Abstract
Photosynthesis by diatoms accounts for roughly one-fifth of global primary production, but despite this, relatively little is known about their plastid genomes. We report the completely sequenced plastid genomes for eight phylogenetically diverse diatoms and show them to be variable in size, gene and foreign sequence content, and gene order. The genomes contain a core set of 122 protein-coding genes, with 15 additional genes exhibiting complex patterns of 1) gene losses at varying phylogenetic scales, 2) functional transfers to the nucleus, 3) gene duplication, divergence, and differential retention of paralogs, and 4) acquisitions of putatively functional recombinase genes from resident plasmids. The newly sequenced genomes also contain several previously unreported genes, highlighting how poorly characterized diatom plastid genomes are overall. Genome size variation reflects major expansions of the inverted repeat region in some cases but, more commonly, large-scale expansions of intergenic regions, many of which contain unique open reading frames of likely foreign origin. Although many gene clusters are conserved across species, rearrangements appear to be frequent in most lineages.
Keywords: chloroplast, diatoms, genomes, plastid, horizontal gene transfer
Introduction
Diatoms are photosynthetic algae within the large and diverse heterokont lineage, which includes brown algae, golden algae, and more distantly related nonphotosynthetic taxa, including the pathogenic water mold, Phytophthora (a small number of diatoms are secondarily nonphotosynthetic too [Li and Volcani 1987]). Like cryptophytes, haptophytes, and most dinoflagellates, the plastids of diatoms—like all plastid-bearing heterokonts—trace their origin to a secondary endosymbiosis with a red alga (Archibald 2009). Primary and secondary “red” lineages are now principal components of marine ecosystems and important contributors to the global cycling of carbon and oxygen (Falkowski et al. 2004). Diatoms, in particular, are prolific photosynthesizers, responsible for roughly 20% of global net primary production (Nelson et al. 1995). By fixing and exporting massive amounts of carbon from the atmosphere to the deep ocean, diatoms are primary drivers of the “biological pump” (Hopkinson et al. 2011). Their photosynthetic output reflects the vast breadth of their ecological and phylogenetic diversity, sheer numerical abundance, and Form ID Rubisco enzyme, which has an unusually high affinity and selectivity for carbon dioxide (Roberts et al. 2007). Moreover, their photosynthetic products include a suite of energy-rich lipids and complex polysaccharides that are a primary entry point of carbon into marine food webs (Kroth et al. 2008).
Plastid genome data from primary and secondary red lineages have revealed substantial differences in genome size, gene content, and gene order. Compared with their counterparts in the green lineage (green algae and land plants), both primary and secondary red plastid genomes tend to have more genes, minimal intergenic space, little repetitive sequence, and few if any introns (Green 2011). To date, the plastid genomes of seven diatoms and two dinoflagellates with diatom-derived plastids have been sequenced. These genomes have a moderate gene content (158–162 genes), intermediate between haptophytes and cryptophytes (Green 2011). Introns are rare, with just one report of an intron in the atpB gene of Seminavis robusta (Brembu et al. 2013). Finally, unlike their primary red algal progenitors, diatom plastid genomes appear to be highly rearranged (Oudot-Le Secq et al. 2007), even between close relatives (Lommer et al. 2010).
Diatoms are an extraordinarily diverse lineage (Mann and Vanormelingen 2013), so the small sample of sequenced plastid genomes has precluded meaningful insights into broad-scale patterns of evolution. We sequenced plastid genomes for eight diverse diatoms, doubling the number of sequenced genomes and filling in several important phylogenetic gaps, including taxa that bracket some of the earliest splits in the phylogeny. This expanded taxonomic sampling showed that diatom plastid genomes are particularly labile in size, structure, and sequence content.
Materials and Methods
Diatom Cultures, DNA Extraction, and Sequencing
Culture information, growth conditions, and sequencing strategies for the eight newly sequenced genomes are summarized in table 1. Didymosphenia could not be cultured, so six individual cells were isolated from a sample collected in Boulder Creek, Colorado, USA, and whole-genome amplification was performed on each cell using the Qiagen REPLI-g Mini Kit. The six amplification products were then pooled for sequencing.
Table 1.
Culturing, Sequencing, and Assembly Information for the Eight Newly Sequenced Diatom Plastid Genomes
| Taxon | GenBank Accession | Culture Collection | Strain ID | Growth Medium | Sequencing Platform | Sequence Assembler |
|---|---|---|---|---|---|---|
| Leptocylindrus danicus | KC509524 | NCMA | CCMP1856 | F/2 | Roche 454, Illumina HiSeq | Newbler, ABySS |
| Coscinodiscus radiatus | KC509521 | NCMA | CCMP310 | F/2 | Roche 454 | Newbler |
| Lithodesmium undulatum | KC509525 | NCMA | CCMP1797 | F/2 | Roche 454 | Newbler |
| Asterionellopsis glacialis | KC509520 | NCMA | CCMP1717 | F/2 | Roche 454 | Newbler |
| Asterionella formosa | KC509519 | CPCC | UTCC605 | COMBO | Roche 454 | Newbler |
| Eunotia naegelii | KF733443 | UTEX | FD354 | COMBO | Illumina MiSeq | ABySS, Ray |
| Cylindrotheca closterium | KC509522 | NCMA | CCMP1855 | F/2 | Roche 454 | Newbler |
| Didymosphenia geminata | KC509523 | NAa | BCCO11 | F/2 | Illumina HiSeq | ABySS, Ray |
Note.—NCMA, Provasoli-Guillard National Center for Marine Algae and Microbiota; CCPC, Canadian Phycological Culture Centre at the University of Toronto; UTEX, The Culture Collection of Algae at The University of Texas at Austin
aEnvironmental sample, Boulder Creek, Colorado, USA, April 2011.
For Eunotia, we disrupted frozen cell pellets by agitating them with glass beads in a Mini-Beadbeater-24 (BioSpec Products) before extracting total genomic DNA with the Qiagen DNeasy Plant Mini Kit. For the remaining species, we isolated plastid DNA by resuspending frozen cells in 10–15 ml of resuspension buffer (50 mM Tris [pH 8.0], 25 mM ethylenediaminetetraacetic acid, and 50 mM NaCl) and disrupting them by nitrogen decompression with a Parr Cell Disruption Bomb at 750–800 psi for 20–30 min. Plastids were lysed by shaking them at 100 rpm for 60 min at 50 °C in a solution containing 250 µl of 20% Triton X-100 and 1 ml Pronase (10 mg/ml) per 10 ml of cell slurry. We then added equal weight cesium chloride (CsCl) and mixed the slurry until the CsCl was fully dissolved and dispensed it into 6 ml PA Ultracrimp tubes (Sorvall) with 50 µl of ethidium bromide (EtBr) (10 mg/ml). After centrifugation at 65,000 rpm in a Sorvall TV-1665 rotor for 12 h, we extracted the DNA bands and removed EtBr with repeated washes in salt-saturated isopropanol. The spin was repeated with 40 µl Hoechst 33258 dye (10 mg/ml H2O). Following the spin, the DNA bands were extracted and Hoechst dye removed by repeated 1:1 washes with salt-saturated isopropanol. We removed the CsCl by dialysis in TE buffer with buffer changes every 12 h for 48 h.
We used three different DNA sequencing platforms, individually or in combination, to generate the data (table 1). Roche 454 GS-FLX sequencing (Titanium reagents) generated 500-bp single-end reads and was carried out at the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois. The Illumina HiSeq 2000 platform generated 100-bp paired-end reads, used libraries of length 300 bp, and was carried out at the Genome Sequencing and Analysis Facility at the University of Texas at Austin. Finally, the Eunotia genome was sequenced using the Illumina MiSeq platform at the Institute for Genomics and Systems Biology at Argonne National Laboratory, using a 300-bp library and 150-bp paired-end reads.
Genome Assembly and Analysis
We used Newbler, ABySS ver. 1.3, or Ray ver. 2.2.0 (Simpson et al. 2009; Boisvert et al. 2010) to assemble the reads (table 1), and Geneious ver. 5.4 (Biomatters Ltd., Auckland, New Zealand) or Sequencher ver. 4.5 (Gene Codes Corporation, Ann Arbor, MI, USA) to guide finishing of the assemblies. Protein genes were annotated with DOGMA (Wyman et al. 2004), and predicted tRNAs and tmRNAs were identified with ARAGORN (Laslett and Canback 2004). Boundaries of the rRNA and ffs genes were delimited by direct comparison to sequenced diatom genomes with NCBI-BLASTN. We identified pseudogenes based on their BLASTN similarity to functional homologs (e-value ≤ 1e−6) and, in most cases, by their conserved positions in the genome.
We used NCBI-BLASTP to search the nuclear genomes of Phaeodactylum tricornutum, Thalassiosira pseudonana, and Thalassiosira oceanica for genes missing from one or more plastid genomes. NCBI's ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/, last accessed March 24, 2014) was used to search intergenic regions for open reading frames (ORFs) ≥100 amino acids in length. Intergenic sequences were considered unique if they had no match to a local database consisting of 37 primary and secondary red plastid genomes and three plastid-localized diatom plasmids, based on a BLAST search with an e-value cutoff of 1e−6 and the following search parameters: word size = 9; reward = 2; mismatch penalty = −3; gap opening = 5, and gap extension = 2. Whole-genome alignments were performed using progressive MAUVE ver. 2.3.1 with default parameters (Darling et al. 2010).
Phylogenetic Analyses
Sequence alignments for acpP and tsf included the diatoms sequenced for this study, all fully sequenced primary and secondary red plastid genomes that contained the gene, and any nuclear homologs found in the sequenced diatom nuclear genomes. All genes were manually aligned using MacClade ver. 4.08 (Maddison and Maddison 2005).
To account for taxon-specific amino acid compositional heterogeneity (e.g., in nuclear vs. plastid genes), tree inference was performed with NH-PhyloBayes ver. 0.2.3 using the time- and site-heterogenous model (CAT-BP) (Blanquart and Lartillot 2006, 2008). We ran two MCMC chains for 2 × 103 (tsf) or 3.2 × 103 (acpP) generations, sampling every tenth cycle. The number of categories was set to 30 (tsf) and 20 (acpP), corresponding to the mean of the posterior distribution of this parameter estimated from a PhyloBayes analysis under the GTR+G+CAT substitution model (Lartillot and Philippe 2004; Lartillot et al. 2013). Convergence and stationarity of runs was assessed through the built-in diagnostics in NH-PhyloBayes after discarding the first 103 samples as the burnin.
Results and Discussion
General Features
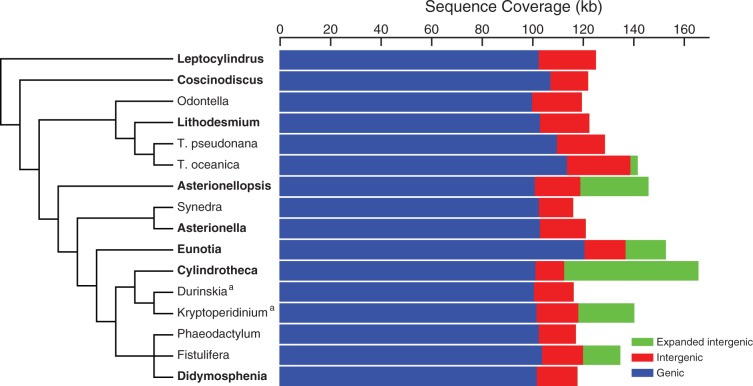
Each plastid genome mapped as a single, circular chromosome with two large inverted repeats (IR) separating small (SSC) and large single-copy (LSC) regions. The eight genomes ranged in size from 118 kb in Didymosphenia to 166 kb in Cylindrotheca (fig. 1). Diatom plastid genomes share a core set of 122 protein-coding genes, 3 rRNAs, 27 tRNAs, and 2 additional RNA genes, tmRNA and ffs (supplementary table S1, Supplementary Material online).
Fig. 1.—
Sequence coverage by protein-coding, intergenic, and expanded (≥1 kb in length) intergenic regions in diatom plastid genomes. Bars are drawn proportional to the genome size and show the fraction of the genome occupied by these three sequence categories. Taxa in boldface identify genomes sequenced for this study. Phylogenetic relationships were redrawn from Theriot et al. (2010) and unpublished data. Taxa marked with a superscript “a” are dinoflagellates with diatom-derived plastids (Imanian et al. 2010).
Nucleotide composition is highly conserved, with G+C (GC) content ranging from 29% to 32% across the eight genomes. GC content of protein-coding genes ranged from 30% to 33%, mirroring that of the overall genome, whereas intergenic values were substantially lower—just 16–20% in most species. The Asterionellopsis, Eunotia, and Cylindrotheca genomes contained large amounts of comparatively GC-“rich” expanded intergenic DNA (but still low: 28% GC), driving up their overall intergenic GC content to as high as 27% in some species (supplementary table S2, Supplementary Material online).
Genome Expansions
Expansions of the Inverted Repeat Region
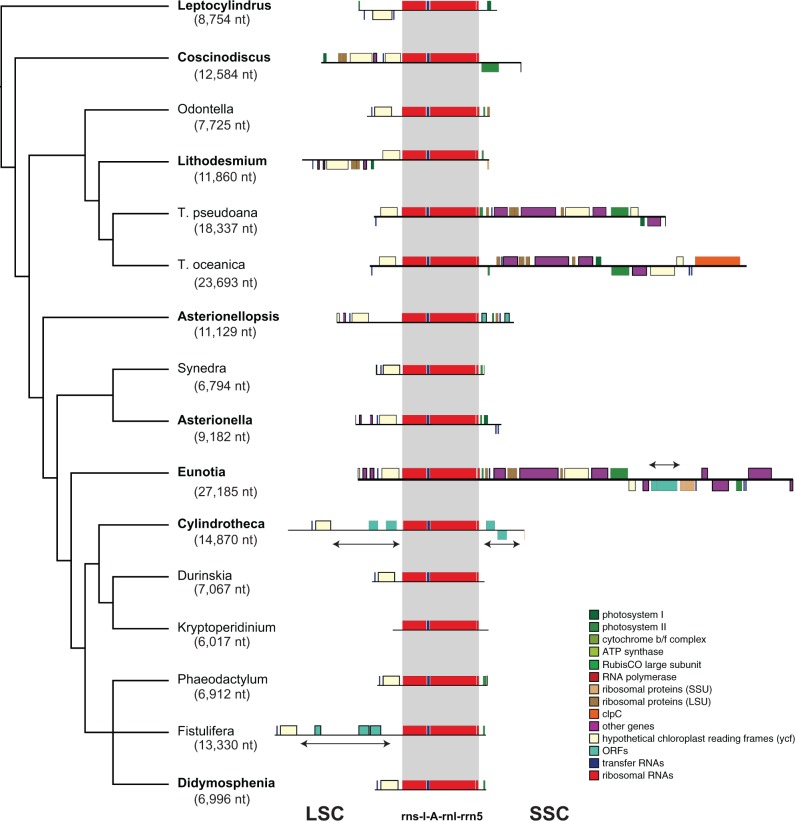
The eight newly sequenced plastid genomes include the largest so-far sequenced from diatoms, substantially expanding their known size range. Expansion and contraction of the IR accounts for most of the size variation in angiosperm plastid genomes (Plunkett and Downie 2000), and similarly, the IR in diatoms varies in length by nearly 4-fold—from 7 kb in Didymosphenia to 27 kb in Eunotia (fig. 2). This variation reflects several independent IR expansions and, very likely, contractions. Expansions have been bi-directional, incorporating parts of one or both of the LSC and SSC regions (fig. 2). In some cases, IR expansions have resulted in a large number of gene duplications. The IR expansions in T. pseudonana and T. oceanica resulted in the duplication of more than a dozen plastid genes (fig. 2). The largest IR expansion occurred in Eunotia, resulting in the duplication of >20 genes and an 18 kb increase in IR length compared with Asterionella (fig. 2). As a result, the IR (27 kb) is now larger than the SSC region (25 kb) in Eunotia.
Fig. 2.—
Size variation of the inverted repeated region (IRa) across 16 fully sequenced plastid genomes in diatoms. Colored boxes circumscribe genes in various functional categories, with those above the line transcribed on the forward strand and vice versa for genes below the line. Maps are drawn to scale, and the gray box demarcates the core rns-trnI-trnA-rnl-rrn5 gene cluster conserved across the 16 genomes. Newly sequenced taxa from this study are in boldface and the nucleotide length of IRa are in parentheses beneath each taxon name. Double arrows delimit large putatively foreign sequence insertions.
Expansions of Intergenic Regions
Variation in plastid genome size primarily reflected differences in the amount of intergenic DNA, which comprises 12–39% (15–65 kb) of the genome in the eight newly sequenced genomes (fig. 1). Diatom plastid genomes are generally compact, with any given intergenic region rarely exceeding 500 bp in length. The plastid genomes of six species—T. oceanica, Asterionellopsis, Eunotia, Cylindrotheca, Kryptoperidinium, and Fistulifera—are, however, larger than average due to the presence of numerous expanded intergenic regions of ≥1 kb in length (fig. 1). These regions are spread across a dozen or so locations in the genomes and range in length from 1 to 10 kb, accounting for anywhere from 3 to 53 kb (2–32%) of the overall genome in these six species (fig. 1).
While a small fraction (<3% in all cases) of these “extra” intergenic sequences can be traced to diatom plasmids, the majority are of unknown origin. Excluding plasmid-derived sequences, roughly 68–99% of the expanded intergenic sequences are species-specific, showing no similarity to sequenced primary or secondary red (including diatoms) plastid or plasmid genomes; roughly a quarter of the large Cylindrotheca plastid genome has no matches to GenBank sequences of any kind. The expanded intergenic sequences have significantly higher GC content (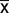 = 29%) compared with the small, highly AT-rich (
= 29%) compared with the small, highly AT-rich (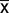 = 19%) intergenic regions ancestrally present in diatom plastid genomes, strongly suggesting that the expanded regions have a different ancestry (Lawrence and Ochman 1997; Ragan et al. 2006). Many of these regions also contain long, unique ORFs. Considering only those ORFs ≥100 amino acids in length and with canonical start and stop codons, we found a total of 64 of them across the six species with expanded intergenic regions (fig. 1). ORFs ranged from 100 to 439 amino acids in length, and notably, just four of them were shared between any two genomes.
= 19%) intergenic regions ancestrally present in diatom plastid genomes, strongly suggesting that the expanded regions have a different ancestry (Lawrence and Ochman 1997; Ragan et al. 2006). Many of these regions also contain long, unique ORFs. Considering only those ORFs ≥100 amino acids in length and with canonical start and stop codons, we found a total of 64 of them across the six species with expanded intergenic regions (fig. 1). ORFs ranged from 100 to 439 amino acids in length, and notably, just four of them were shared between any two genomes.
Similar types of anonymous stretches of intergenic DNA have been found in other primary and secondary red plastid genomes, though not to this extent (Cattolico et al. 2008; Janouškovec et al. 2013). Additional comparative genomic data will help winnow in on the timing of these acquisitions and, hopefully, show whether they reflect an extreme case of differential loss of ancestral sequences or acquisitions of foreign DNA. A foreign origin seems much more plausible, however, considering that the differential loss model requires that the ORFs—found in no other primary or secondary red plastid genomes—were present in the ancestral diatom plastid genome, maintained or subsequently evolved an aberrantly high GC content, and experienced an exceptional pattern of repeated loss.
Gene Acquisitions, Losses, and Functional Transfers to the Nucleus
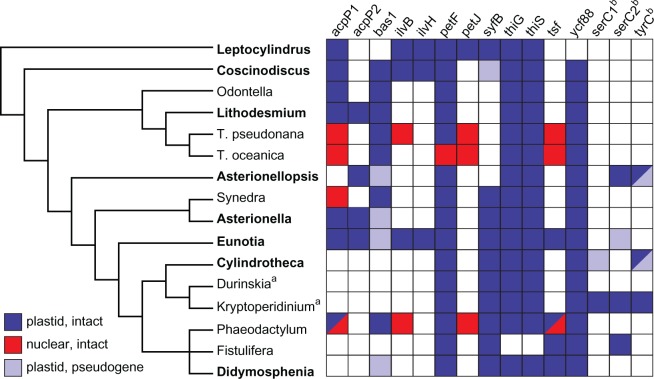
A total of 15 genes were variably present across the 16 species in our analysis (fig. 3). This pattern reflects 1) a dynamic history of gene losses and functional transfers to the nucleus across a broad range of phylogenetic depths, 2) gene duplications followed by differential losses of paralogs, and 3) acquisitions of foreign genes. In some cases, the small number of sequenced diatom nuclear genomes limited our ability to distinguish between gene losses and functional transfers to the nucleus. Likewise, the number of genes dually resident in the plastid and nuclear genomes has almost certainly been underestimated. For example, the psb28 gene is present in the nuclear genome of T. pseudonana (Jiroutová et al. 2010), a transfer one would not have predicted based on the universal presence of psb28 in diatom plastid genomes, including that of T. pseudonana. We expect, therefore, that the patterns inferred here will be continuously refined in the coming years as more plastid and nuclear genome sequences become available.
Fig. 3.—
Evolutionary patterns of pseudogenization, loss, and gain of genes in diatom plastid genomes. The matrix shows 15 genes variably present among the sequenced genomes. The presence of nuclear gene copies is almost certainly underreported for lack of nuclear genome data in most species. Taxa in boldface identify genomes sequenced for this study. Phylogenetic relationships were redrawn from Theriot et al. (2010) and unpublished data. Taxa marked with a superscript “a” are dinoflagellates with diatom-derived plastids (Imanian et al. 2010), and genes marked with a superscript “b” are of plasmid (serC) or unknown (tyrC) origin.
Widespread and Ongoing Gene Loss
Although some genes have been lost or transferred to the nucleus just once, most of the variably present genes showed considerably more complex patterns involving independent losses across a broad range of phylogenetic depths. For example, the peroxiredoxin gene, bas1, has been lost repeatedly from both red algal and chromalveolate plastid genomes (Douglas and Penny 1999; Glöckner et al. 2000; Sánchez-Puerta et al. 2005), and this pattern extends to diatoms as well. Assuming bas1 was present in the ancestral diatom plastid genome, the gene has been lost at least six separate times in taxa spanning the entire phylogeny (fig. 3). Although most of the genomes show no remaining trace of bas1, four distantly related taxa have retained what appear to be independently ameliorated pseudogene fragments, indicating that losses are ongoing in several lineages (fig. 3). Additional nuclear genomic data will help clarify whether the system of antioxidative protection provided by bas1 to plastids (Baier and Dietz 1997) has been lost, replaced, or handed over to the nucleus in some diatoms.
The tRNA synthetase gene, syfB, has a similar history of repeated loss—in Asterionellopsis, deep within the Odontella+Thalassiosira clade, and in Coscinodiscus, which retains a highly degenerated pseudogene (fig. 3). The syfB and syfH genes are the last remaining tRNA synthetase genes in primary (B and H) and secondary (B only) red plastids, so their mere persistence in diatom plastid genomes is probably more noteworthy than the seemingly inevitable losses recorded here. The syfB gene typically encodes the β subunit of Phenylalanyl-tRNA synthetase (PheRS), a heterotetramer with α- and β-subunits often encoded by separate genes (Safro et al. 2000). Organellar PheRS can function, however, as a single chimerically structured monomer with α- and β-domains encoded within a single gene (Safro et al. 2000; Duchêne et al. 2009). Thalassiosira pseudonana and T. oceanica both lack the plastid syfB gene but have a nuclear-encoded PheRS gene with this chimeric structure as well as signal and target peptides that predict plastid localization of the product (not shown). Thus, tRNA-Phe in diatom plastids, at least those lacking a syfB gene, appear to be loaded by a monomeric PheRS. The plastid-targeted PheRS gene appears to have been ancestrally present in diatoms but is missing from Phaeodactylum, which might account for the conservation of plastid syfB in araphid and raphid pennates (fig. 3).
Several genes showed a pattern of recent, lineage-specific loss. For example, losses of the thiamine biosynthesis genes, thiG and thiS, were restricted to a single lineage, represented here by Fistulifera (fig. 3). Likewise, ycf88 is missing only from Leptocylindrus (fig. 3). This conserved hypothetical protein is known only from diatom plastid genomes. If ycf88 was present in the ancestral diatom plastid genome, its absence in Leptocylindrus represents a lineage-specific loss. Alternatively, ycf88 might have originated after the split between Leptocylindrus and the rest of the diatoms.
Functional Transfers to the Nucleus
The early stages of establishment of an organelle are characterized by massive gene losses and functional transfers from the endosymbiont to the host nuclear genome (Kleine et al. 2009). Although this process has all but ceased in many organelles (e.g., animal mitochondria, Boore 1999), gene losses are ongoing in several lineages, including the mitochondrial genomes of land plants (Adams and Palmer 2003). Despite many potential obstacles (Martin and Herrmann 1998; Gruber et al. 2007), intracellular gene transfers from the plastid to the nuclear genome are quite common in diatoms (Oudot-Le Secq et al. 2007; Lommer et al. 2010; this study).
A total of five plastid genes have been either functionally transferred to the nucleus or maintain dual residency in the plastid and nuclear genomes (fig. 3). Two of these transfers, involving petF and petJ, were previously known (Kilian and Kroth 2004; Lommer et al. 2010). The petF case is a special ecologically driven transfer restricted to a single species (Lommer et al. 2010), and it is now clear that petJ was transferred to the nucleus early on in diatom evolution, sometime after the split between Leptocylindrus and all other diatoms (fig. 3).
Two genes involved in amino acid biosynthesis, ilvB and ilvH (the large and small subunits of acetolactate synthase) are widespread in primary and secondary red plastid genomes, absent only from haptophytes and previously sequenced diatoms (Sánchez-Puerta et al. 2005; Wang et al. 2013). The highly disjunct distribution of these genes in diatom plastid genomes reflects a history of repeated loss, at least four of them among our small sample of diatom diversity (fig. 3). The nuclear genomes of T. pseudonana and Phaeodactylum contain plastid-like ilvB genes with signal and target peptides that predict plastid localization of the protein, so losses of ilvB from the plastid genome likely coincided with functional transfers into the nucleus. Unlike ilvB, the apparently single, deep loss of the other acetolactate synthase subunit, ilvH, was not accompanied by a functional transfer to the nuclear genome.
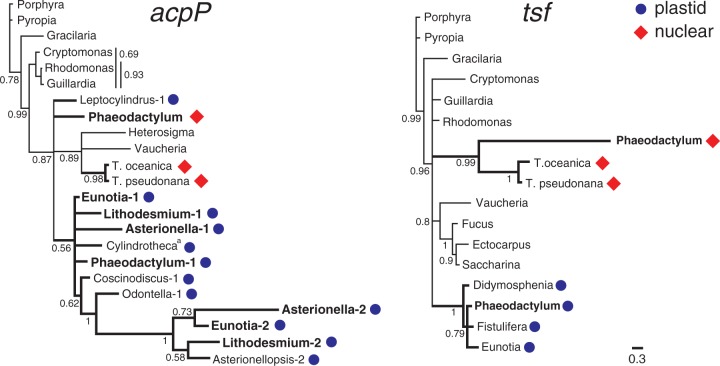
Dual residency of a gene in the organelle and nuclear genomes is common in the early stages of intracellular transfer, but the transfer generally resolves with loss of the organellar or, in some cases, the nuclear copy of the gene (Adams et al. 1999). The translation factor gene, tsf, appears to represent an altogether different phenomenon (fig. 3). The gene is present in the plastid genomes of two distantly related raphid pennates (Eunotia and Fistulifera), the nuclear genomes of T. pseudonana and T. oceanica, and both the nuclear and plastid genomes of Phaeodactylum (Oudot-Le Secq et al. 2007; Tanaka et al. 2011). Although the nuclear tsf genes in both Thalassiosira species have signal and transit peptides that predict targeting to the plastid, the nuclear copy in Phaeodactylum lacks both of these. While this would seem to suggest separate plastid-to-nuclear transfers in these two lineages, phylogenetic analysis resolved all nuclear copies into a strongly supported clade (fig. 4). The plastid-encoded tsf copies are also monophyletic and show levels of sequence divergence on par with other chromalveolates (fig. 4). Taken together, these results are consistent with a single deep plastid-to-nuclear transfer event followed by long-term conservation of both the plastid and nuclear copies (for tens of millions of years), with repeated losses of the plastid copy (fig. 3)—at least ten of them when mapped onto a representative sample of diatom diversity (Theriot et al. 2010). Long-term maintenance of the plastid and nuclear copies may have led to functional differentiation of the nuclear copy in Phaeodactylum, which is highly divergent (fig. 4) and apparently no longer targeted to the plastid. Experimental data are necessary to determine the exact localization of the nuclear-encoded product and show whether it has, in fact, assumed a new or modified function in Phaeodactylum.
Fig. 4.—
Gene phylogenies for acpP and tsf, both of which exhibit complex phylogenetic distributions across diatoms. Numbers are Bayesian posterior probability values. Genes located in the diatom plastid genome are marked with a circle, whereas genes in the nuclear genome are marked with a diamond. Taxa whose plastid genomes have multiple copies of the gene are boldfaced, with the −1 and −2 suffix denoting the different ortholog groups. Taxon marked with a superscript “a” is not the same Cylindrotheca strain sequenced as part of this study.
Gene Duplication
Although gene duplication and divergence provide an important source of new genetic variation in nuclear genomes (Conant and Wolfe 2008) and a smattering of animal mitochondrial genomes (Milani et al. 2013), divergent gene duplicates are rare in plastid genomes. Most duplicated plastid genes maintain their sequence identity through active recombination and gene conversion involving either duplicate copies of the genome within a cell or the recombinationally active IR (Chumley et al. 2006). Thus, gene duplicates in the plastid tend either to remain identical in sequence (Wakasugi et al. 1994; Haberle et al. 2008; Guisinger et al. 2011) or suffer deterioration and loss of one copy (Poczai and Hyvönen 2013).
Within this context then, the presence of two highly divergent copies of the fatty acid biosynthesis gene, acpP, in several plastid genomes is exceptional, reflecting a history that is more characteristic of a dynamically evolving nuclear gene family than a typical organelle gene. Although relationships within the acpP gene tree were generally unsupported, phylogenetic analysis recovered a strongly supported acpP2 clade, to the exclusion of counterpart acpP1 duplicates in Lithodesmium, Asterionella, and Eunotia (fig. 4)—a result that points to a relatively ancient (tandem) duplication followed by at least seven separate losses of one or both paralogs in the descendant lineages (fig. 3). These losses left some plastid genomes with both copies and others with one or, in some cases, none (fig. 3). Despite highly divergent amino acid sequences between plastid acpP paralogs (28–35% amino acid identity), differential retention of just the acpP2 copy in Asterionellopsis suggests that the two genes are functionally equivalent, a hypothesis that would be further supported if acpP1 is not found in the Asterionellopsis nuclear genome. The plastid acpP gene was, in fact, also duplicated into the nucleus, possibly around the time of the plastid gene duplication (fig. 3). A few species have retained only the nuclear copy of the gene (fig. 3), which has signal and target peptides consistent with plastid localization of the product.
Genes of Foreign or Uncertain Origin
Although foreign sequence acquisitions by plastid genomes are rare, horizontal transfer has introduced novel genes and introns into a few algal plastid genomes, including the diatom, Seminavis (Brouard et al. 2008; Khan and Archibald 2008; Brembu et al. 2013). Some of these foreign sequences were acquired from plasmids (Imanian et al. 2010; Brembu et al. 2013; Wang et al. 2013), whose cellular localization in diatoms includes both the nucleus and plastid (Hildebrand et al. 1992). Most notably, plasmids have introduced intact and putatively functional site-specific recombinase genes into the plastid genomes of several diatoms (fig. 3). Recombinases enzymatically break and rejoin DNA and fall into two unrelated families based on their DNA break–religate mechanism and the amino acid (serine or tyrosine) that mediates DNA cleavage (Grindley et al. 2006). They are essential for bacterial genome replication and differentiation (Nash 1996) and play important roles in the movement of transposons, plasmids, and bacteriophages within and between bacterial genomes (Smith and Thorpe 2002), making them highly plausible candidates for horizontal transfer.
The plastid genomes of five species contain one or two plasmid-derived serine recombinase (serC) genes or pseudogenes (fig. 3). Although a previous survey did not find plasmids outside of the raphid pennate lineage (Hildebrand et al. 1991), the discovery of serC in Asterionellopsis predicts that araphid pennates contain plasmids as well. In addition to a serC pseudogene, the Cylindrotheca plastid genome also contains a short fragment with similarity to a newly discovered plasmid from our assembly (supplementary fig. S1, Supplementary Material online). Asterionellopsis, Eunotia, and Cylindrotheca also contain sequences matching noncoding sequences and ORFs from known diatom plasmids (Hildebrand et al. 1991, 1992).
Although tyrC shares similar recombinase functions with serC, the origins of tyrC in select diatom (Imanian et al. 2010), raphidophyte (Cattolico et al. 2008), and green algal (Brouard et al. 2008) plastid genomes are less clear. Like serC, however, tyrC appears to be restricted to the pennate diatom lineage (the Asterionellopsis+Didymosphenia clade in fig. 3). Moreover, in both Asterionellopsis and Heterosigma (another heterokont), tyrC is adjacent to ORFs with low similarity to known plasmid ORFs, pointing to a probable plasmid origin for tyrC in diatom plastid genomes. Still, tyrC is common in bacterial genomes and plasmids (Leplae et al. 2006; Van Houdt et al. 2012), and in light of the close associations between diatoms and bacteria (Bowler et al. 2008; Amin et al. 2012), a direct bacterial origin of tyrC cannot be ruled out. Indeed, bacterial HGT has introduced novel foreign genes into both primary (Janouškovec et al. 2013) and secondary (Khan et al. 2007) red algal plastid genomes.
Genome Rearrangements
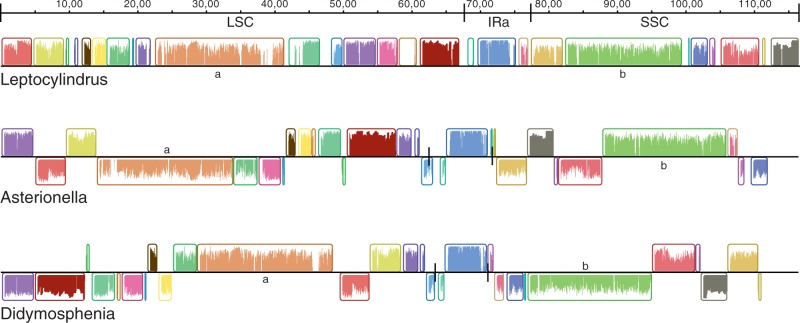
Aside from sharing the common quadripartite plastid genome architecture, diatom plastid genomes are otherwise highly rearranged (Oudot-Le Secq et al. 2007)—a finding underscored by the eight newly sequenced genomes. Illustrative of this, the plastid genomes of three representative diatoms had to be subdivided into 32 colinear gene blocks to create a whole-genome alignment (fig. 5). Some lineages have experienced a higher frequency of rearrangements than others. For example, the genomes of two Thalassiosirales, T. pseudonana and T. oceanica, are highly rearranged relative to one another, whereas the genomes two raphid pennates, Didymosphenia and Phaeodactylum, are perfectly collinear (not shown). Because the single-copy regions of the genome are so highly rearranged, shifts in the IR boundaries result in the annexation or loss of different sets of genes in different diatom lineages (fig. 2). Dense, focused sampling within particular lineages will show whether rearrangements are associated with rare recombination events across tRNAs or other small repetitive sequences (Turmel et al. 2002; Weng et al. 2013).
Fig. 5.—
Plastid-genome alignments for three representative diatoms. One copy of the inverted repeat region was removed from each genome prior to aligning the genomes. Colored blocks indicate gene clusters with conserved gene order across the three taxa. Blocks below black center line are inverted relative to the Leptocylindrus reference genome. Two blocks (a and b) contain the largest conserved gene clusters, consisting of the RNA polymerase genes (a) or the large ribosomal operon in the small single copy region (b).
Conclusions
Despite their ecological importance and substantial contribution to global primary production, surprisingly little is known about the plastid genomes of diatoms. Our goal was to help fill this gap by doubling the number of fully sequenced plastid genomes and greatly expanding the phylogenetic breadth of sampled species. Our increased taxon sampling revealed levels of variation in plastid gene content, genome size, and genome architecture exceeding those in many other plastid-bearing lineages. Angiosperms, for example, are similar to diatoms in both taxonomic diversity and geologic age, but with just a few noteworthy exceptions (Cai et al. 2008; Sloan et al. 2012), their plastid genomes are characterized by long-term evolutionary stasis (Jansen and Ruhlman 2012). Diatom plastid genomes, by contrast, exhibit complex patterns of gene gains and losses and, more compelling still, a propensity to acquire and retain foreign DNA.
In many cases, our inferences, especially with respect to gene gains and losses, hinged heavily on our taxonomic sampling. For example, the Eunotia plastid genome is a hoarder, holding onto genes that have been tossed out in most other species. This single genome highlighted patterns of loss far more complex than would have been evident if it had not been sequenced. In light of this, and given that diversity estimates for diatoms number into the hundreds of thousands of species, we expect that diatom plastid genomes hold many more surprises, that the full pan-genome is still unknown for diatom plastids, and that the inferences made here will be substantially modified in the coming years. Finally, important phylogenetic relationships within diatoms remain unresolved or poorly supported (Theriot et al. 2010), severely constraining current and future comparative genomic studies. Efforts to better characterize the phylogenetic relationships of diatoms will pay great dividends to these and other emergent fields of research on this diverse and ecologically important lineage.
Supplementary Material
Supplementary tables S1 and S2 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Jeff Palmer and three anonymous reviewers for critical comments on an earlier version of this manuscript. Genome analyses were carried out using resources available through the Texas Advanced Computing Center (TACC) at The University of Texas at Austin and the Arkansas High Performance Computing Center (AHPCC) at the University of Arkansas. Resources managed by AHPCC are supported in part by NSF grants MRI 0722625, MRI-R2 0959124, and a grant from the Arkansas Science and Technology Authority. This research was funded by NSF grant EF062410 to E.C.T. and R.K.J., USGS/NPS NRPP 141338 to E.C.T., and start-up funds from the University of Arkansas to A.J.A.
Literature Cited
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Adams KL, et al. Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc Natl Acad Sci U S A. 1999;96:13863–13868. doi: 10.1073/pnas.96.24.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Baier M, Dietz KJ. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 1997;12:179–190. doi: 10.1046/j.1365-313x.1997.12010179.x. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol Biol Evol. 2006;23:2058–2071. doi: 10.1093/molbev/msl091. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A site-and time-heterogeneous model of amino acid replacement. Mol Biol Evol. 2008;25:842–858. doi: 10.1093/molbev/msn018. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Brembu T, et al. The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar Genomics. 2013 doi: 10.1016/j.margen.2013.12.002. Advance Access published December 21, 2013, doi/10.1016/j.margem/2013.12.002. [DOI] [PubMed] [Google Scholar]
- Brouard J-S, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, et al. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 2008;67:696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- Cattolico R, et al. Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics. 2008;9:211. doi: 10.1186/1471-2164-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley TW, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48:236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- Duchêne A-M, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- Glöckner G, Rosenthal A, Valentin K. The structure and gene repertoire of an ancient red algal plastid genome. J Mol Evol. 2000;51:382–390. doi: 10.1007/s002390010101. [DOI] [PubMed] [Google Scholar]
- Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Gruber A, et al. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66:350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Hildebrand M, et al. Plasmids in diatom species. J Bacteriol. 1991;173:5924–5927. doi: 10.1128/jb.173.18.5924-5927.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M, et al. Nucleotide sequence of diatom plasmids: identification of open reading frames with similarity to site-specific recombinases. Plant Mol Biol. 1992;19:759–770. doi: 10.1007/BF00027072. [DOI] [PubMed] [Google Scholar]
- Hopkinson BM, Dupont CL, Allen AE, Morel FM. Efficiency of the CO2-concentrating mechanism of diatoms. Proc Natl Acad Sci U S A. 2011;108:3830–3837. doi: 10.1073/pnas.1018062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Pombert J-F, Keeling PJ. The complete plastid genomes of the two ‘dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One. 2010;5:e10711. doi: 10.1371/journal.pone.0010711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS One. 2013;8:e59001. doi: 10.1371/journal.pone.0059001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA. Plastid genomes of seed plants. In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Advances in photosynthesis and respiration. Dordrecht (The Netherlands): Springer; 2012. pp. 103–126. [Google Scholar]
- Jiroutová K, Kořený L, Bowler C, Oborník M. A gene in the process of endosymbiotic transfer. PLoS One. 2010;5:e13234. doi: 10.1371/journal.pone.0013234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Archibald JM. Lateral transfer of introns in the cryptophyte plastid genome. Nucleic Acids Res. 2008;36:3043–3053. doi: 10.1093/nar/gkn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24:1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Presequence acquisition during secondary endocytobiosis and the possible role of introns. J Mol Evol. 2004;58:712–721. doi: 10.1007/s00239-004-2593-z. [DOI] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- Kroth PG, et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One. 2008;3:e1426. doi: 10.1371/journal.pone.0001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Leplae R, Lima-Mendez G, Toussaint A. A first global analysis of plasmid encoded proteins in the ACLAME database. FEMS Microbiol Rev. 2006;30:980–994. doi: 10.1111/j.1574-6976.2006.00044.x. [DOI] [PubMed] [Google Scholar]
- Li C-W, Volcani BE. Four new apochlorotic diatoms. Br Phycol J. 1987;22:375–382. [Google Scholar]
- Lommer M, et al. Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics. 2010;11:718. doi: 10.1186/1471-2164-11-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison D, Maddison W. Sunderland (MA): Sinauer Associates; 2005. MacClade v. 4.08. [Google Scholar]
- Mann DG, Vanormelingen P. An inordinate fondness? The number, distributions, and origins of diatom species. J Eukaryot Microbiol. 2013;60:414–420. doi: 10.1111/jeu.12047. [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M. A comparative analysis of mitochondrial ORFans: new clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biol Evol. 2013;5:1408–1434. doi: 10.1093/gbe/evt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments. Escherichia coli and Salmonella. Cell Mol Biol. 1996;2:2363–2376. [Google Scholar]
- Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cy. 1995;9:359–372. [Google Scholar]
- Oudot-Le Secq M-P, et al. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol Genet Genomics. 2007;277:427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- Plunkett GM, Downie SR. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst Bot. 2000;25:648–667. [Google Scholar]
- Poczai P, Hyvönen J. Plastid trnF pseudogenes are present in Jaltomata, the sister genus of Solanum (Solanaceae): molecular evolution of tandemly repeated structural mutations. Gene. 2013;530:143–150. doi: 10.1016/j.gene.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Ragan MA, Harlow TJ, Beiko RG. Do different surrogate methods detect lateral genetic transfer events of different relative ages? Trends Microbiol. 2006;14:4–8. doi: 10.1016/j.tim.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Roberts K, Granum E, Leegood RC, Raven JA. Carbon acquisition by diatoms. Photosynth Res. 2007;93:79–88. doi: 10.1007/s11120-007-9172-2. [DOI] [PubMed] [Google Scholar]
- Safro M, Moor N, Lavrik O. 2000. Phenylalanyl-tRNA synthetases. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience. Available from: http://www.ncbi.nlm.nih.gov/books/NBK6321/ [DOI] [PubMed] [Google Scholar]
- Sánchez-Puerta MV, Bachvaroff TR, Delwiche CF. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res. 2005;12:151–156. doi: 10.1093/dnares/12.2.151. [DOI] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol Evol. 2012;4:294–306. doi: 10.1093/gbe/evs006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Thorpe HM. Diversity in the serine recombinases. Mol Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- Tanaka T, et al. High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth Res. 2011;109:223–229. doi: 10.1007/s11120-011-9622-8. [DOI] [PubMed] [Google Scholar]
- Theriot EC, Ashworth M, Ruck E, Nakov T, Jansen RK. A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol Evol. 2010;143:278–296. [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci U S A. 2002;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R, Leplae R, Lima-Mendez G, Mergeay M, Toussaint A. Towards a more accurate annotation of tyrosine-based site-specific recombinases in bacterial genomes. Mob DNA. 2012;3:1–11. doi: 10.1186/1759-8753-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, et al. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci U S A. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. Complete sequence and analysis of plastid genomes of two economically important red algae: Pyropia haitanensis and Pyropia yezoensis. PLoS One. 2013;8:e65902. doi: 10.1371/journal.pone.0065902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M-L, Blazier JC, Govindu M, Jansen RK. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 2013;31:645–659. doi: 10.1093/molbev/mst257. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.