Abstract
Nonretroviral integrated RNA viruses (NIRVs) are genes of nonretroviral RNA viruses found in the genomes of many eukaryotic organisms. NIRVs are thought to sometimes confer virus resistance, meaning that they could impact spread of the virus in the host population. However, a NIRV that is expressed may also impact the evolution of virus populations within host organisms. Here, we experimentally addressed the evolution of a virus in a host expressing a NIRV using Tobacco etch virus (TEV), a plant RNA virus, and transgenic tobacco plants expressing its replicase, NIb. We found that a virus missing the NIb gene, TEV-ΔNIb, which is incapable of autonomous replication in wild-type plants, had a higher fitness than the full-length TEV in the transgenic plants. Moreover, when the full-length TEV was evolved by serial passages in transgenic plants, we observed genomic deletions within NIb—and in some cases the adjacent cistrons—starting from the first passage. When we passaged TEV and TEV-ΔNIb in transgenic plants, we found mutations in proteolytic sites, but these only occurred in TEV-ΔNIb lineages, suggesting the adaptation of polyprotein processing to altered NIb expression. These results raise the possibility that NIRV expression can indeed induce the deletion of the corresponding genes in the viral genome, resulting in the formation of viruses that are replication defective in hosts that do not express the same NIRV. Moreover, virus genome evolution was contingent upon the deletion of the viral replicase, suggesting NIRV expression could also alter patterns of virus evolution.
Keywords: experimental evolution, genome evolution, nonretroviral integrated RNA viruses, plant virus, RNA virus, virus evolution
Introduction
The genome size of RNA viruses is highly variable, ranging from less than 4 kb for members of the Narnaviridae to more than 30 kb for some Coronaviridae. However, most RNA viruses have genome sizes in the 5–15-kb range (Holmes 2009). What could explain this wide range of genome sizes in RNA viruses? Small genomes probably replicate faster than large ones, while genome size limits enzymatic complexity. This limited complexity results in lower-fidelity replication enzymes, which in turn leads to higher variability (Smith and Szathmáry 1995). On the other hand, larger genomes will replicate more slowly, while being less constrained in the complexity of the proteins they code for. Therefore, larger genomes allow for evolutionary innovations such as polymerases with greater replication fidelity (Denison et al. 2011) or the modulation of host immune responses (Shackelton and Holmes 2004). However, the evolution of larger genomes and greater complexity may be impeded by Eigen’s paradox: to obtain proofreading activity, an RNA virus must already have a large genome (Eigen 1971; Smith and Szathmáry 1995). It has therefore been suggested that RNA virus genome size evolves to the upper limit imposed by mutation rate (Swetina and Schuster 1982).
If genome size of RNA viruses is limited by a tradeoff between RNA-dependent RNA polymerases (RdRp) fidelity and decreased replication rates, then examples of enlargement in genome size should be scarce for RNA viruses. There are, however, some examples of the integration of host genetic material into viral RNA genomes, leading to the largest known RNA viruses. For instance, the genome of coronaviruses contains homologous sequences of at least five cellular enzymes that are associated with RNA processing and that may be involved in modulating mutation rate (Snijder et al. 2003). Acquisition of a host’s ubiquitin gene by certain strains of bovine viral diarrhoea pestivirus resulted in increased pathogenicity (Meyers et al. 1989). Plant closterovirus genomes encode for a homologue of cellular molecular chaperone HSP70 (Dolja et al. 2006) that is required for virion assembly (Satyanarayana et al. 2000). Finally, a particularly interesting case is the ubiquitous presence of DNA/RNA repair AlkB-like proteins in several groups of plant viruses, including closterovirus, flexivirus, tymovirus, and even one potyvirus (Blackberry virus Y; Susaimuthu et al. 2008), whose function has been postulated to be both repair of nucleic acid alkylation and suppression of posttranscriptional gene silencing (Aravind and Koonin 2001; Bratlie and Drabløs 2005). Similarly, the insertion of sequences into a viral genome generally leads to their rapid removal, as has been seen in the case of heterologous genes (Chapman et al. 1992; Guo et al. 1998; Chung et al 2007; Paar et al. 2007; Zwart et al 2014) and repetitions of endogenous noncoding sequences (Nagai et al. 2003; Gritsun and Gould 2006). Finally, most viruses have a tendency to evolve defective interfering viruses under conditions promoting cellular coinfection (Huang 1973; Roux et al. 1991), again suggesting a link between genome size and replication. Although demographic conditions may allow for the maintenance of extraneous sequences (Dolja et al. 1993; Zwart et al. 2014), overall it appears as if genome size is limited and under purifying selection, and that the incorporation or maintenance of foreign sequences comes at a cost to viruses (Sakai et al. 1999; Marks et al. 2005; Holmes 2009; Zwart et al. 2014).
On the other hand, genome size does not appear to be under the same selective pressure in eukaryotes, probably due to the smaller effective population sizes in higher organisms (Lynch et al. 2006). Although the existence of many retroviral sequences in eukaryotic genomes was already well documented (Gifford and Tristem 2003), it has been recently found that there are also many nonretroviral RNA virus genes in eukaryotic genomes. These nonretroviral integrated RNA viruses (NIRVs) have been found in mammals (Geuking et al. 2009; Belyi et al. 2010; Horie et al. 2010; Katzourakis and Gifford 2010), plants (Hong et al. 1998; Chiba et al. 2011), and fungi (Taylor and Bruenn 2009). In fungi, NIRVs are very common and have in some cases become functional genes (Koonin 2010). In one case, the entire viral RdRp was integrated and conserved, probably conferring viral resistance to the host (Taylor and Bruenn 2009). It has also been suggested that NIRVs can confer resistance (Bertsch et al. 2009) or tolerance (Flegel 2009) against the original virus to plant or animal hosts. Indeed, it has been a common practice to generate commercial transgenic plants resistant to viruses by inserting full or partial viral genes into the genome of the plants of agronomic interest (reviewed in Kundu and Mandal [2001]); these transgenic plants can be considered as artificial cases of NIRVs.
Because the reading frame is conserved in many NIRVs (Taylor and Bruenn 2009) they can in principle be expressed. In practice, this will depend on their location in the genome and the state of the host cell. Some NIRVs are, however, clearly expressed by the host (Taylor and Bruenn 2009). Host NIRV expression has been proposed to mediate host immunity against the cognate virus (Bertsch et al. 2009; Flegel 2009; Taylor and Bruenn 2009). However, host NIRV expression may also have important implications for the evolution of the cognate virus. The absence of an effective NIRV-induced host immune response and sufficiently high NIRV expression levels could lead to complementation of host proteins, making the NIRV-expressed genes in the cognate virus functionally redundant. What would be the impact of NIRV expression on viral replication? Is there any selective advantage to using in trans the protein produced from the NIRV as a functional viral protein? How will a virus population evolve in such a situation of functional redundancy? Finally, how do such large-scale genomic perturbations affect the regulation of expression and accumulation of all other viral components? To our knowledge these questions have not been addressed in any study, while given the prevalence of NIRVs in natural host populations and biosafety issues linked to the use of transgenic plants expressing viral genes for resistance, it is a highly relevant topic.
Here, we evaluated the patterns of genomic evolution of Tobacco etch virus (TEV, genus Potyvirus, family Potyviridae) in transgenic Nicotiana tabacum plants expressing NIb, the TEV RdRp (Li and Carrington 1995). This system is an experimental model for exploring what the effects of high levels of NIRV expression are on virus infection and evolution. During infection of these transgenic plants, plant-derived NIb expression makes virus-derived NIb-expression redundant (Li and Carrington 1995). We therefore expect that there may be relaxed selection for maintenance of the NIb cistron in the viral genome, which might lead to selection of variants with genomic deletions at the NIb locus, due to the replicative advantage associated with a smaller genome size. Ultimately, the expression of NIb might therefore induce genome shrinkage and result in a virus population that can no longer autonomously infect wild-type plants. We first determined whether the deletion of viral NIb led to an increase in viral fitness in NIb-expressing plants. Then, we passaged a full-length wild-type virus in NIb-expressing plants and used next-generation sequencing (NGS) to check for deletions in TEV NIb locus. Finally, we preempted the complete deletion of TEV NIb in NIb-expressing plants and studied the effects of NIb complementation in trans on genomic changes.
Materials and Methods
Virus Genotypes
The pTEV7DA infectious clone (Dolja et al. 1992) was used as a source for TEV. A TEV genotype was produced that lacked the full replicase gene (ΔNIb) by inverse polymerase chain reaction (PCR) using Pfu turbo DNA polymerase (Stratagene) and primers conserving the proteolytic NIa/NIb (ΔNIb-F) and NIb/CP (ΔNIb-R) sites (supplementary table S1, Supplementary Material online). The resulting clone was named pTEV7DA-ΔNIb (Tromas and Elena 2010).
Direct Competition between TEV and TEV-ΔNIb Genotypes
Concentrated saps of TEV and TEV-ΔNIb were obtained by grinding 500 mg of 7 days postinoculation (dpi) infected tissue in a mortar with 500 μl of grinding buffer. Nicotiana tabacum 35S::NIb plants were inoculated with a 1:1 mixture of homogenates of both viruses. Twenty plants were inoculated by abrasion of the third true leaf with 30 μl mixed homogenate, and 20 plants were taken as a mock-inoculated control. After 7, 21, and 63 dpi, total RNA was extracted from the youngest leaves of five plants. The region flanking the NIb cistron was reverse transcribed using Moloney Murine leukemia virus (M-MLV) reverse transcriptase (RT) (Fermentas) and primer 97-101-R (supplementary table S1, Supplementary Material online). PCR amplification was performed using the high-fidelity Phusion DNA polymerase (Finnzymes) and the 73-80-F/92-96-R primers (supplementary table S1, Supplementary Material online). PCR products were resolved on a 1% agarose gel to verify the presence of both genotypes. RT-qPCR was performed to compare viral accumulation in case of direct competition between the two genotypes. RT-qPCR for the CP cistron was used to determine the total level of viral accumulation (i.e., accumulation of both genotypes), using the specific primers TEV-CP-qPCR-F and TEV-CP-qPCR-R (supplementary table S1, Supplementary Material online). RT-qPCRs were performed using One-Step SYBR PrimeScript RT-PCR kit II (Takara) following the instructions provided by the manufacturer. CP was chosen because it locates in the 3′ end of TEV genome and hence would only quantify complete genomes but not partial incomplete amplicons. Each RNA sample was quantified three times in independent experiments. Amplifications were done using the ABI PRISM Sequence Analyzer 7000 (Applied Biosystems). The thermal profile was as follows: RT phase consisted of 5 min at 42 °C followed by 10 s at 95 °C; and PCR phase of 40 cycles of 5 s at 95 °C and 31 s at 60 °C. Quantification results were examined using SDS7000 software v. 1.2.3 (Applied Biosystems). RT-qPCR for NIaPro/NIb cistrons was then performed to determine viral accumulation of the TEV genotype only, using specific primers TEV-NIaPro/NIb-F and TEV-NIaPro/NIb-R (supplementary table S1, Supplementary Material online) and otherwise identical conditions when compared with the quantification of the CP. The ratio of TEV-ΔNIb to TEV (R) is then R = (nCP−nNIaPro/NIb)/nNIaPro/NIb, where nCP and nNIaPro/NIb are the copy numbers of the CP and NIaPro/NIb, respectively, as measured by RT-qPCR. To estimate the replicative advantage W (Carrasco et al. 2007) of TEV-ΔNIb, we performed linear regression on the infection time in days and the log-transformed ratio data. The antilog-transformed slope is W.
Experimental Evolution Protocol
Infectious plasmids pTEV7DA and pTEV7DA-ΔNIb were linearized with BglII (Takara) and transcribed into 5′-capped RNAs using the SP6 mMESSAGE mMACHINE kit (Ambion Inc). Transcripts were prepared as described previously (Carrasco et al. 2007). Two batches of five 4-week-old N. tabacum 35S::NIb plants were inoculated mechanically on the third true leaf either with TEV transcripts (4–7 µg) in inoculation buffer (3% polyethyleneglycol 8000, pH = 7.0) or TEV-ΔNIb transcripts (20–25 µg) in the same inoculation buffer. In all cases, TEV symptoms appeared 6–7 dpi.
Two evolution experiments were performed with these viruses. In the first experiment, conceived to study the appearance of genomic deletions de novo, four 3-week passages of TEV and TEV-ΔNIb were performed in N. tabacum 35S::NIb plants. At 21 dpi, all symptomatic leaves of each of the five lineages per virus were collected. Five hundred milligrams of symptomatic tissue was ground in 500 µl of grinding buffer (50 mM potassium phosphate pH = 7.0, 3% polyethyleneglycol 6000). Ten microliters of the resulting sap was then mixed with inoculation buffer and used to infect a batch of five healthy transgenic plants by abrasion for each TEV genotype. In the second experiment, conceived to evaluate the subsequent evolution and fitness optimization of genotypes missing the NIb cistron, 30 one-week passages of TEV and TEV-ΔNIb were performed in N. tabacum 35S::NIb plants. At 7 dpi, symptomatic leaves were collected for each of the five lineages per virus. Infectious sap was prepared as described earlier.
TEV Genomic RNA Extraction and Purification for Sequence Analysis
To analyze the lineages of TEV-ΔNIb and TEV evolved in N. tabacum 35S::NIb (second experiment above), total RNA was extracted using RNAeasy Plant Mini Kit (Qiagen) from symptomatic leaves of five N. tabacum 35S::NIb infected by the evolved lineages of TEV and five N. tabacum 35S::NIb infected by the evolved lineages of TEV-ΔNIb. To verify the presence of deletions, the full genome of each genotype was reverse transcribed using M-MLV RT (Fermentas) and primer 97-101-R (supplementary table S1, Supplementary Material online), then PCR amplified using the Ultra High-Fidelity DNA polymerase Phusion (Finnzymes) and primers 73-80-F and 97-101-R (supplementary table S1, Supplementary Material online). Deletions were confirmed by resolution of PCR products on 1% agarose gels.
Subsequently, the full genome was reverse transcribed using M-MLV RT (Fermentas) and primers 97-101-R, 67-77-R, and 40-45-R and PCR amplified using the high-fidelity DNA polymerase Phusion (Finnzymes) and three pairs of primers 2-10-F/40-45-R, 45-48-F/67-77-R, and 73-80-F/97-101-R (supplementary table S1, Supplementary Material online). By using these pairs, we ensured that the mRNA from the transgene was not amplified and that we obtained only TEV sequences from viral genomes. PCR products were resolved on 1% agarose gels to verify the presence of deletions. For both genotypes, TEV and TEV-ΔNIb, the three amplicons were purified and sequenced by GenoScreen (Lille, France) using BIGDYE 3.1 and a 96-capillars ABI3730XL sequencing system (Applied Biosystems) with overlapping readouts using the same set of primers as in Agudelo-Romero et al. (2008). Contigs were assembled using GENEIOUS version 4.8.
High-Throughput Sequencing and Data Analysis
To analyze the diversity of deletions that could occur in NIb cistron during the evolution of TEV in N. tabacum 35S::NIb (first experiment described above), the full genome of each genotype was reverse transcribed using M-MLV RT (Fermentas) and primer R97-101, then PCR amplified using the Ultra High-Fidelity DNA polymerase Phusion (Finnzymes) and primers F73-80 and R97-101 (supplementary table S1, Supplementary Material online). Seven independent RT-PCR replicates were performed for each reaction and then pooled, to limit possible PCR artifacts. These PCR amplicons were sequenced by GenoScreen (Lille, France) with an Illumina HiSeq2000 equipment (Illumina Inc.) using a paired-end (2 × 100 b) protocol. The reads were already demultiplexed and adapters removed. Data were then analyzed with GSNAP (Wu and Nacu 2010). GSNAP aligned both single-end and paired-end reads. GSNAP is designed to detect short and long-distance splicing, but in our case, we used it to detect deletions instead of introns. GSNAP was used with its default set of parameters, and results were parsed using custom Perl scripts: 1) we excluded reads that are mapped on 100 consecutive bases on TEV genome; 2) for the remaining sequences, we identified sequences that mapped in two different virus genome regions with a minimum of eight bases in the 5′ and 3′ ends; 3) we excluded reads poorly represented (<10) and showing a low CIGAR (concise idiosyncratic gapped alignment report) string diversity (<10) in order to eliminate possible artifacts; 4) we calculated the distance between the two different regions to define deletion size and position; 5) we calculated the frequency of each deletion detected; and 6) finally, we obtained the number of reads for each deletion variant in R1 and R2 and we compared it to the total number of reads for each lineage.
Analysis of TEV-ΔNIb Mutants
Each mutation was introduced in the pMTEV infectious clone (Bedoya and Daròs 2010) by site-directed mutagenesis using the Quickchange II XL kit (Stratagene) and following the indications given by the manufacturer. To minimize unwanted errors during the mutagenesis process, the kit incorporates Pfu Ultra High-Fidelity Turbo DNA polymerase (Stratagene). The amplification conditions were 1 min at 95 °C (initial denaturation), followed by 18 cycles consisting of 30 s at 95 °C, 45 s at 65 °C and 18 min at 68 °C, and a final extension step of 28 min at 68 °C.
Infectious plasmids of each mutant were linearized with BglII (Takara) and transcribed into 5′-capped RNAs. Transcripts were prepared as described previously. Two batches of five 4-week-old N. tabacum 35S::NIb plants were, respectively, inoculated mechanically on the third true leaf with TEV transcripts (20–30 µg) in inoculation buffer. In all cases, the first symptoms appeared 6–7 dpi. Seven dpi, infectious saps were obtained by grinding 500 mg of TEV mutant-infected tissue in a mortar with 500 µl of grinding buffer; then, three 4-week-old N. tabacum 35S::NIb plants for every TEV mutants were, respectively, inoculated mechanically on the third true leaf with 15 µl of the corresponding sap.
Total RNA was extracted from the youngest leaves of each of the three plants per TEV mutant 2.67 dpi and then RNAs were purified. RT-qPCR with the CP primers was performed to compare viral accumulation of each mutant to the TEV-ΔNIb ancestor as described previously. For each genotype, the Malthusian growth rate per day was computed as m = (1/t)log Qt, where Qt is amount of TEV RNA (in pg) per 100 ng of total plant RNA quantified at t = 2.67 dpi. Absolute fitness was then defined as W = em (Crow and Kimura 1970).
Results and Discussion
TEV-ΔNIb Has Greater Within-Host Fitness than TEV in N. tabacum 35S::NIb
Li and Carrington (1995) generated transgenic N. tabacum cv. Xanthi plants expressing TEV NIb under control of the Cauliflower mosaic virus (CaMV) 35 S promoter, referred to here as N. tabacum 35S::NIb. TEV variants with disruption of the polymerase GDD amino acid motif, the nuclear localization signal, or the NIb N-terminal cleavage site could replicate in these transgenic plants, whereas replication was not supported in wild-type plants. Moreover, a TEV variant missing NIb altogether could also infect these transgenic plants and induce symptoms after 7 dpi, similar to the wild-type virus. Tromas and Elena (2010) also generated a TEV variant missing the NIb cistron, referred to here as TEV-ΔNIb. TEV-ΔNIb can infect N. tabacum 35S::NIb, but not the wild-type N. tabacum, in agreement with previous results (Li and Carrington 1995). TEV is the full-length wild-type virus, capable of infecting both wild-type N. tabacum and 35S::NIb transgenic plants. When wild-type N. tabacum plants were coinoculated with different mixtures of TEV:TEV-ΔNIb (1:100 to 100:1), we detected only infection by TEV. Coinfected wild-type plants were probably not observed because the low cellular multiplicity of infection of TEV in N. tabacum (Tromas et al. 2014) precludes cellular coinfection, especially early in infection.
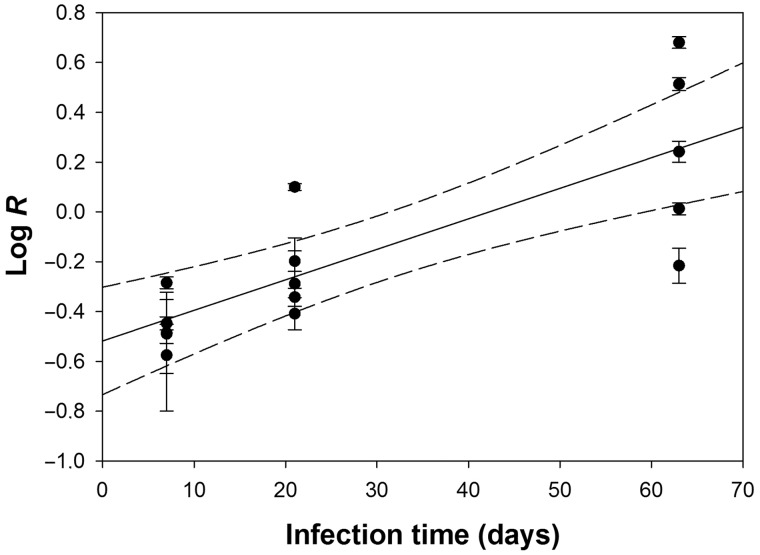
TEV-ΔNIb has a 16% smaller genome than TEV. If 1) a relationship between genome size and virus replication exists (Sakai et al. 1999; Zwart et al. 2014) and 2) dependence on in trans NIb expression for replication does not impose a large fitness burden to the virus, then NIb deletion in the virus genome will cause a faster replication and a selective advantage over a full-length wild-type virus. We therefore inoculated 15 N. tabacum 35S::NIb plants with a 1:1 mixture of ground infectious tissue with TEV and TEV-ΔNIb and measured the ratio of the two variants at 7, 21, and 63 dpi using quantitative reverse transcription PCR (RT-qPCR). Five plants were analyzed for each time point, meaning that each data point represents an independent measurement. On the basis of these data, we estimated the selective advantage for TEV as the per day selection rate constant W (Materials and Methods). A significant, upward-sloping linear regression was observed between time and the ratio of the TEV-ΔNIb:TEV (fig. 1; r2 = 0.632, F1,14 = 22.330, P < 0.001), meaning that the ratio of the viruses shifts significantly toward TEV-ΔNIb, and this virus therefore has a significantly higher competitive fitness. We estimated that W = 1.028 ± 0.006 d−1 (±1 SD), suggesting that TEV-ΔNIb will slowly displace the wild-type virus in transgenic plants. If genome size was inversely proportional to fitness and the 16% reduction in genome size were the only determinant of fitness for TEV-ΔNIb, we would expect W = 1/(1–0.16) = 1.19 d−1. The 95% confidence interval for our estimate of W is 1.016–1.040 d−1, suggesting that 1) there is a cost to depend on in trans NIb expression, but 2) this cost is partially compensated for by faster replication of TEV-ΔNIb, resulting in a higher W than the wild-type virus. Our data therefore support the idea of a relationship between genome size and fitness, and suggest that viruses harboring deletions in NIb will outcompete the wild-type virus in NIb-expressing plants. Therefore, NIRV expression might also lead to conditions favoring the replication of virus variants, with deletions in the gene(s) encoded for by the NIRV. However, the accumulation of viruses with large genomic deletions due to in trans expression of virus’ genes will also depend on mutational supply, specifically how often recombination generates viruses with deletions in NIb that conserve the reading frame in downstream genes.
Fig. 1.—
Plot of the log-transformed ratio of TEV:TEV-ΔNIb (ordinate) over time (abscissa), given in weeks postinoculation. Error bars represent ± 1 standard deviation, the solid line represents the linear regression and the dashed lines the 95% CI of regression.
Full-Length Genomes Were Not Recovered by Recombination between TEV-ΔNIb and the Transcripts from the 35::NIb Transgene
Borja et al. (1999) reported that inoculation of transgenic N. benthamiana plants expressing the CP protein of tomato bushy stunt tombusvirus (TBSV) with TBSV mutants carrying defective CP resulted in the restoration of wild-type virus throughout a double-recombination event between the viral genome and the mRNA transcribed from the transgene. In sharp contrast, several authors have failed to detect recombination events between transgenes and potyviruses (Chung et al. 2007; Dietrich et al. 2007). To test for possible recombination events in our system, we performed an evolution experiment consisting of five independent lineages of TEV-ΔNIb serially passaged four times in N. tabacum 35S::NIb transgenic plants, with each passage being 3 weeks long. At the end of the evolution experiment, we analyzed the evolved lineages by RT-PCR using a combination of primers that will specifically amplify full-genome viruses (supplementary table S1, Supplementary Material online). We found no evidence for recombination between TEV-ΔNIb and host-derived NIb transcripts. Therefore, we conclude that, in our experimental set up, regeneration of the wild-type genome by recombination is an unlikely phenomenon. This outcome is congruent with the estimates of within-host fitness for TEV and TEV-ΔNIb; recombinants analogous to the wild-type virus would be displaced by TEV-ΔNIb, and the process could occur rapidly as de novo variation will initially be present at low frequencies in the virus population. On the other hand, this result may reflect the possibility that recombination between TEV-ΔNIb and the transgene transcripts is very rare, as shown for other potyviruses (Chung et al. 2007; Dietrich et al. 2007).
Rapid Occurrence of Large Genomic Mutations in TEV upon Passaging in N. tabacum 35S::NIb Plants
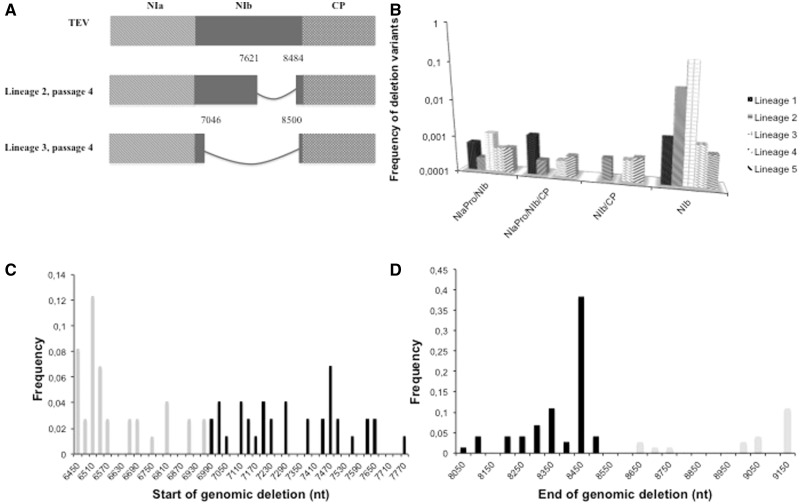
To test whether there will indeed be rapid accumulation of virus variants with large deletions as suggested by estimates of W, we performed serial passage experiments. Here, we chose to perform four serial passages, with each passage having 3 weeks of duration, because genome size appears to be under stronger selection in long passages (Zwart et al. 2014). Five independent lineages of TEV were evolved. After four passages, deletions were detected by RT-PCR in two lineages: Lineage 2 and Lineage 3 (fig. 2A). These two deletions (nucleotide positions 7621–8484 and 7046–8500) conserved the reading frame and were situated within the NIb cistron (positions 6982–8517 of the wild-type genome).
Fig. 2.—
(A) TEV and deletion variants detected by RT-PCR in different lineages at passages one and four. After four 3-week passages, NIb deletion variants are observed in different lineages as, for example, Lineage 4 and Lineage 5. All deletions observed after RT-PCR are situated within NIb and retain the reading frame. (B) Histogram of the frequency of different deletion variants observed in the five evolved lines. (C) Frequency distribution of the start nucleotide position of genomic deletions in the evolved lineages. (D) Frequency distribution of the end nucleotide position of genomic deletions in the evolved lineages. In (C) and (D), black bars indicate positions within the NIb cistron, gray bars indicate regions outside NIb, that is, in (C) within the NIaPro cistron and in (D) within the CP cistron.
To detect virus deletion variants with much greater sensitivity, Illumina NGS was performed on these five TEV lineages evolved for four 3-week passages. A TEV genome region encompassing the NIaPro, NIb, and CP cistrons (positions 6377–9473 of the wild-type genome) was amplified and sequenced (Materials and Methods). We developed a new approach for identifying deletions, adapting GSNAP (Wu and Nacu 2010) to detect deletions instead of introns (Materials and Methods). As a negative control, the same region was analyzed using NGS data for five lineages of TEV-eGFP evolved for three 9-week passages in wild-type N. tabacum plants (Zwart et al. 2014). No deletions were detected in the entire region, suggesting that our NGS analysis does not generate artifacts. The frequencies of deletion variants (see supplementary table S1, Supplementary Material online) in systemically infected leaves of transgenic plants were significantly higher than the frequencies found in our negative controls (Fisher’s exact test, P < 0.001). The NGS results were consistent with RT-PCR data; the deletion variant detected by RT-PCR in Lineage 2 (positions 7621–8484) and Lineage 3 (positions 7046–8500) was the variant found at the highest frequency by NGS, 0.249 and 0.246, respectively. Sixty-four different deletion variants were observed in total, ranging in frequency from 8.44 × 10−5 (positions 6538–8478; Lineage 4) to 0.249 (positions 7621–8484; Lineage 2). Interestingly, 6.25% of deletion variants disrupted the reading frame of the coding sequence. Moreover, 43.75% of deletion variants extended into genes flanking NIb, meaning that probably only the remaining 56.25% variants could be viable when transcomplemented by the NIb expressed by the plant.
In 17 deletion variants, the 5′ end of the deletion extended into NIaPro; the frequencies of these variants in the virus populations range from 8.44 × 10−5 (positions 6538–8478; Lineage 4) to 1.73 × 10−3 (positions 6460–8478; Lineage 4). Furthermore, in eight other variants the deletion started within NIaPro (upmost 5′ position being 6516) and ended within CP (downmost 3′ position being 9163), varying in frequency from 1.02 × 10−4 (positions 6923–9013; Lineage 4) to 1.88×10−3 (positions 6569–8746; Lineage 1). The 43.75% of genotypes carrying deletions larger than the NIb cistron will produce viruses with truncated cistrons NIaPro, CP, or both. Moreover, deletions beyond the NIb cistron will lead to the loss of proteolytic cleavage sites and result in fusion of the truncated proteins. These genomes will probably be defective particles and some may additionally interfere with replication of the full-length virus. These defective particles can only replicate if they are complemented by a full-length TEV or possibly other defective genomes, and their spread may therefore be limited. The highest frequency of putatively defective deletion variants does not exceed 1%, in line with our expectations: two NIaPro-to-CP deletion variants (positions 6569–8746 and 6774–8795; Lineage 1) show frequencies of 1.88 × 10−3 and 1.58 × 10−3, respectively. All lineages are dominated by variants with NIb deletion only, which is consistent with the fact that these variants are viable. Although defective viruses are often maintained in virus populations by complementation (Roux et al. 1991; Moreno et al. 1997; Froissart et al. 2004; García-Arriaza et al. 2004), the low level of coinfection (Dietrich and Maiss 2003; Tromas et al. 2014) may rapidly eliminate defective viruses from potyvirus populations.
The ends of the genomic deletions found by NGS (fig. 2C and D) were not uniformly distributed (Kolmogorov–Smirnov tests: 5′ end n = 42, P < 0.001; 3′ end n = 23, P < 0.001). Surprisingly, the 5′ start of the genomic deletion was clustered in NIaPro, suggesting a possible recombination hotspot outside of the NIb cistron. For the 3′ end of the deletion, ending sites clustered toward the 3′ end of NIb cistron. This clustering is consistent with the idea that we are more likely to detect deletion variants that only affect part of NIb and are, therefore, likely viable by complementation in trans.
Our results demonstrate that rapid deletions of NIb occurred upon passaging in N. tabacum 35S::NIb. In two lineages, these variants reached high frequencies, comprising approximately a fourth of the population. These results are therefore a proof of principle that NIRV expression could have a similar effect on a virus population: allowing deletion mutants to be maintained in the virus populations, while also providing conditions in which these mutants could reach high frequencies. The immediate effect of these deletions will be that a part of the virus population is replication defective in host individuals that do not express the NIRV. This could have important ramifications for viral spread, and virus–host coevolution. On the other hand, the further evolution of a virus population in a host expressing virus genes may also be contingent upon the presence of genomic deletions in the viral genome.
TEV Evolution in N. tabacum 35S::NIb Plants Is Contingent upon the Presence of NIb in the Viral Genome
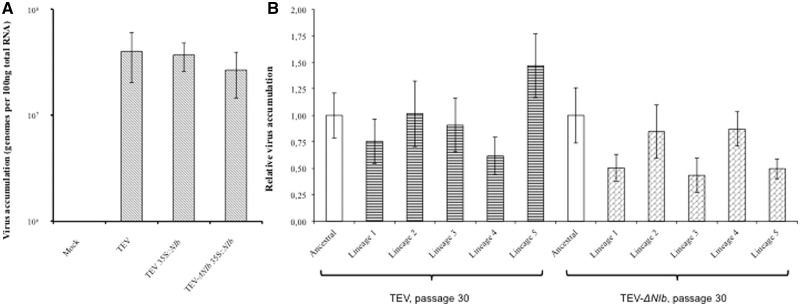
As we just described, serial passages of TEV in N. tabacum 35S::NIb led to the rapid occurrence of a whole spectrum of deletion variants. How would these deletion mutants further evolve in transgenic plants, and would their evolution be different from that of a full-length TEV? To address this question and gain insight into the long-term evolution of viruses harboring NIRV-induced deletions, we performed a second evolution experiment for both TEV and TEV-ΔNIb passaged in N. tabacum 35S::NIb, with five independent lineages for each virus. Li and Carrington (1995) showed that beta-glucuronidase (GUS) activity in leaves systemically infected by the TEV-ΔNIb-GUS mutant was comparable to that of the parental TEV-GUS. We found a similar result for the accumulation of TEV and TEV-ΔNIb in N. tabacum 35S::NIb using RT-qPCR (fig. 3A): there was not a significant difference in virus accumulation at 7 dpi (t8 = 1.394, P = 0.201). The infection dynamics of the two viruses should therefore be similar, allowing us to compare the evolutionary outcomes. However, to compare the evolution of the full-length TEV and TEV-ΔNIb, we had to devise a setup in which the integrity of the NIb gene is ensured for TEV. Two previous studies have found that marker genes were highly stable in the viral genome when many 1-week passages were performed, although the markers were quickly lost in longer passages (Dolja et al. 1993; Zwart et al. 2014). By performing 30 one-week passages with TEV and TEV-ΔNIb in N. tabacum 35S::NIb, we therefore hoped to avoid the occurrence of large genomic deletions in TEV. On the other hand, we realize that selection may not operate as effectively as during longer passages, due to short passage duration and frequent bottleneck associated to the mechanical transmissions among plants.
Fig. 3.—
(A) TEV and TEV-ΔNIb accumulation evaluated by absolute RT-qPCR after one passage in N. tabacum 35S::NIb transgenic plants. No significant difference in the accumulation of TEV and TEV-ΔNIb was observed. (B) Relative TEV and TEV-ΔNIb virus accumulation in N. tabacum 35S::NIb after 30 passages.
After 30 weeks of evolution, we measured virus accumulation of the evolved lineages (fig. 3B). The data were analyzed by ANOVA (table 1), and we found no effect of the virus type (TEV or TEV-ΔNIb) on accumulation, although there was a strong effect of lineage. The significant lineage-dependent effects suggest that random factors have played an important role during evolution, implicating genetic drift effects due to short passage duration. When pairwise comparisons were made between the evolved lineages and their respective ancestral viruses, Lineage 1 of TEV-ΔNIb had a significantly lower accumulation (t-test with Holm–Bonferroni correction: t7 = 4.001, P = 0.005). Accumulation of all other lineages was not significantly different from the ancestral virus, although the data suggest Lineage 3 and Lineage 5 of TEV-ΔNIb also have low accumulation (fig. 3B).
Table 1.
Nested ANOVA for Virus Accumulation of Evolved TEV and TEV-ΔNIb
| Source of Variation | SS | d.f. | MS | F | P |
|---|---|---|---|---|---|
| Virus | 0.768 | 1 | 0.768 | 1.663 | 0.233 |
| Lineage within virus | 3.721 | 8 | 0.465 | 3.779 | 0.003 |
| Error | 4.431 | 36 | 0.123 |
Note.—Viral accumulation data were normalized by the accumulation of the ancestral virus. SS is the sum of squares, d.f. is the degrees of freedom and MS is the mean square.
All evolved lineages of TEV and TEV-ΔNIb were fully sequenced to determine whether there were differences in their patterns of molecular evolution. We were particularly interested in understanding whether the use of NIb in trans may have affected the regulation of expression and accumulation of all other viral components. Mutational spectra for both viral genotypes were dominated by point mutations as not a single insertion or deletion was observed (table 2). As anticipated, the passaging setup used leads to the retention of NIb cistron by all TEV lineages, and by contrasting the evolution of TEV and TEV-ΔNIb, we are therefore considering the evolutionary effects of having a full-length or truncated genome. There were no differences between ratios of transitions to transversions; the ratio was 8.5 for TEV and 6.0 for TEV-ΔNIb (Fisher’s exact test: P = 1.000). Given that it is biochemically easier to produce transitions than transversions, it is not surprising to observe these biased ratios. Likewise, there was not a significant difference in the ratio of synonymous to nonsynonymous substitutions between the two viruses (Fisher’s exact test: P = 0.330); for TEV, this ratio was 1.375 whereas for TEV-ΔNIb it was 0.636. For both genotypes, mutations were evenly distributed along the genome (fig. 4A; Kolgomorov–Smirnov test: TEV n = 19, P = 0.330; TEV-ΔNIb n = 21, P = 0.688). An interesting example of convergent evolution was mutation A6805G (K2269E) that appeared in 80% of TEV lineages and in 40% of TEV-ΔNIb lineages. This nonsynonymous mutation is localized in a superficial domain of NIaPro (Nunn et al. 2005), and it could affect the proteinase function or modulate the interactions of NIaPro with other proteins. This same mutation has also been previously reported in serial-passage evolution experiments of TEV in N. tabacum: in 10% of lineages after 15 one-week passages (Bedhomme et al. 2012), and in 30% of lineages after three 9-week passages (Zwart et al. 2014).
Table 2.
Mutation Spectrum and Substitution Matrix for TEV and TEV-ΔNIb Evolved Genome after 30 one-week Passages in N. tabacum 35S::NIb Transgenic Plants
| Type of Mutations |
||
|---|---|---|
| TEV | TEV-ΔNIb | |
| Total | 19 | 21 |
| Base substitution | 19 | 21 |
| Transitions | 17 | 18 |
| Transversions | 2 | 3 |
| Synonymous | 11 | 7 |
| Nonsynonymous | 8 | 11 |
| Untranslated regions | 0 | 3 |
| Substitution matrices | ||||||||||
| A | U | G | C | A | U | G | C | |||
| A | — | 0 | 5 | 0 | A | — | 0 | 9 | 0 | |
| U | 2 | — | 0 | 2 | U | 0 | — | 0 | 3 | |
| G | 6 | 0 | — | 0 | G | 4 | 1 | — | 0 | |
| C | 0 | 4 | 0 | — | C | 1 | 3 | 0 | — | |
Fig. 4.—
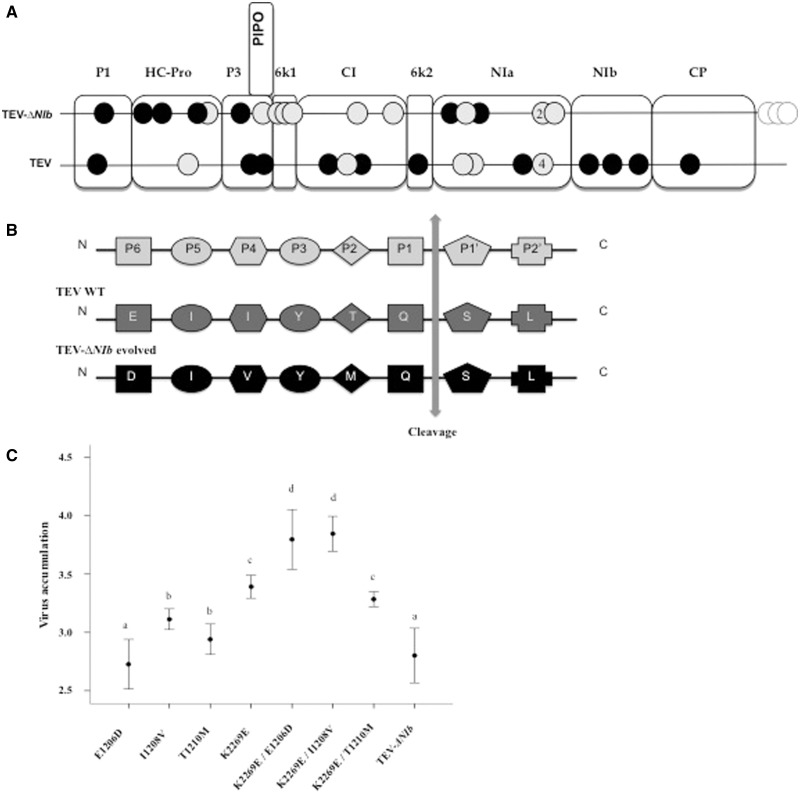
(A) Mutations found in TEV-ΔNIb and TEV consensus genome after 30 weekly passages in N. tabacum 35S::NIb transgenic plants. Black circles represent synonymous mutation, gray circles nonsynonymous mutation, and white circle mutations localized in UTRs. (B) TEV-ΔNIb mutations on the NIaPro proteolytic 6K1/CI site. P6 to P2′ (E-X-X-Y-X-Q-G/S) are key amino acids constituting the NIaPro binding site. Cleavage site is between P1 and P1′ (vertical arrow). (E) (26.5%) and (D) (12.8%) are the most frequent amino acids at P6 position in the family Potyviridae; V (80%) is the most frequent in position P4; H (40%) and F (15%) are the most common in position P2 among the Potyviridae, M is represented with a frequency of only 2%. Generally, mutations at positions P2/P4 may involve regulation of the cleavage rate (Dougherty et al. 1989). (C) Accumulation of mutants E1206D, I1208V, T1210M, K2269E, and the combinations K2269E/E1206D, K2269E/I1208V, and K2269E/T1210M. Mutant viruses belong to four groups (A–D) with homogeneous accumulation (Holm-Bonferroi post hoc test; P < 0.05).
In three lineages of TEV-ΔNIb, we observed three nonsynonymous mutations localized in the 6K1/CI junction (fig. 4B): G3618U (E1206D) (Lineage 1), A3622G (I1208V) (Lineage 5), and C3629U (T1210M) (Lineage 3), affecting residues P6, P4, and P2 of the proteolytic site, respectively (Adams et al. 2005). A conserved heptapeptide sequence has been identified at this site [E-X-X-Y-X-Q/S (or G)] at the 6K1/CI junction (Carrington and Dougherty 1987, 1988; Carrington et al. 1988; Adams et al. 2005). Each mutation was observed in a single lineage, and combinations thereof were not observed. The E1206D mutation at the P6 site may only weakly affect the proteolytic processing as amino acids E and D are widely represented in Potyviridae family at this site (Adams et al. 2005). On the other hand, I1208V at position P4 should have a stronger impact on processing activity; in only 7% of all Potyviridae is I present at this site, whereas V is present in 80% (Adams et al. 2005). Similarly, T1210M in P2 may also have a strong impact on processivity: T is found at the P2 site in only 2% of the Potyviridae (2%). Generally, mutation at sites P2/P4 may involve regulation of the cleavage rate (Dougherty et al. 1989).
We conclude that the evolution of TEV and TEV-ΔNIb in N. tabacum 35S::NIb was similar when transmission bottlenecks are close in time. Although there was a significant lineage-dependent effect on accumulation, only one evolved lineage of TEV-ΔNIb had a significantly lower accumulation than its ancestral virus. Moreover, patterns of molecular evolution were similar. There were no significant differences in the ratios of transitions to transversions, synonymous to nonsynonymous mutations, and the overall distribution of mutations. The one mutation that occurred in multiple lineages, A6805G, occurred in both lineages of TEV and TEV-ΔNIb. Therefore, the only conspicuous difference between the two viruses is mutations localized in the 6K1/CI junction.
Analysis of Mutations in the 6K1/CI Junction of TEV-ΔNIb
The K2269E, E1206D, I1208V, and T1210M mutations were introduced in the pMTEV infectious clone by site-directed mutagenesis. As K2269E is common among the different lineages, we also generated clones containing this mutation in combination with the other three. As a proxy for within-host fitness, we measured viral accumulation of each simple and double mutant at 2.67 dpi by RT-qPCR in three biological replicates. Data were fitted to a GLM with a log-link function and gamma distributed error structure, using replicate as factor nested within virus genotype. A significant effect of virus genotype on the observed viral accumulation was found (χ2 = 134.648, 7 d.f., P < 0.001), and a Holm–Bonferroni post hoc test identified four different homogenous groups (fig. 4C), with a being the group with the lowest viral accumulation value and d the highest. The results are generally in agreement with our expectations based on the prevalence of residues in the Potyviridae. E1206D has the same accumulation as the ancestral virus. The other single mutations all increased accumulation, being represented in groups b and c. The double mutations resulted in the highest accumulation levels, with two being the sole members of group d and one being represented in group c.
Thus, we conclude that all mutations except E1206D increase accumulation of TEV-ΔNIb early in infection and that this effect was even stronger for double mutations. High levels of early accumulation are likely to occur for a virus variant that expands rapidly, and therefore suggest an increase in the within-host fitness of the evolved lineages. The mutations in the 6K1/CI proteolytic site were not observed in the wild-type virus, suggesting that this is an instance of adaptive evolution contingent upon deletion of NIb.
TEV is translated into a large polyprotein, thus the cleavage process may be a mechanism to regulate the effective concentration of viral proteins in infected cells (Merits et al. 2002). Most of these mutations analyzed in this section showed a significant higher viral accumulation than the ancestral genotype. The mutations we found in evolved lineages may positively regulate the cleavage 6K1/CI in order to produce more mature 6K1 and CI proteins. NIb and CI are constituents of the viral replication complex (Li et al. 1997), and thus increasing the effective CI concentration may counterbalance the lower production of NIb from the transgene.
Concluding Remarks
NIRVs are thought to impact viruses by—in some cases—conferring virus resistance to host organisms (Koonin, 2010). However, these virus-derived sequences could also impact the evolution of virus populations. Here, we used an experimental model system to study the effects of NIRV expression on virus populations, using TEV and transgenic tobacco plants expressing its replicase, NIb. We found that 1) the loss of NIb cistron leads to higher within-host fitness in transgenic plants, 2) that genomic deletions quickly accumulate in a full-length virus passaged in transgenic plants, and 3) that the evolution of a virus population can be contingent upon the deletion of virus genes, although many aspects of phenotypic and molecular evolution were unchanged. Overall, our results therefore suggest that the expression of NIRVs could have far-reaching consequences for virus evolution. Moreover, these effects can occur on relatively short timescales (i.e., deletion variants were generated de novo and reached frequencies of ∼ 0.25 within four 3-week passages). These changes on short timescales support the notion that plausible epidemiological models of virus spread in NIRV-expressing host populations need to take virus evolution into consideration. For example, if large genomic deletions occur rapidly in NIRV-expressing hosts, then eventually many virus-infected hosts will only be infectious to those hosts expressing the NIRV. The generation of various classes of infectious hosts could in this case decrease viral transmission between hosts, an effect that could be further augmented by the clustering of NIRV-expressing hosts.
Our results provide further support for the notion of a relationship between genome size and replication/within-host competitive fitness (Sakai et al. 1999; Marks et al. 2005; Zwart et al. 2010, 2014), although there are clearly limitations to this idea (Bull et al. 2004). Not only could TEV-ΔNIb outcompete TEV in transgenic plants, also all independent TEV lineages accumulate various NIb deletions upon passaging. We identified 64 different deletion variants, most of which conserved the reading frame. In 17 deletion variants the deletion extended far into the upstream cistron, NIaPro, while for eight deletion variants, the deletion also extended into the downstream cistron, CP. Because both NIaPro and CP proteins are essential for virus replication and these deletions removed one or more proteolytic cleavage sites, the virus variants probably cannot replicate autonomously, even in the transgenic plants. Therefore, the virus population in the simulated NIRV-expressing host appears to have generated two types of defective viruses: 1) those with deletions in NIb only and can replicate autonomously in the transgenic plants but not in the wild-type plants and 2) those with deletions extending beyond the NIb cistron, which cannot replicate autonomously in either host. On the basis of these observations, we speculate that the NIRV-expressing hosts may be a source of defective genotypes in natural virus populations. If a deletion genotype arises de novo in a NIRV-expressing host, it will not require coinfection with a full-length virus to be maintained in the population. Consequently, it may more easily reach intermediate to high frequencies in a NIRV-expressing plant, allowing the defective virus to be sustained even in non-NIRV hosts by coinfection with full-length viruses (Montville et al. 2005). Complementation can slow down the rate at which deleterious alleles are eliminated from the virus population (Froissart et al. 2004; Sardanyés and Elena 2010) and promote selection for defective-interfering particles as cheater genotypes that reduce mean population fitness (Turner and Chao 1999).
Our study has important implications for the development and agronomic use of transgenic plants resistant to viruses generated by inserting complete viral genes into the plant genome, in particular by inserting viral replicase genes (reviewed in Kundu and Mandal [2001]). From a biosafety standpoint, it has been known for several years that recombination between viral sequences transgenically expressed by plants and virus genomes may occur (Turturo et al. 2008; Morroni et al. 2009), raising concerns about whether this transgenic strategy would favor the emergence of new viruses (Tepfer 2002; Thompson and Tepfer 2010). Here, we have illustrated a different evolutionary scenario: the expression by the plant of functional viral proteins creates the conditions in which viruses with long or even full deletions of the complemented protein may dominate the population, thus precluding their spread in nontransgenic plants.
Finally, it is fair to mention that our experimental system remains artificial to some extent. Although many examples of DNA viruses using host enzymes for replication exist (e.g., geminiviruses and polydnaviruses), no RNA virus has been described so far that does not encode for its own functional RdRp, even though certain plants encode functional RdRps of viral origin in their genomes, for example, the narnavirus-like RdRp found in the mitochondria of Arabidopsis thaliana (Hong et al. 1998). In this respect, a critical reader may argue that RdRp was not the best choice for studying the pseudogenization of viral genes in the presence of a homologous NIRV. However, our results should be strong enough to convince such a reader that even a highly constrained gene, such as RdRp can be lost from viral genomes if provided in trans by the host.
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Francisca de la Iglesia, Paula Agudo, and Àngels Pròsper for excellent technical support, Dr José-Antonio Daròs for gifting pMTEV, and the Bioinformatics Core Service at the IBMCP for the support provided. This project was supported by grant 22371 from the John Templeton Foundation to S.F.E. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation. Additional support was provided by the Spanish Ministerio de Economía y Competitividad (MINECO) grant BFU2009-06993 and European Commission FP7-ICT program grant 610427-EvoEvo to S.F.E., by a predoctoral fellowship from MINECO to N.T., and by MINECO grant JCI2011-10379 and Rubicon grant from the Netherlands Organization for Scientific Research (www.nwo.nl) to M.P.Z.
Literature Cited
- Adams MJ, Antoniw JF, Beaudoin F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol. 2005;6:471–487. doi: 10.1111/j.1364-3703.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- Agudelo-Romero P, Carbonell P, Pérez-Amador MA, Elena SF. Virus adaptation by manipulation of host’s gene expression. PLoS One. 2008;3:e2397. doi: 10.1371/journal.pone.0002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001 doi: 10.1186/gb-2001-2-3-research0007. 2:research0007.1–research0007.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme S, Lafforgue G, Elena SF. Multihost experimental evolution of a plant RNA virus reveals local adaptation and host-specific mutations. Mol Biol Evol. 2012;29:1481–1492. doi: 10.1093/molbev/msr314. [DOI] [PubMed] [Google Scholar]
- Bedoya LC, Daròs JA. Stability of Tobacco etch virus infectious clones in plasmid vectors. Virus Res. 2010;149:234–240. doi: 10.1016/j.virusres.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch C, et al. Retention of the virus-derived sequences in the nuclear genome of grapevine as a potential pathway to virus resistance. Biol Direct. 2009;4:21. doi: 10.1186/1745-6150-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borja M, Rubio T, Scholthof HB, Jackson AO. Restoration of wild-type virus by double recombination of tombusvirus mutants with a host transgene. Mol Plant Microbe Interact. 1999;12:153–162. doi: 10.1094/MPMI.1999.12.2.153. [DOI] [PubMed] [Google Scholar]
- Bratlie MS, Drabløs F. Bioinformatic mapping of AlkB homology domains in viruses. BMC Genomics. 2005;6:1. doi: 10.1186/1471-2164-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Springman R, Molineux IJ. Genome properties and the limits of adaptation in bacteriophages. Evolution. 2004;58:692–701. doi: 10.1111/j.0014-3820.2004.tb00402.x. [DOI] [PubMed] [Google Scholar]
- Carrasco P, de la Iglesia F, Elena SF. Distribution of fitness and virulence effects caused by single-nucleotide substitutions in Tobacco etch virus. J Virol. 2007;81:12979–12984. doi: 10.1128/JVI.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Cary SM, Dougherty WG. Mutational analysis of Tobacco etch virus polyprotein processing: cis and trans proteolytic activities of polyproteins containing the 49-kilodalton proteinase. J Virol. 1988;62:2313–2320. doi: 10.1128/jvi.62.7.2313-2320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Dougherty WG. Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. J Virol. 1987;61:2540–2548. doi: 10.1128/jvi.61.8.2540-2548.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Dougherty WG. A viral cleavage site cassette: identification of amino acid sequences required for Tobacco etch virus polyprotein processing. Proc Natl Acad Sci U S A. 1988;85:3391–3395. doi: 10.1073/pnas.85.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Kavanagh T, Baulcombe D. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- Chiba S, et al. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011;7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BN, Canto T, Palukaitis P. Stability of recombinant plant viruses containing genes of unrelated plant viruses. J Gen Virol. 2007;88:1347–1355. doi: 10.1099/vir.0.82477-0. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. New York: Harper and Row; 1970. [Google Scholar]
- Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Maiss E. Fluorescent labeling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J Gen Virol. 2003;84:2871–2876. doi: 10.1099/vir.0.19245-0. [DOI] [PubMed] [Google Scholar]
- Dietrich C, et al. No recombination detected in artificial potyvirus mixed infections and between potyvirus derived transgenes and heterologous challenging potyviruses. Environ Biosafety Res. 2007;6:207–218. doi: 10.1051/ebr:2007042. [DOI] [PubMed] [Google Scholar]
- Dolja VV, Herndon KL, Pirone TP, Carrington JC. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J Virol. 1993;67:5968–5975. doi: 10.1128/jvi.67.10.5968-5975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Kreuze JF, Valkonen JP. Comparative and functional genomics of closteroviruses. Virus Res. 2006;117:38–51. doi: 10.1016/j.virusres.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, McBride HJ, Carrington JC. Tagging of plant potyvirus replication and movement by insertion of beta-glucuronidase into the viral polyprotein. Proc Natl Acad Sci U S A. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty WG, Cary SM, Parks TD. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Eigen M. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Flegel TW. Hypothesis for heritable, anti-viral immunity in crustaceans and insects. Biol Direct. 2009;4:32. doi: 10.1186/1745-6150-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froissart R, et al. Co-infection weakens selection against epistatic mutations in RNA viruses. Genetics. 2004;168:9–19. doi: 10.1534/genetics.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arriaza J, Manrubia SC, Toja M, Domingo E, Escarmís C. Evolutionary transition toward defective RNAs that are infectious by complementation. J Virol. 2004;78:11678–11685. doi: 10.1128/JVI.78.21.11678-11685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking P, Narasimamurthy R, Lemaitre B, Basler K, Leulier F. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PLoS One. 2009;4:e1007709. doi: 10.1371/journal.pone.0007709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- Gritsun TS, Gould EA. Direct repeats in the 3' untranslated regions of mosquito-borne flaviviruses: possible implications for virus transmission. J Gen Virol. 2006;87:3297–3305. doi: 10.1099/vir.0.82235-0. [DOI] [PubMed] [Google Scholar]
- Guo HS, López-Moya JJ, García JA. Susceptibility to recombination rearrangement of a chimeric Plum pox potyvirus genome after insertion of a foreign gene. Virus Res. 1998;57:183–195. doi: 10.1016/s0168-1702(98)00100-2. [DOI] [PubMed] [Google Scholar]
- Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford series in ecology and evolution (OSEE). Oxford: Oxford University Press. [Google Scholar]
- Hong Y, Cole TE, Brasier CM, Buck KW. Evolutionary relationships between putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology. 1998;246:158–169. doi: 10.1006/viro.1998.9178. [DOI] [PubMed] [Google Scholar]
- Horie M, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AS. Defective interfering viruses. Annu Rev Microbiol. 1973;27:101–117. doi: 10.1146/annurev.mi.27.100173.000533. [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC Biol. 2010;8:2. doi: 10.1186/1741-7007-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Mandal RK. Transgenic approaches for producing virus resistant plants. Proc Indian Natl Sci Acad B. 2001;67:53–80. [Google Scholar]
- Li XH, Carrington JC. Complementation of Tobacco etch potyvirus mutants by active RNA polymerase expressed in transgenic cells. Proc Natl Acad Sci U S A. 1995;92:457–461. doi: 10.1073/pnas.92.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, Valdez P, Olvera RE, Carrington JC. Functions of the Tobacco etch virus RNA polymerase (NIb): subcellular transport and protein–protein interaction with VPg/proteinase (NIa) J Virol. 1997;71:1598–1607. doi: 10.1128/jvi.71.2.1598-1607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Marks H, van Duijse JJ, Zuidema D, van Hulten MCW, Vlak JM. Fitness and virulence of an ancestral white spot syndrome virus isolate from shrimp. Virus Res. 2005;110:9–20. doi: 10.1016/j.virusres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Merits A, et al. Proteolytic processing of potyviral proteins and polyprotein processing intermediates in insect and plant cells. J Gen Virol. 2002;83:1211–1221. doi: 10.1099/0022-1317-83-5-1211. [DOI] [PubMed] [Google Scholar]
- Meyers G, Rümenapf T, Thiel HJ. Ubiquitin in a togavirus. Nature. 1989;341:491. doi: 10.1038/341491a0. [DOI] [PubMed] [Google Scholar]
- Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of mutational robustness in an RNA virus. PLoS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno IM, Malpica JM, Rodríguez-Cerezo E, García-Arenal F. A mutation in Tomato aspermy cucumovirus that abolishes cell-to-cell movement is maintained to high levels in the viral population by complementation. J Virol. 1997;71:9157–9162. doi: 10.1128/jvi.71.12.9157-9162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morroni M, Thompson JR, Tepfer M. Analysis of recombination between viral RNAs and transgene mRNA under conditions of high selection pressure in favor of recombinants. J Gen Virol. 2009;90:2798–2807. doi: 10.1099/vir.0.013771-0. [DOI] [PubMed] [Google Scholar]
- Nagai M, et al. Insertion of cellular sequence and RNA recombination in the structural protein coding region of cytopathogenic Bovine viral diarrhea virus. J Gen Virol. 2003;84:447–452. doi: 10.1099/vir.0.18773-0. [DOI] [PubMed] [Google Scholar]
- Nunn CM, et al. Crystal structure of Tobacco etch virus protease shows the protein C terminus bound within the active site. J Mol Biol. 2005;350:145–155. doi: 10.1016/j.jmb.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Paar M, et al. Effects of viral strain, transgene position, and target cell type on replication kinetics, genomic stability, and transgene expression of replication-competent Murine leukemia virus-based vectors. J Virol. 2007;81:6973–6983. doi: 10.1128/JVI.02470-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L, Simon AE, Holland JJ. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, et al. Accommodation of foreign genes into the Sendai virus genome: sizes of inserted genes and viral replication. FEBS Lett. 1999;456:221–226. doi: 10.1016/s0014-5793(99)00960-6. [DOI] [PubMed] [Google Scholar]
- Sardanyés J, Elena SF. Error threshold in RNA quasispecies models with complementation. J Theor Biol. 2010;265:278–286. doi: 10.1016/j.jtbi.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Satyanarayana T, et al. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology. 2000;278:253–265. doi: 10.1006/viro.2000.0638. [DOI] [PubMed] [Google Scholar]
- Shackelton LA, Holmes EC. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends Microbiol. 2004;12:458–465. doi: 10.1016/j.tim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Smith JM, Szathmáry E. The major evolutionary transitions. Nature. 1995;374:227–232. doi: 10.1038/374227a0. [DOI] [PubMed] [Google Scholar]
- Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the Coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaimuthu J, Tzanetakis IE, Gergerich RC, Martin RR. A member of a new genus in the Potyviridae infects Rubus. Virus Res. 2008;131:145–151. doi: 10.1016/j.virusres.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Swetina J, Schuster P. Self-replication with errors. A model for polynucleotide replication. Biophys Chem. 1982;16:329–345. doi: 10.1016/0301-4622(82)87037-3. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Bruenn J. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 2009;7:88. doi: 10.1186/1741-7007-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepfer M. Risk assessment of virus-resistant transgenic plants. Annu Rev Phytopathol. 2002;40:467–491. doi: 10.1146/annurev.phyto.40.120301.093728. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Tepfer M. Assessment of the benefit and risks for engineered virus resistance. Adv Virus Res. 2010;76:33–56. doi: 10.1016/S0065-3527(10)76002-4. [DOI] [PubMed] [Google Scholar]
- Tromas N, Elena SF. The rate and spectrum of spontaneous mutations in a plant RNA virus. Genetics. 2010;185:983–989. doi: 10.1534/genetics.110.115915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas N, Zwart MP, Lafforgue G, Elena SF. Within-host spatiotemporal dynamics of plant virus infection at the cellular level. PLoS Genet. 2014;10:e1004186. doi: 10.1371/journal.pgen.1004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Chao L. Prisoner’s dilemma in an RNA virus. Nature. 1999;398:441–443. doi: 10.1038/18913. [DOI] [PubMed] [Google Scholar]
- Turturo C, et al. Evaluation of potential risks associated with recombination in transgenic plants expressing viral sequences. J Gen Virol. 2008;89:327–335. doi: 10.1099/vir.0.83339-0. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Dieu BTM, Hemerik L, Vlak JM. Evolutionary trajectory of White spot syndrome virus (WSSV) genome shrinkage during spread in Asia. PLoS One. 2010;5:e13400. doi: 10.1371/journal.pone.0013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Willemsen A, Daròs JA, Elena SF. Experimental evolution of pseudogenization and gene loss in a plant RNA virus. Mol Biol Evol. 2014;31:121–134. doi: 10.1093/molbev/mst175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.