the sequence in each species. ZNFs are numbered at top in N- to C-terminal orientation in the protein. (B) Maps of the gene cluster including ZNF282, ZNF777, and ZNF783 in human chromosome 7 (hg19 sequence build, chr7), dog (canfFam3) chr15 and chr16, mouse (mm9) chr6, opossum (monDom5) chr8, and the fragmented genomes of chicken (galGal3), zebra finch (taeGut1), and lizard (AnoCar 2.0). Colored dotted lines connect orthologs in the different species. chrUn is assigned to genes in fragmented assemblies that have not been assigned to specific chromosomes in some species. ZNF786, ZNF425, ZNF398, ZNF212, and ZNF746 are HUB- and KRAB-containing ZNF genes that are closely related to ZNF282, ZNF777, and ZNF783. ZNF467, a deeply conserved ZNF-only gene that is also found clustered with the KRAB-ZNF genes in mammals, is also shown.
Notably, ZNF282, ZNF777, and ZNF783 are clustered as neighbors in the distal end of human chromosome 7 (chr7; cytogenetic band 7q36.1; fig. 1B). These three genes and their cluster neighbors, ZNF398, ZNF212, ZNF746, and ZNF767, correspond to 7 of the 17 total KRAB-ZNF genes that are conserved between human and opossum. The orthologous opossum genes are also clustered in chr8 and although the bird and lizard genomes are mostly too fragmented to assess clustering, ZNF777 and ZNF783 are also found clustered in zebra finch chr2 (fig. 1B).
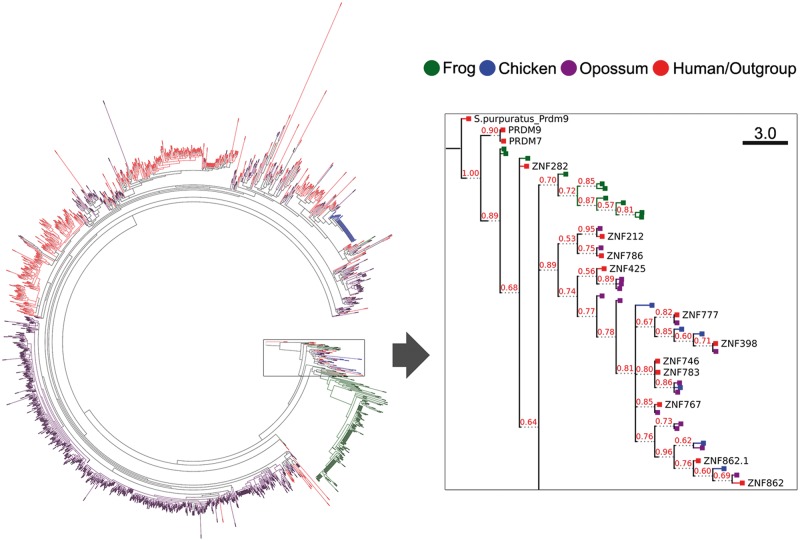
We predicted 158 intact KRAB-ZNF genes in the frog genome including 112 that overlap with ENSEMBL gene models (table 1 and supplementary table S1, Supplementary Material online). However, none of these or other predicted frog ZNF genes had a significant fingerprint match with a mammalian KRAB-ZNF gene. To test another measure of relatedness, we examined KRAB-A sequence similarity, an approach we have used successfully to assess KRAB-ZNF subfamily relationships in the past (Dehal et al. 2001; Huntley et al. 2006). We aligned all ZNF-associated KRAB-A sequences from human, opossum, chicken, and frog and created a maximum likelihood tree of these sequences. We rooted this diverse collection of KRAB-A domains on the KRAB domain of the sea urchin (Strongylocentrotus purpuratus) PRDM9 protein to gain a more global view of lineage-specific expansions and distant relationships between subfamily groups (fig. 2A).
Fig. 2.—
Evolutionary tree showing relationships between KRAB domain sequences from human, opossum, chicken, and frog. (A) A circular tree showing clustering of sequences including all ZNF-associated KRAB domains from human (red), opossum (purple), chicken (blue), and frog (green) KRAB-ZNF gene models (see Materials and Methods). The boxed region is expanded and shown as a rectangular tree in (B). The KRAB domain of the Strongylocentrotus purpuratus PRDM9 protein was included to root the tree. The human ZNF282-related cluster dominates this branch of the tree which also includes isoforms of ZNF862, a KRAB-containing TTF-finger ZNF gene that also maps to the cluster region in human chromosome 7q36.1.
The tree yielded several interesting results that together shed new light on the early history of KRAB-ZNF subfamily. In particular, after a branch leading to human PRDM9 and its primate-specific paralog, PRDM7, the second branch to emerge from the S. purpuratus PRDM9 root includes KRAB domains from human ZNF282, ZNF777, ZNF783, and other members of human chr7q36.1 cluster (fig. 2B). Curiously, this clade also includes the KRAB domain of ZNF862, which contains TTF-type fingers rather than the C2H2 type and is unrelated to the ZNF282 family but nevertheless clusters with them in human, other eutherians, and in the opossum genomes (not shown).
KRAB domains from different species were otherwise largely segregated in the tree with only a few clades including sequences from more than one lineage. As expected, one large clade mostly comprised human KRAB sequences (red bars in fig. 3A), but we also identified one large clade comprised only of genes from opossum (429 opossum genes; purple bars) and another isolated clade from frog (142 genes; green bars). These groups suggest that the KRAB-ZNF genes we observed in those species are derived from lineage-specific expansions in amphibians and marsupials that are very similar to those that have been documented in detail for eutherians.
Fig. 3.—
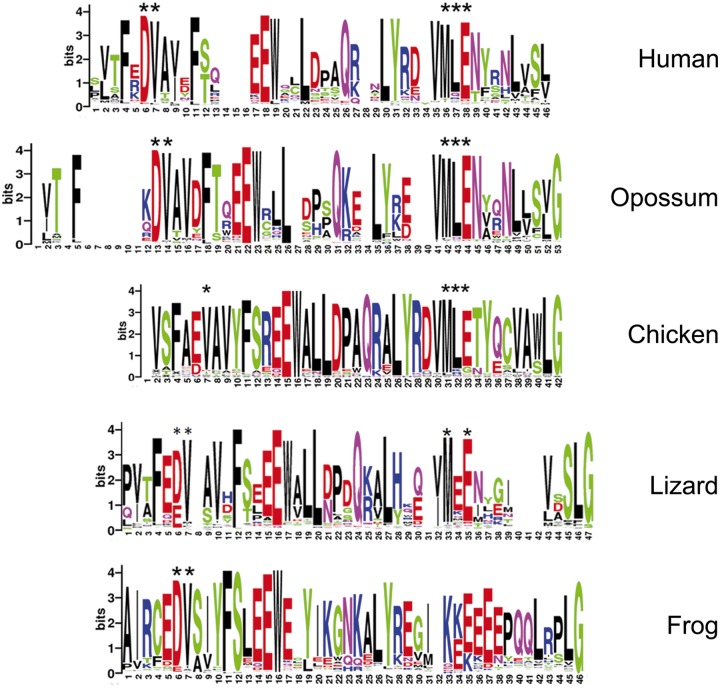
Consensus sequences of KRAB domains from human, opossum, chicken, and frog. Numbers under the x axis in each panel represent amino acid positions, in N- to C-terminal orientation, in the consensus derived from all ZNF-linked KRAB domain sequences in each species; the y axis represents information content (bits) at each position. The height of each letter represents the frequency with which amino acids represented by the letters are found at each position. Asterisks above certain letters at each position indicate agreement with the sequence that has been determined to be necessary for KAP1 binding in human KRAB sequences, at positions 6,7 (DV) and 36–38 (MLE).
The KRAB A domain has diverged significantly from the PRDM9 root in all species examined. But the PRDM9 (not shown) and ZNF282-related KRAB domains (Okumura et al. 1997) lack the amino acid sequences that are known to be essential for KAP1 interaction. Specifically, five amino acids, conserved in two clusters within canonical mammalian KRAB A domains (DV at positions 6,7 in the human consensus in fig. 4 and MLE in positions 36–38), have been shown to be essential for KAP1 binding (Margolin et al. 1994; Urrutia 2003). If all KRAB A domains from each species are assembled into a consensus sequence, it is clear that the C-terminal MLE cluster is also absent from the majority of frog KRAB A sequences (fig. 3). The opossum consensus includes both clusters of KAP-binding amino acid sequences spaced similar to the canonical human domain; these KRAB sequences are thus likely capable of binding KAP1. Chicken genes also include conserved amino acids in most positions although spacing between the two clusters is relatively condensed compared with the human consensus, and the lizard consensus sequence lacks the central leucine in the essential MLE cluster (fig. 3). The avian and lizard KRAB domains may interact with the KAP1 corepressor, although this function cannot be assumed without experimental testing.
Fig. 4.—
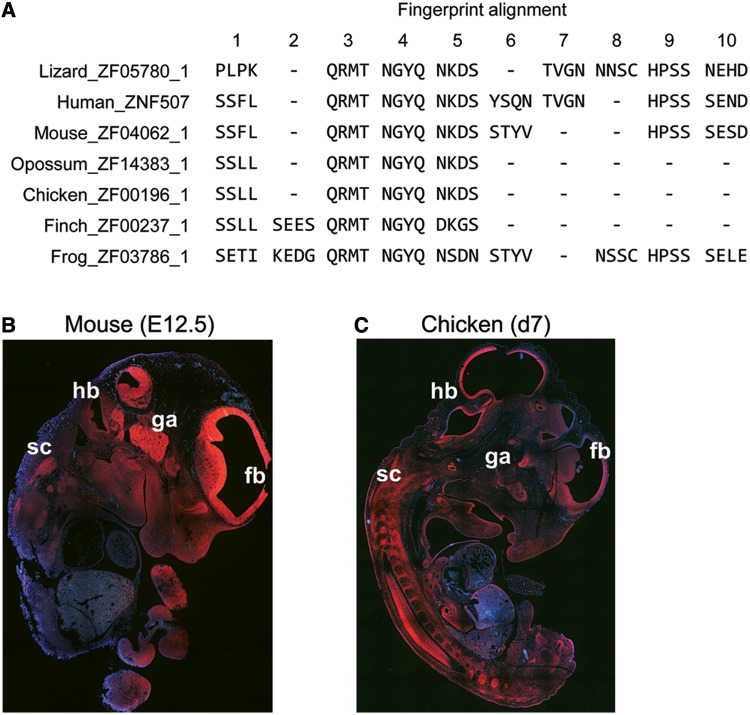
Structural divergence but expression conservation for ZNF507 orthologs. (A) Alignment of fingerprints from the ZNF507 proteins of different vertebrate species. Numbers at top denote ZNF motif positions as they occur in the overall alignment in N-terminal to C-terminal orientation. Dashes denote deletions of ZNF motifs at particular aligned positions in each species. ZNFs 3, 4, and 5 in the alignment are deeply conserved, whereas other positions vary from species to species. (B and C) RNA in situ hybridization in sectioned mouse (B) and chicken (C) embryos at embryonic day 12.5 (E12.5) or 7 (d7), respectively. Despite differences in the pace of development of brain compared with other tissues in these specials, expression in forebrain (fb), hindbrain (hb), ganglia (ga), spinal cord (sc) and in the developing structures of the face is very similar in the two species.
SCAN- and SCAN-KRAB-ZNF Subfamilies
The SCAN domain was exapted from a Gypsy retrotransposon element and incorporated into ZNF-containing gene structures in tetrapods, most likely during or just preceding the emergence of amniotes (Emerson and Thomas 2011). Although we predict a single frog SCAN-ZNF gene (ZF02611) which overlaps well with a X. tropicalis ENSEMBL model, ENSXETT00000023617, this gene was interpreted by the previous authors as a misassembly or erroneous gene prediction, and without experimental evidence we cannot comment further on its validity. The genome of the lizard, A. carolinensis, was shown previously to include many polydactyl ZNF genes that include either SCAN or an ancestral version of the domain (Emerson and Thomas 2011), and we did predict more than 300 SCAN-containing ZNF genes in this species (table 1 and supplementary table S1, Supplementary Material online). However, none of these lizard genes were identified, either by reciprocal Blast or by fingerprint matches, as convincing candidate orthologs for ZNF genes of any type in any of the mammalian groups. This suggests that like the KRAB-ZNF subtype in frogs and other species, SCAN-ZNF loci expanded independently in reptilian lineages.
BTB/POZ-Containing and ZNF-Only Genes
The majority of BTB/POZ-ZNF genes are quite ancient, with orthologs in most or all species examined (table 1 and supplementary table S1, Supplementary Material online). Four BTB/POZ-ZNF gene copies appear to have been acquired as novel genes in amniotes. However, alignments suggest that genes in this subfamily have evolved under some pressure for ZNF divergence (supplementary table S3, Supplementary Material online, and discussed later). For example, one gene that would be counted as “primate-specific” based on fingerprint matches alone, ZBTB48, is actually well conserved in overall protein sequence and syntenic location in amniotes, but proteins from the different species bear no recognizable similarities in DNA-contacting amino acid residues. Indeed, based on reciprocal best-Blast and conserved genome locations, none of the human BTB/POZ-ZNF genes appear to be specific to primates, although several of the genes including ZBTB48, ZBTB41, ZBTB44, and ZBTB49 display highly diverged fingerprint sequences across the different vertebrate lineages (supplementary table S2, Supplementary Material online). Most BTB/POZ-ZNF genes we identified have only two or very few fingers, and mutations in one finger can thus have a dramatic effect on fingerprint similarity scores and presumably, overall protein function.
The 212 human “ZNF-only” models we counted are distributed throughout primate-specific, eutherian, mammalian, amniote, and tetrapod evolutionary groups, with the largest number of genes showing evidence of tetrapod (or earlier) origins (table 4). This group, together with the BTB/POZ genes, includes most of the deeply conserved polydactyl ZNF genes.
Fingerprints in Many Orthologous ZNF Groups Are Evolutionarily Divergent
Combining Blast and fingerprint alignment and focusing particularly on human–mouse comparisons, we identified a selection of genes with strong Blast identities but significant fingerprint diversity, and also a few genes with the opposite pattern (supplementary table S3, Supplementary Material online). One example of an ancient, highly conserved gene encoding a protein with a divergent fingerprint pattern is ZNF507, a gene that has recently been implicated as a novel risk factor for human neurodevelopmental disorders (Talkowski et al. 2012). The ZNF507 fingerprint patterns suggest a complex history, with certain ZNF positions having been selectively deleted or diverged through missense mutations in certain lineages, while retained strictly in order and sequence in other evolutionary groups (fig. 4A). For example, human and lizard retain the exact pattern of four amino acids (TVGN) in ZNF 7 in the alignment, but this ZNF has been lost in other species, including mouse; the mouse protein also differs from human in fingerprint sequence in other ZNF positions. Frog and lizard share sequence in ZNF alignment position 8, suggesting that the motif was ancestral and lost in mammals. A core of three fingers (positions 3, 4, and 5 in the alignment, fig. 4A) is strictly conserved in this ancient protein for every species examined, suggesting an especially important functional role.
Despite this structural divergence, ZNF507 orthologs have retained very similar patterns of developmental expression, as evidenced by ISH in sectioned midgestation mouse and chicken embryos (fig. 4B and C). Expression of mouse Zfp507 was particularly high at embryonic day 12.5 (E12.5), with intense expression in the developing brain, in the spinal and facial ganglia, and developing facial structures (E12.5 corresponds to Theiler stage [TS] 16). The organ systems of birdsand mammals do not develop apace, but we saw remarkable similarities in the pattern of neural expression in TS20 chicken embryos (fig. 4C). The very high levels of neural and craniofacial expression in embryos of these two species fit the predicted neurodevelopmental role of this human gene very well.
As illustrated well by ZNF507, most of the species differences we noted involved the in-phase insertion or deletion (indel) of ZNF motifs. We also detected groups in which orthologs had similar number and arrangement of ZNFs but divergence in fingerprint sequence, and many cases with a mixture of both types of mutation. These patterns have been noted previously as being common paths to divergence for KRAB-ZNF paralogs and orthologs (Looman et al. 2002b; Hamilton et al. 2003; Shannon et al. 2003; Krebs et al. 2005; Huntley et al. 2006; Nowick et al. 2010). However, our alignments show clearly that these same patterns occur frequently in orthologous groups of polydactyl ZNF genes of all types (supplementary table S2, Supplementary Material online). We identified only a handful of genes that, like PRDM9 (Thomas et al. 2009), vary so dramatically in fingerprint sequence that ZNFs could not be aligned. These cases include five human KRAB-ZNF genes for which orthologs were detected only in mouse: ZNF160 and mouse Zfp160, ZNF780B/Zfp780B, ZNF658/Zfp658, and ZNF84/Zfp84, and the previously reported pair of ZNF226/Zfp61 (Shannon et al. 2003) (supplementary table S2, Supplementary Material online). Nothing is known about the functions of any of these strikingly divergent genes.
At the other extreme, we found 170 human genes that, like ZNF282 and ZNF777, encode fingerprint patterns that have been rigidly conserved since their inception. This group includes 65 tetrapod-, 29 amniote-, 26 marsupial-, and 50 eutherian-conserved genes (supplementary tables S2 and S3, Supplementary Material online). The human genes of this type that are conserved in amniotes or tetrapods include many with well-studied developmental functions, including members of the SP1 and KLF families (Berg 1992; Swamynathan 2010). However, this most highly conserved group also includes many genes for which no functional information is currently available. The rigid conservation of the DBDs in proteins encoded by these genes suggests that they have been selected to maintain essential regulatory roles.
Both Conserved and Primate-Specific Polydactyl ZNF Genes Are Most Highly Expressed in Evolutionarily Divergent Tissues
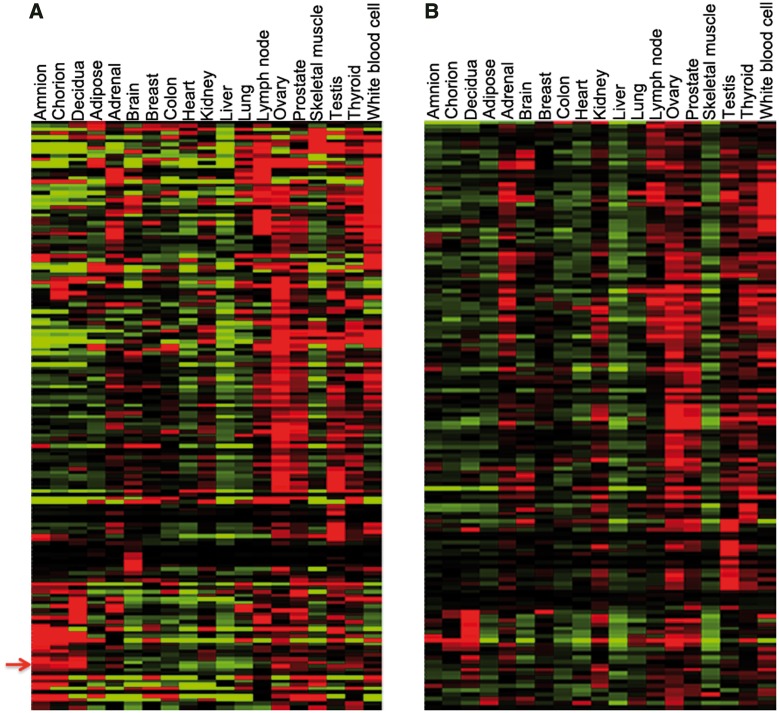
To gain clues to the functions of the most conserved genes, we examined public gene expression (RNA-seq) data from human adult tissues (Illumina Human Body Map 2.0 or HBM2.0), and including more recent data gained from dissected human term placental tissues (Kim et al. 2012). From expression values calculated for uniquely mapped sequence reads by Kim et al. in this published article, we extracted expression data for ZNF genes conserved to tetrapods and amniote species and clustered the data to view gene expression patterns as heat maps. Expression patterns vary significantly over this group, but clusters of genes showed similar expression with highest mRNA levels in 1) lymph nodes and white blood cells, 2) ovary, prostate, and testis or 3) placenta. Interestingly, among the genes expressed at highest levels in placenta are the most ancient members of the KRAB-ZNF family: ZNF777 and ZNF282 (the two genes cluster as indicated by the arrow in fig. 5A).
Fig. 5.—
RNA-seq expression patterns for deeply conserved (A) or primate-specific (B) polydactyl ZNF genes in adult human tissues. For both groups, expression is especially high in reproductive and immune tissues. Arrow to the left of panel (A) is between the adjacent positions of ZNF282 and ZNF777, which are tightly clustered in their expression. Expanded versions of both panels with gene names associated are provided as supplementary figures S1 and S2, Supplementary Material online. Sk.Muscle: skeletal muscle; Wh.BloodCell: white blood cells.
For comparison, we also examined expression patterns of the primate-specific polydactyl ZNF genes in the same RNA-seq data set (fig. 5B). These recently duplicated genes also displayed enrichment for expression in immune and reproductive tissues, with ovary being the most common site of highest expression. Expression patterns for the conserved and the primate-specific gene sets were thus very similar, although primate-specific genes are relatively more enriched in adrenal gland and relative few primate-specific genes are expressed highly in skeletal muscle or in the amniotic and chorionic components of the placenta (fig. 5A and B).
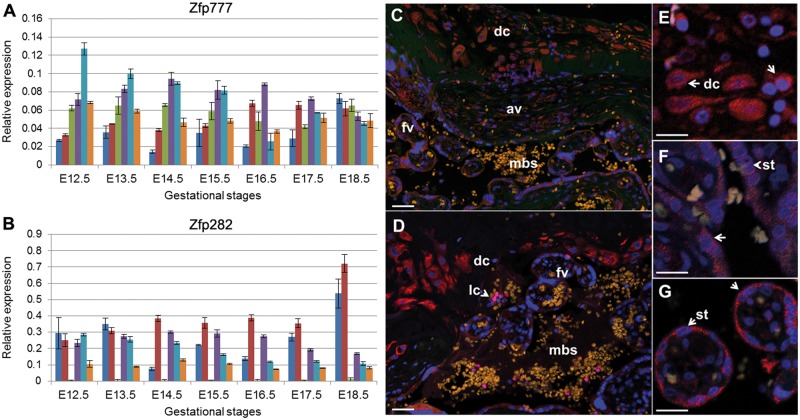
For further information regarding the functions of ZNF282 and ZNF777, we carried out two sets of additional experiments. First, we used quantitative RT-PCR (qRT-PCR) to measure expression levels in RNA isolated from dissected mouse embryos and placenta collected at successive days from embryonic day 12.5 (E12.5) to E18.5 (which is just before birth in mice). The transcripts were expressed with distinct patterns in the dissected decidua, fetal placenta, yolk sac, fetal head, fetal body, and fetal liver of mouse embryos across midgestation development (fig. 6A and B; tissues presented in the order listed above for each gestational stage). Placenta expression for both ZNF genes were high in both fetal and maternal components and fetal membranes at the latest gestational stages (E18.5), concordant with the high levels of expression seen in human term placenta (fig. 5A).
Fig. 6.—
Expression of Zfp282 and Zfp777 genes in embryonic mouse tissues and ZNF282 and ZNF777 proteins in human term placenta. Mouse Zfp777 (A) and Zfp282 (B) transcripts were measured in RNA extracted from dissected embryos and extraembryonic tissues isolated from embryonic day 12.5 (E12.5) to E18.5. Relative expression, measured against the average levels of two ubiquitous genes, Sdha and Ywhaz, are plotted across 2-day gestational intervals in decidua (blue bars), fetal placenta (red), yolk sac (green), fetal heads (purple), fetal bodies (turquoise), and fetal liver (gold). Antibodies specific to the human ZNF777 and ZNF282 proteins (stained in red) were also used to track cell-type specific protein expression in sectioned human term placenta. The sections were counterstained with Hoechst dye (blue) to highlight locations of nuclei. Panels (C) (ZNF777) and (D) (ZNF282) show lower resolution views of placental regions near the maternal:fetal interface including maternal decidual cells (dc), fetal anchoring villi (av), and floating villi (fv) surrounding maternal blood spaces (mbs). An arrowhead in panel (D) highlights the location of a maternal lymphocyte (lc) that is brightly stained by the ZNF282 antibody. Panels (E–G) show higher magnification images from the ZNF777 IHC highlighting decidual cells (dc, panel E) and fetal syncytiotrophoblasts (st) lining anchoring (F) and floating villi (G). White bar in each image represents 25 µm.
To extend expression analysis for these two genes to the level of cell type in placenta, we performed IHC experiments using commercial antibodies in paraffin-embedded sectioned human tissues. Concordant with RNA-seq experiments, ZNF777 and ZNF282 are both highly expressed in multiple cell types in both fetal and maternal components of the human term placenta (fig. 6C and D). More specifically, high levels of expression for both proteins were detected in decidual cells in the maternal compartment (fig. 6E) and in the syncytiotrophoblast cells lining the anchoring and floating chorionic villi (fig. 6F and G). Unlike ZNF777, ZNF282 is also very highly expressed in a subset of lymphocytes within the maternal blood spaces (lc in fig. 6D).
Discussion
With their predicted involvement in transcriptional regulation and their unusually dynamic evolutionary histories, vertebrate polydactyl ZNF genes have commanded a substantial amount of analytical attention. However, despite these efforts, much about their evolution and function has remained unclear. This includes their family history, their patterns of conservation or nonconservation, and specifically their orthology relationships and functional similarities across species. The present study adds new clarity to the ZNF family picture in several respects. First, by creating gene models de novo we were able to compare not only established gene models as several other studies have done (e.g., Ding et al. 2009; Emerson and Thomas 2009; Corsinotti et al. 2013) but also to include recent lineage-specific pseudogenes and novel, especially unannotated protein-coding genes. These include at least 122 novel mouse gene models, all of which are supported by EST evidence; intriguingly, the supporting ESTs are overwhelmingly derived from early embryonic sources. The expression of these novel mouse genes in early embryos would fit well with recent data tying known polydactyl ZNF genes to cell fate decision making and early development (Quenneville et al. 2012; Santoni de Sio, Barde et al. 2012; Santoni de Sio, Massacand et al. 2012; Barde et al. 2013; Corsinotti et al. 2013; Schep and Adryan 2013). Extrapolating this information to other species, it seems likely that many more of the novel models we found will represent functional, developmentally active genes.
Second, by examining both Blast-based and DNA-binding amino acid fingerprint similarities, we identified clear cases of orthologous groups in which there has nonetheless been significant divergence in fingerprint sequence over evolutionary time. In a small number of cases, the exemplar of which is PRDM9, we found clearly orthologous genes with no discernible fingerprint similarity across species. However, fingerprint divergence for most orthologous groups involves the in-phase deletion or tandem duplications within the ZNF array, similar to the pattern we and others have noted for KRAB-ZNF orthologs in the past (Hamilton et al. 2003; Krebs et al. 2005; Huntley et al. 2006; Nowick et al. 2011). Neuronally expressed ZNF-only gene, ZNF507, provides an excellent example (fig. 4). The ZNF507 fingerprint alignments could suggest that the protein has evolved to favor different DNA-binding motifs in each species; alternatively (or perhaps additionally), they could point plainly to three conserved ZNF motifs as having the most essential regulatory roles. In either case, the alignments we present may provide a useful resource as members of this large TF family are targeted for functional characterization.
This ZNF “indel” pattern of divergence was observed for all subfamilies of polydactyl ZNF genes, even within orthologous groups (like ZNF507) that are otherwise relatively well conserved. It thus seems likely that it is the tandem arrangement of ZNF-encoding motifs, per se, that confers a propensity for ZNF indel generation, possibly through a replication slippage mechanism (Krebs et al. 2005). If this model is correct, strong selection pressure would be required to maintain rigid conservation of the number and order of motifs within ZNF arrays. It is therefore especially noteworthy that the ZNF motifs in hundreds of genes have been strictly conserved in number and sequence over millions of years of vertebrate evolution.
The group of genes showing this highly conserved pattern is dominated by ZNF-only and BTB/POZ-ZNF gene subtypes, including many that are known to regulate critical steps in differentiation and development (Swamynathan 2010; Hui and Angers 2011; Ali et al. 2012; Siggs and Beutler 2012). Intriguingly, however, many unstudied genes, including members of the exceptionally dynamic KRAB-ZNF subfamily, are also highly conserved. The most ancient of these conserved human KRAB-ZNF genes, ZNF282 and ZNF777, a more diverged but ancient relative, ZNF783, and more recently derived cluster neighbors stand out in mammals for their inclusion of an unusually structured KRAB domain that does not bind KAP1 and functions as a transcriptional activator (Okumura et al. 1997; Conroy et al. 2002). Evolutionary analysis supports the ancient provenance of this activating KRAB and reveals that KRAB-ZNF genes with similar KRAB domains have expanded independently in amphibians and reptiles. The canonically structured, KAP1-binding repressive version of the KRAB A domain is dominant only in mammals, although a similar (and possibly still KAP1-binding) sequence is also the dominant version of the ZNF-linked KRAB A domain in birds.
The dramatic expansion and rapid divergence of repressive KRAB-ZNF genes in mammals has suggested their participation in an “arms race,” with the need to silence endogenous retroviruses (ERVs) hypothesized as the dominant driver (Thomas and Schneider 2011). Supporting this notion, a handful of KRAB-ZNF genes have been shown to silence retroviral sequences by binding to motifs within their flanking long terminal repeats (LTRs). Intriguingly, human ZNF282 (also called HUB-1) is one of this very small number of verified LTR-binding KRAB-ZNF proteins, recognizing a motif within the U5RE regulatory region of the human T-cell leukemia virus (HTLV-I) LTR and repressing viral activity. Although the KRAB domain in ZNF282 and other cluster relatives activates transcription, these proteins also include a second domain, called HUR, which confers repressive activity. Rather than acting simply as HTLV-I inhibitor, ZNF282 has been proposed to facilitate an alternative path for the virus by promoting latent infection (Okumura et al. 1997). Interestingly, one cluster relative, ZNF398, can generate HUB-containing repressive or HUB-minus activating isoforms through alternative splicing (Conroy et al. 2002), and ZNF282 may be able to do the same. Thus, ZNF282 may have evolved to regulate retroviral sequences but has a complex relationship with the virus that cannot be described in simple arms race terms. Indeed, it may be possible that preestablished ZNF282 binding motif, which evolved for other purposes, was captured and domesticated by HTLV-I. Genome-wide DNA binding assays in cells and tissues of different species should allow us to dissect the history of this intriguing interaction.
Whatever the model, the rigid conservation of fingerprint patterns in polydactyl ZNF proteins suggests that their DNA-binding activities have evolved essential biological roles. Indeed, naturally occurring or targeted mutations in many deeply conserved polydactyl ZNF genes confirm essential roles in differentiation and development in humans and model organisms (Zhao and Meng 2005; Swamynathan 2010; Hui and Angers 2011; Ali et al. 2012; Siggs and Beutler 2012). We hypothesize that other coexpressed genes in this highly conserved cohort are also associated with unstudied and important developmental functions, albeit functions that in many cases may be challenging to discern. For example, in light of their antiquity and very tight DNA-binding conservation, the high expression of ZNF777 and ZNF282 in placenta—within cell types that vary significantly even between humans and mice including some that do not exist in lizards and birds—is particularly puzzling. Although these placental cells are lineage-unique, they evolved from fetal membranes and uterine cell types that are common to all amniotes (Black et al. 2010; Lindenfors and Tullberg 2011; Elso et al. 2013). However, these cell types and structures have continued to evolve independently in every species, making placenta the most evolutionarily divergent of all mammalian tissue types (Krebs et al. 2005).
In fact, the rapid pace of placental divergence reflects another type of arms race—that between the interests of the mother and developing fetus—which is a defining feature of mammalian biology (Moore 2012). Similar types of evolutionary battles have played major roles in shaping vertebrate reproductive tissues and cell types, with wide impact on species-specific morphology, metabolism, and behavior (Lindenfors and Tullberg 2011; McPherson and Chenoweth 2012). Given the very high levels at which both conserved and primate-specific polydactyl ZNF genes are expressed in reproductive tissues, we propose that this larger evolutionary arms race has been the real driver of polydactyl ZNF expansion and divergence in vertebrate history. The facility with which polydactyl ZNF genes can diverge to generate opportunities for DNA-binding diversity makes them ideal raw materials for crafting novelty in gene regulatory pathways. The data provided here identify prime targets, in the form of deeply conserved and unique genes and proteins, for testing these hypotheses in future studies.
Supplementary Material
Supplementary tables S1–S3 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Elbert Branscomb for many useful discussions, Elbert Branscomb, Joseph Troy, and Derek Caetano-Anolles for critical comments on the manuscript, and Yi Xing for providing the processed RNA-seq data set. They also thank Stuart Huntley for carrying out the early pilot work that inspired this project. This study was supported by the National Institutes of Health, NIGMS grant number RO1 GM078368 (awarded to L.S.).
Literature Cited
- Ali RG, Bellchambers HM, Arkell RM. Zinc fingers of the cerebellum (Zic): transcription factors and co-factors. Int J Biochem Cell Biol. 2012;44:2065–2068. doi: 10.1016/j.biocel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Barde I, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid EJ, et al. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 1993;12:1363–1374. doi: 10.1002/j.1460-2075.1993.tb05781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM. Sp1 and the subfamily of zinc finger proteins with guanine-rich binding sites. Proc Natl Acad Sci U S A. 1992;89:11109–11110. doi: 10.1073/pnas.89.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtle Z, Ponting CP. Meisetz and the birth of the KRAB motif. Bioinformatics. 2006;22:2841–2845. doi: 10.1093/bioinformatics/btl498. [DOI] [PubMed] [Google Scholar]
- Black SG, Arnaud F, Palmarini M, Spencer TE. Endogenous retroviruses in trophoblast differentiation and placental development. Am J Reprod Immunol. 2010;64:255–264. doi: 10.1111/j.1600-0897.2010.00860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy AT, et al. A novel zinc finger transcription factor with two isoforms that are differentially repressed by estrogen receptor-alpha. J Biol Chem. 2002;277:9326–9334. doi: 10.1074/jbc.M107702200. [DOI] [PubMed] [Google Scholar]
- Constantinou-Deltas CD, et al. The identification and characterization of KRAB-domain-containing zinc finger proteins. Genomics. 1992;12:581–589. doi: 10.1016/0888-7543(92)90451-w. [DOI] [PubMed] [Google Scholar]
- Corsinotti A, et al. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS One. 2013;8:e56721. doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dehal P, et al. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Ding G, Lorenz P, Kreutzer M, Li Y, Thiesen HJ. SysZNF: the C2H2 zinc finger gene database. Nucleic Acids Res. 2009;37:D267–D273. doi: 10.1093/nar/gkn782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition. Structure. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Elso C, et al. A reciprocal translocation dissects roles of Pax6 alternative promoters and upstream regulatory elements in the development of pancreas, brain, and eye. Genesis. 2013;51:630–646. doi: 10.1002/dvg.22409. [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Gypsy and the birth of the SCAN domain. J Virol. 2011;85:12043–12052. doi: 10.1128/JVI.00867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Kim J, Branscomb E, Stubbs L. Lineage-specific expansion of KRAB zinc-finger transcription factor genes: implications for the evolution of vertebrate regulatory networks. Cold Spring Harb Symp Quant Biol. 2003;68:131–140. doi: 10.1101/sqb.2003.68.131. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Huntley S, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci U S A. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Berg JM. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. Transcriptome landscape of the human placenta. BMC Genomics. 2012;13:115. doi: 10.1186/1471-2164-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ, Larkins LK, Khan SM, Robins DM. Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2. Genomics. 2005;85:752–761. doi: 10.1016/j.ygeno.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lindenfors P, Tullberg BS. Evolutionary aspects of aggression the importance of sexual selection. Adv Genet. 2011;75:7–22. doi: 10.1016/B978-0-12-380858-5.00009-5. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Looman C, Abrink M, Mark C, Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol Biol Evol. 2002a;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- Looman C, Abrink M, Mark C, Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol Biol Evol. 2002b;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- Margolin JF, et al. Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci U S A. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson FJ, Chenoweth PJ. Mammalian sexual dimorphism. Anim Reprod Sci. 2012;131:109–122. doi: 10.1016/j.anireprosci.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T. Review: Parent-offspring conflict and the control of placental function. Placenta. 2012;33(Suppl):S33–S36. doi: 10.1016/j.placenta.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, et al. Gain, loss and divergence in primate zinc-finger genes: a rich resource for evolution of gene regulatory differences between species. PLoS One. 2011;6:e21553. doi: 10.1371/journal.pone.0021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Hamilton AT, Zhang H, Stubbs L. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol. 2010;27:2606–2617. doi: 10.1093/molbev/msq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Sakaguchi G, Naito K, Tamura T, Igarashi H. HUB1, a novel Kruppel type zinc finger protein, represses the human T cell leukemia virus type I long terminal repeat-mediated expression. Nucleic Acids Res. 1997;25:5025–5032. doi: 10.1093/nar/25.24.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pengue G, Calabro V, Bartoli PC, Pagliuca A, Lania L. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 1994;22:2908–2914. doi: 10.1093/nar/22.15.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, et al. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Barde I, et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012;26: 4561–4575. doi: 10.1096/fj.12-206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Massacand J, et al. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood. 2012;119:4675–4685. doi: 10.1182/blood-2011-12-401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Adryan B. A comparative analysis of transcription factor expression during metazoan embryonic development. PLoS One. 2013;8:e66826. doi: 10.1371/journal.pone.0066826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggs OM, Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L, Sun Y, Caetano-Anolles D. Function and evolution of C2H2 zinc finger arrays. Subcell Biochem. 2011;52:75–94. doi: 10.1007/978-90-481-9069-0_4. [DOI] [PubMed] [Google Scholar]
- Swamynathan SK. Kruppel-like factors: three fingers in control. Hum Genomics. 2010;4:263–270. doi: 10.1186/1479-7364-4-4-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Emerson RO, Shendure J. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009;4:e8505. doi: 10.1371/journal.pone.0008505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Golding MC. Selection of stable reference genes for quantitative rt-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS One. 2011;6:e27592. doi: 10.1371/journal.pone.0027592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissing H, Meyer WK, Aagaard L, Tommerup N, Thiesen HJ. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153–157. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- Witzgall R, O'Leary E, Leaf A, Onaldi D, Bonventre JV. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci U S A. 1994;91:4514–4518. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Zhao C, Meng A. Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev Growth Differ. 2005;47:201–211. doi: 10.1111/j.1440-169X.2005.00797.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.