Abstract
Chlorophylls (Chls) play pivotal roles in energy absorption and transduction and also in charge separation in reaction centers in all photosynthetic organisms. In Chl biosynthesis steps, only a step for the enzymatic reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide) is mediated by both nuclear- and chloroplast-encoded genes in land plants. Many plants encode the genes for light-dependent Pchlide reductase (LPOR) and light-independent Pchlide reductase (DPOR) in the nucleus and chloroplast genome, respectively. During the diversification of land plants, the reduction step of Pchlide to Chlide has become solely dependent on LPOR, and the genes for DPOR have been lost from chloroplast genome. It remains unclear why DPOR persists in some land plants, how they were eliminated from chloroplast genomes during the diversification of land plants, and under what environmental conditions DPOR was required. We demonstrate that DPOR is functional in liverwort (Marchantia polymorpha L.) and plays an important role in Chl biosynthesis. Having established a plastid transformation system in liverwort, we disrupted chlB, which encodes a subunit of DPOR in the M. polymorpha chloroplast genome. Morphological and Chl content analysis of a chlB mutant grown under different photoperiods revealed that DPOR is particularly required for Chl biosynthesis under short-day conditions. Our findings suggest that an environmental condition in the form of photoperiod is an important factor that determines the loss or retention of chloroplast-encoded genes mediating Pchlide reduction to Chlide.
Keywords: plastid transformation, chloroplast genome evolution, bryophyte, chlorophyll metabolism
Introduction
Chloroplasts are the main site for photosynthesis and are considered to be descendants of cyanobacteria that were engulfed by an ancestral eukaryotic cell. After this engulfment event, cyanobacterial genes were either lost or transferred into the nuclear genome. Alternatively, genes of different origin or de novo genes in the nuclear genome replaced organelle-encoded genes (Mereschkowski 1905; Gray 1992; Martin and Kowallik 1999; Ueda and Kadowaki 2012). In this study, we focused on genes involved in chlorophyll (Chl) synthesis to elucidate the driving force of chloroplast genome evolution.
Chls play pivotal roles in energy absorption and transduction and also in charge separation in reaction centers. In angiosperms represented by Arabidopsis thaliana, there are 15 steps in the pathway from glutamyl-tRNA to Chls a and b, and the genes involved in these steps have been identified (Beale 2005; Nagata et al. 2005). Among them, Pchlide oxidoreductase, which is responsible for the reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), is characteristic, given that genes for the enzymes are encoded in both nuclear and chloroplast genomes in most plants (Armstrong 1998; Schoefs and Franck 2003).
There are two distinct Pchlide oxidoreductases: light-dependent NADPH-Pchlide oxidoreductase (LPOR), which requires photoenergy for catalysis and is encoded in the nucleus, and light-independent Pchlide oxidoreductase (DPOR), which requires no photoenergy and whose subunits are encoded in the chloroplast genome. LPOR is a monomeric protein, whereas DPOR is a multimeric protein consisting of three subunits encoded by the chlB, chlL, and chlN genes (Reinbothe et al. 2010).
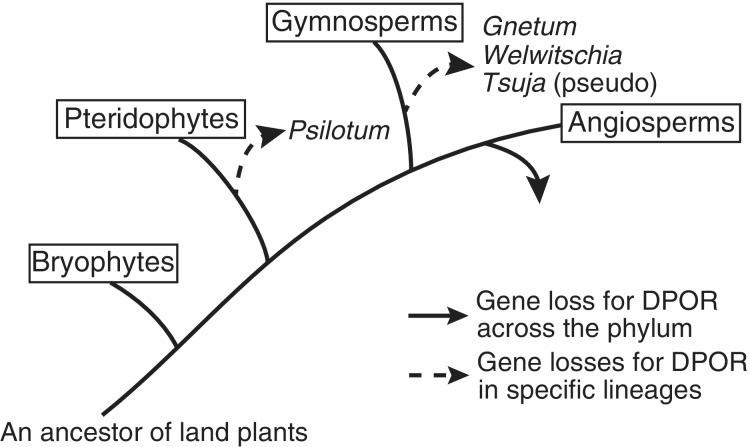
DPOR has been lost and the reduction step of Pchlide to Chlide has become solely dependent on LPOR in angiosperms, several gymnosperms, and some Pteridophytes (fig. 1). Based on LPOR and DPOR distribution in the plant, eubacterial, and archaebacterial kingdoms and the oxygen sensitivity of DPOR, a model has been proposed to explain the gene transfer of LPOR and the substitution of LPOR for DPOR during land plant evolution: 1) oxygen-sensitive DPOR initially emerged when the atmosphere of Archaean Earth was anaerobic, presumably from nitrogenase-like genes (Fujita et al. 1991) and 2) LPOR, which is an oxygen-insensitive and light- and NADPH-dependent enzyme belonging to a short-chain dehydrogenase/reductase superfamily, evolved during the transition from an anaerobic to aerobic atmosphere (Reinbothe et al. 1996, 2010). Both enzymes seem to have emerged before the endosymbiosis of cyanobacteria, which are believed to be the ancestors of chloroplasts, given that Chlides are biosynthesized using both DPOR and LPOR in modern cyanobacteria (Schoefs and Franck 2003). 3) Upon the establishment of chloroplasts, it is likely that LPOR was transferred to the nucleus, whereas the genes for DPOR were retained in chloroplast genomes. Despite their apparent functional redundancy, both genes have been strictly retained in several algae, lower plants, and gymnosperms. 4) As a final step of “gene replacement,” DPOR loss independently occurred in land plants.
Fig. 1.—
Schematic diagram illustrating the distribution of DPOR in land plants. The distribution of DPOR among land plant chloroplast genomes is described by Armstrong (1998), Wicke et al. (2011), and Wu et al. (2013). Tsuja species have lost the greening ability in darkness, although they appear to have conventional open reading frames for genes of DPOR subunits (Kusumi et al. 2006).
Why are LPOR and DPOR preserved together in a large number of plants, and what was the force driving the loss of DPOR during the evolution of land plants? Previous studies suggest that DPOR plays a key role in greening of algae and vascular plants under heterotrophic growth conditions, such as in chemoheterotrophic-grown Leptolyngbya boryana (formerly Plectonema boryanum), dark-grown Chlamydomonas reinhardii cell cultures, and dark-grown gymnosperm seedlings (Suzuki and Bauer 1992; Li et al. 1993; Fujita et al. 1996; Armstrong 1998). These reports suggest that the light environment is one of the critical factors that determine the fate of DPOR.
The liverwort Marchantia polymorpha L. may be a key organism for elucidating the aforementioned questions. For studying the early evolution of land plants, particularly the water-to-land transition that dramatically changed the light environment, M. polymorpha is emerging as a premier model system because it is considered to be the earliest lineage of land plants derived from green algal ancestors. In particular, liverworts occupy a critical position in land plant evolution, forming the sister group to all extant land plants (Crandall-Stotler et al. 2008).
Gene losses from the chloroplast genome have repeatedly occurred even after the divergence of land plants (Martin et al. 2002; Jansen et al. 2007; Wicke et al. 2011). The Marchantia chloroplast genome has the largest number of genes of all land plants (Ohyama et al. 1986; Martin et al. 2002; Ohyama et al. 2009), suggesting that liverworts retain ancestral characters in terms of the gene content of chloroplast genome. This peculiarity raises the possibility of identifying roles of genes encoded in the liverwort chloroplast genome, that is, genes for DPOR in this study, which have been lost from the chloroplast genomes of other land plants.
Liverworts possess both LPOR and DPOR (Suzuki et al. 2001). Having established the plastid transformation system in M. polymorpha (Chiyoda et al. 2007; Ueda et al. 2012), we prepared a DPOR-deficient mutant (a chlB knockout mutant). We obtained evidence that Chl biosynthesis operates in the dark through DPOR in M. polymorpha. Mutant analysis revealed that DPOR is still required for Chl biosynthesis in M. polymorpha, particularly under short-day conditions. Our results suggest that DPOR is required for at least M. polymorpha to adapt to environmental conditions (photoperiod) that may lead to the persistence of DPOR in the M. polymorpha plastid genome. The establishment of a regulation system for Chl biosynthesis without DPOR during the evolution of land plants is discussed, with reference to the comparison of genes, involving the suppression of Chl biosynthesis between A. thaliana and M. polymorpha.
Materials and Methods
Plant Materials and Growth Conditions
Male accession Takaragaike-1 (Tak1) and female accession Takaragaike-2 (Tak2) liverworts (M. polymorpha L.) were asexually maintained and propagated via gemmae as described previously (Chiyoda et al. 2008). Plants were usually grown on 1/2 Gamborg’s B5 media, containing 1% sucrose in a growth chamber at 20 °C under continuous light. Fourteen-day-old plants grown under a 14-h light (L) and 10-h dark (D) cycle (70–90 μmol photons m−2 s−1) at 20 °C were used for immunoblotting. To monitor morphological difference and measure Chl content and Pchlide under different photoperiod, plants were grown in a Biomulti incubator with cold cathode fluorescent lamps (170–200 μmol photons m−2 s−1) at 20 °C for 2 weeks (Nihon-ika, Osaka, Japan).
Cloning of a Plastid Genome Fragment Containing chlB for Plastid Transformation
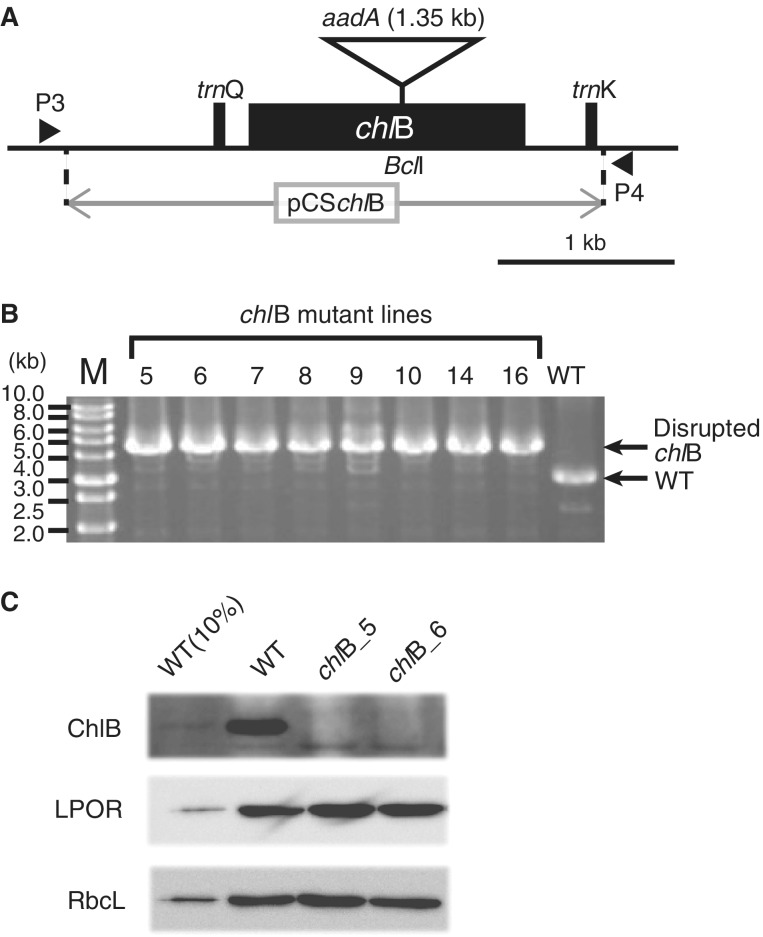
Total DNA was isolated from thalli of plants using Isoplant II (Nippon gene, Tokyo, Japan). Genomic polymerase chain reaction (PCR) to amplify the chlB fragment for the preparation of the plastid transformation vector was performed using KOD-plus-Neo DNA polymerase (TOYOBO, Osaka, Japan) with P1 and P2 primers. All the primers used in this article are shown in supplementary tables S1 and S2, Supplementary Material online. The resulting products were cloned into the pTAC-2 vector (BioDynamics Laboratory, Tokyo, Japan) after adenosine addition, according to the manufacturer’s instructions, and the clones were sequenced using universal or randomly designed primers. The aadA cassette, which confers spectinomycin resistance to chloroplasts (Shikanai et al. 1998), was inserted into the BclI site present in the chlB exon, so that the gene was inactivated (fig. 2A). The resulting plasmid (pCSchlB vector) was sequenced again and digested with NotI to linearize them for plastid transformation.
Fig. 2.—
Preparation of chlB mutant in Marchantia polymorpha. (A) Gene structure around chlB in the M. polymorpha chloroplast genome. Black boxes and horizontal line indicate exons and intergenic region, respectively. Solid triangles show primer positions designed for the confirmation of the aadA cassette insertion. A restriction enzyme (BclI) site within the chlB coding sequences shown with a triangle is the position of the aadA cassette insertion. Black angles and the triangle are not to scale. (B) Genotyping for the confirmation of homoplasmic transplastome in the chlB mutant. P3 and P4 primer pairs detected WT plastome and/or transplastome defective in the chlB (3,165 bp in WT and 4,520 bp in chlB mutant). (C) Immunoblotting analysis of ChlB protein accumulation. Samples containing 0.5 μg chlorophyll (Chl) crude chloroplast fraction proteins were loaded. In the case of WT, 0.05 μg Chl crude chloroplast fraction proteins (10% WT) were loaded to confirm the limit of detection by each antibody. The proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the blots were probed with specific antibodies against ChlB (a subunit of DPOR), LPOR, and rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit).
Plastid Transformation to Knock Out chlB
Spores used for plastid transformation were prepared as described by Chiyoda et al. (2008). Plastids were transformed using 7-day-old sporelings, as described previously (Chiyoda et al. 2007; Ueda et al. 2012). Homoplasmic transplastomic lines were obtained by asexual reproduction via gemmae on selective 0-M51C agar medium (Takenaka et al. 2000), containing 300 µg/ml spectinomycin without sucrose, for 3 months. The consistent difference in Chl and Pchlide accumulation between the chlB_5 and chlB_6 mutants that are plastid transformants responsive to day length may be because of genetic differences between male (Tak-1) and female (Tak-2) lines in M. polymorpha. This is because M. polymorpha is a dioecious plant and has sex chromosomes from which it is difficult to prepare genetically pure lines.
Genotyping of Transplastomic Plants
Total DNA was isolated from thalli using conventional methods and diluted in 200 µl of Tris–EDTA Buffer (10 mM Tris–HCl [pH 8.0], 1 mM ethylenediaminetetraacetic acid [pH 8.0]). To find homotransplastomic plants, 1 µl of the extract was used for a genomic PCR with primers P3 and P4. Primer positions and primer combinations for genotyping both types are shown in figure 2A. PCR was performed with KOD-FX DNA polymerase (TOYOBO).
Chl Measurement
Extraction and measurement of Chl contents were performed according to the method by Arnon (1949). A sample for measurement of Chl contents was extracted from frozen thalli ground to a fine powder with a mortar and pestle in liquid nitrogen. The average value for each experiment was obtained from four independent samples.
Immunoblot Analysis
Crude chloroplast proteins were loaded onto a sodium dodecyl sulfate–polyacrylamide gel on an equal Chl basis. Samples of 0.5 µg Chl proteins from the crude chloroplast fraction were used for immunoblotting. Immunoblotting was performed with anti-ChlB (Fujita et al. 1996), anti-RbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit) (Agrisera, Vännäs, Sweden), and anti-L-POR (Agrisera) antibodies. Signals were detected with an ECL Plus Western Blotting Detection Kit (GE Healthcare UK Ltd, Buckinghamshire, UK) and visualized with an LAS3000 chemiluminescence analyzer (Fuji Film, Tokyo, Japan). The results presented represent two independent experiments.
Pchlide Measurement
Pchlide measurement was performed by high pressure liquid chromatography (HPLC), according to Nagata et al. (2007). Plants grown under different light/dark cycles were frozen with liquid nitrogen at end of light and dark periods, respectively. Sampling was performed in a darkroom with safety light (Nagatani laboratory, Kyoto Univ.).
Database Analysis in M. polymorpha
All similarity searches were conducted using the default parameters of the stand-alone Basic Local Alignment Search Tool (Blast) available through the National Center for Biotechnology Information as described in the previous study (Ueda et al. 2012).
Quantitative Reverse-Transcription PCR
The transcript levels were assayed using the Mx30000p qPCR instrument (Stratagene, La Jolla, CA) and FastStart Universal SYBR Green Master (Rox) reaction mix (Roche Diagnostics GmbH, Mannheim, Germany). The error bars show the standard deviations calculated from three repeated experiments. To standardize the data, the ratio of the absolute transcript level of each gene to the absolute transcript level of elongation factor 1-a (EF-1a) was calculated for each sample. The transcript abundance of genes is shown as the expression relative to the wild-type (WT) plant. The primer sequences and scores in quantitative reverse-transcription PCR (qRT-PCR) experiments are listed in supplementary tables S2 and S3, Supplementary Material online.
Results
Knockout of the Plastid chlB Gene in M. polymorpha
To determine whether chlB is functional in M. polymorpha, we took advantage of the established technique of plastid transformation to knock out the gene (Chiyoda et al. 2007; Ueda et al. 2012). The pCSchlB vector encodes a 3.0-kb region including trnQ, chlB, and trnK genes cloned from the M. polymorpha plastid genome, and chlB was disrupted by the insertion of a chimeric aadA cassette (fig. 2A). To confirm the homoplasmic state of the genome, in which chlB had completely segregated out, the chloroplast DNA was analyzed using genomic PCR. The primer pair of P3 and P4 distinguishes transplastomic DNA from WT DNA. The 4.5-kb fragment that originated in the transplastomic copy was preferentially amplified, and the 3.2-kb fragment that originated in the WT copy was not detected (fig. 2B), indicating that chlB was knocked out in the mutant lines.
To evaluate the impact of the chlB knockout on protein accumulation, a protein blot of crude chloroplast extracts from WT plants and two independent chlB mutant lines was probed with antisera against ChlB, LPOR, and RbcL. Consistent with the knockout of the gene (fig. 2B), ChlB protein was below the detection limit in the chlB mutants. The absence of ChlB did not affect LPOR accumulation (fig. 2C).
chlB Contributes to Chl Synthesis Particularly under Short-Day Conditions
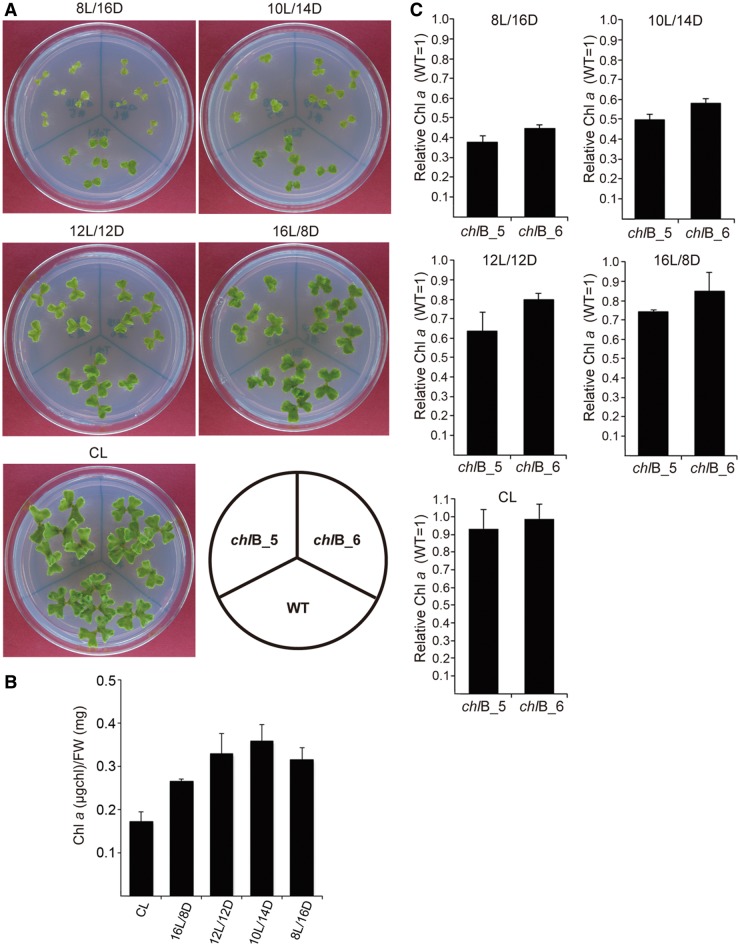
The chlB mutant showed no visible phenotype under continuous light conditions (fig. 3A). DPOR most likely functions in the dark because of the inhibition of its enzymatic activity by oxygen produced in photosystem II in the light (Armstrong 1998; Reinbothe et al. 2010). In a continuous-light condition, DPOR may be inactive and Chl synthesis does not depend on chlB in M. polymorpha. We accordingly grew the chlB mutant under different day-length conditions and observed its morphological variation. An obvious morphological phenotype (pale green and slow growth) was observed in the chlB mutants, particularly when they were grown under 10-h L/14-h D or 8-h L/16-h D conditions (fig. 3A).
Fig. 3.—
Morphological observation and comparison of Chl content in chlB mutants grown under different day-length conditions. (A) Morphological difference between WT plants and chlB mutants grown in plastic petri dish (9 cm diameter). Plants were grown on 1/2 Gamborg’s B5 media without sucrose in a Biomulti incubator with cold cathode fluorescent lamps (170–200 μmol photons m−2 s−1) at 20 °C (Nihon-ika, Osaka, Japan). (B) Quantification of Chl a content in WT plants under different day-length conditions. Chl a contents (μg of Chl per fresh weight mg) were measured according to Arnon (1949). Two-week-old plants were used under different day-length conditions in these experiments. Standard deviations (n = 3) are indicated by lines extending from the bars. (C) Levels of Chl a accumulation in the chlB mutants. Standard deviations (n = 3) are indicated by lines extending from the bars. Each mean represents the ratio of Chl a accumulation level in the chlB mutants to that of WT plants.
Consistent with the morphological phenotype, Chl a accumulation in the chlB mutant was drastically lower in short-day conditions than in the WT; Chl a accumulation was reduced to 37.8% (chlB_5 mutant line) and 44.6% (chlB_6 mutant line) in chlB mutant lines compared with WT plants, when they were grown under a 8-h L/16-h D cycle (fig. 3B and C). In contrast to short-day conditions, drastic differences in Chl a accumulation between the chlB mutant and WT plants were not observed under long-day periods. chlB mutants showed Chl a accumulations of 74.3% (chlB_5 mutant line) and 84.7% (chlB_6 mutant line) under a 16-h L/8-h D cycle and 93.2% (chlB_5 mutant line) and 98.8% (chlB_6 mutant line) under continuous light compared with the respective WT plants (fig. 3B and C).
These results suggest that the M. polymorpha chloroplast genome retains DPOR as necessary for Chl biosynthesis, particularly under short-day conditions.
The chlB Mutant Accumulates Monovinyl-Pchlide a in the Dark
DPOR mediates the reduction of Pchlide (a phototoxic Chl precursor) to Chlide in the dark. The chlB mutant may accumulate Pchlide in the dark. To test this possibility, the ratio of Pchlide to Chl a was evaluated using HPLC at the ends of both light and dark periods in plants grown under different photoperiods. Pchlide consists of a mixture of monovinyl (MV) and divinyl forms. We measured MV-Pchlide a.
At the end of the light period, no MV-Pchlide a accumulation was observed in either WT plants or the chlB mutant under any light/dark cycle condition (table 1), suggesting that LPOR primarily reduces Pchlide to Chlide in the light. In contrast, at the end of the dark period, MV-Pchlide a accumulation in the chlB mutant was detected (table 1). WT plants did not accumulate MV-Pchlide a, suggesting that DPOR-dependent reduction of MV-Pchlide a operates in the dark (table 1). MV-Pchlide a levels were approximately 5-fold higher under 8-h L/16-h D than in 16-h L/8-h D plants (table 1). Chl a content in plants grown under an 8-h L/16-h D cycle accumulates to at least 1.2-fold relative to those grown under a 16-h L/8-h D cycle (fig. 3B), suggesting that MV-Pchlide a accumulation increased with longer dark period from a 16-h L/8-h D to a 8-h L/16-h D cycle.
Table 1.
Pchlide Accumulation in Plants Grown under Different Day-Length Conditions at End of Light and Dark Periods
| Day Length | Plant Materials | MV-Pchlide/Chl a |
|---|---|---|
| Continuous light | WT | 0 |
| chlB_5 | 0 | |
| chlB_6 | 0 | |
| 16-h light/8-h dark (16L/8D) EL | WT | 0 |
| chlB_5 | 0 | |
| chlB_6 | 0 | |
| 16L/8D ED | WT | 0 |
| chlB_5 | 0.0030 | |
| chlB_6 | 0.0027 | |
| 8-h light/16-h dark (8L/16D) EL | WT | 0 |
| chlB_5 | 0 | |
| chlB_6 | 0 | |
| 8L/16D ED | WT | 0 |
| chlB_5 | 0.015 | |
| chlB_6 | 0.017 |
Note.—The results presented represent three independent experiments. Pchlide consists of a mixture of MV and divinyl forms. We measured MV-Pchlide at ends of light and dark periods under different light cycles. EL, end of light period; ED, end of dark period.
The reduction in Chl a content and MV-Pchlide a accumulation in the dark suggests that DPOR is functional and contributes to Chl biosynthesis in the dark and that its function is particularly important under short-day conditions in M. polymorpha.
DPOR-Deficiency Influenced Interconnection of Regulatory Control Circuits for the Chl Biosynthesis Pathways
There are negative and positive feedback regulations in Chl synthesis to avoid photo-oxidative damage caused by Chl intermediates in plants (fig. 4A, see Discussion). To check the impact of DPOR deficiency on the feedback regulations in mRNA level, we measured the relative transcript abundance of five genes (MpHEMA, MpDELLA, MpFLU, MpCHL27, and MpLPOR) involved in Chl biosynthesis in the WT plant and the chlB mutants grown under an 8-h L/16-h D cycle.
Fig. 4.—
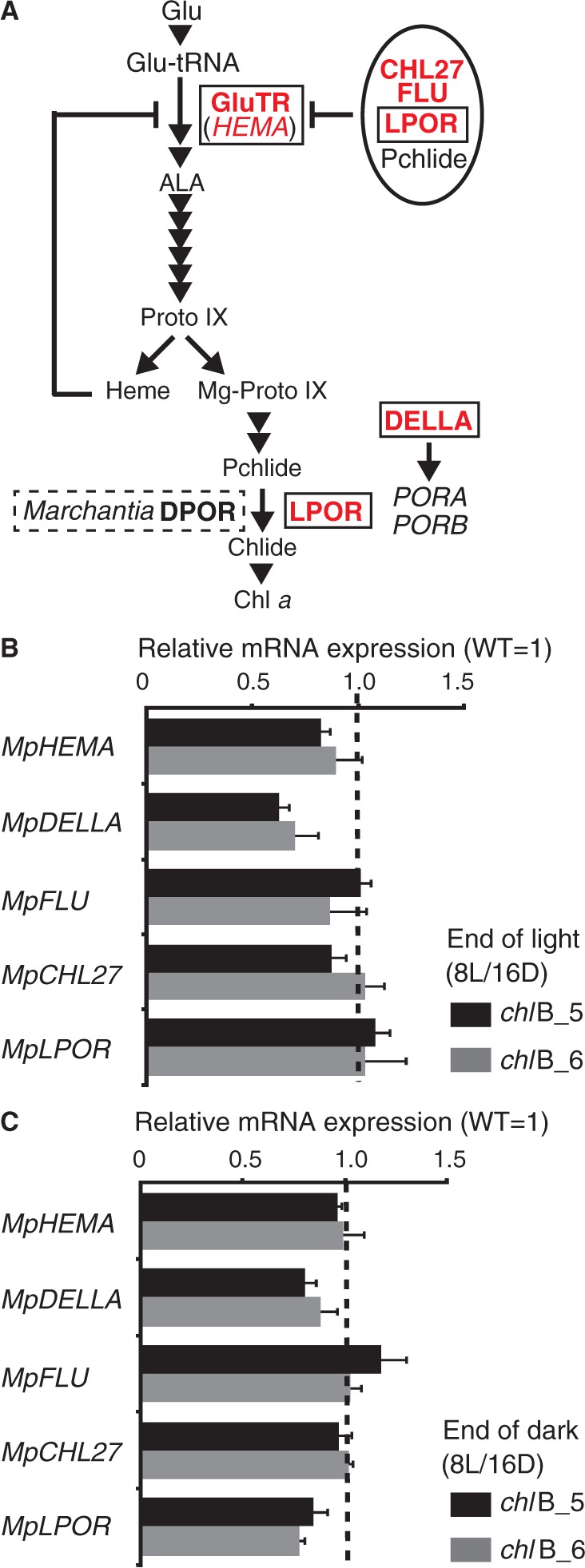
Influence of DPOR deficiency on interconnection of regulatory control circuits for the Chl biosynthesis pathways. (A) Simplified schematic representation of regulation in Chl biosynthesis steps in Arabidopsis thaliana. Genes duplicated during the diversification of land plants are surrounded by a box. Genes involving the regulation of Chl biosynthesis in the dark are shown in red letters. Their gene IDs are listed in table 2. LPORA and LPORB expressions are positively regulated in darkness. DPOR is encoded in the Marchantia polymorpha chloroplast genome but not in A. thaliana. FLU bound to the CHL27–POR–Pchlide complex is able to inhibit glutamyl-tRNA reductase (GluTR) activity. (B) The levels of transcripts in the chlB mutants at the end of the light period. (C) The levels of transcripts in the chlB mutants at end of the dark period. The standard deviations (n = 3, n stands for technical replicates) are indicated by lines extending from the bars. Each mean represents the ratio of the expression level of transcripts in the chlB mutants compared with that of WT plants. The means are depicted by black and gray bars for the chlB_5 and the chlB_6 mutant lines, respectively.
Gene expression of MpDELLA was decreased in the chlB mutants at the end of the light and dark conditions (fig. 4B and C). In addition, gene expression of MpLPOR was mildly reduced only in darkness in the chlB mutants (fig. 4C). These results suggest that DPOR deficiency influenced the regulatory control circuits for the Chl biosynthesis pathway or at least the mRNA expression level in both dark and light periods under short-day conditions.
Discussion
Plants accumulating free tetrapyrroles are particularly prone to photo-oxidative damage. Chl intermediates cause photo-oxidative damage to the cell. In particular, free Pchlide acts as a strong photosensitizer and produces singlet oxygen, leading to cell death (Kim et al. 2008). Tight control of this pathway is essential for survival, particularly for land plants that must tolerate intense and rapidly fluctuating light environments, which may have been the basis for the evolution of several different mechanisms that achieve this goal in land plants.
In our study, M. polymorpha, lacking DPOR, showed pale green and slow growth phenotypes under the 8-h L/16-h D condition, probably due to aberrant Pchlide accumulation during the dark period (fig. 3; table 1). This result indicates that DPOR in M. polymorpha plays an important role in Pchlide reduction and prevents Pchlide accumulation under short-day conditions. This behavior may have been one of the major driving forces for DPOR retention in the of M. polymorpha chloroplast genome. In general, bryophytes, including M. polymorpha, prefer shady conditions (Glime 2007). From an ecological perspective, bryophytes occupy light-limited regions (short-day conditions) such as the Arctic and show a pattern of increasing abundance with increasing latitude (Vitt and Pakarinen 1977; Jagerbrand et al. 2006). DPOR retention may also be important for M. polymorpha to adapt to these natural habitats.
One of the interesting aspects of the genus Marchantia is that it grows all over the world from tropical to polar regions. Therefore, in the future, it would be possible to dissect their strategies to adapt to various ecological environments by analyzing various ecotypes of Marchantia. At present, chloroplast genome sequences are only available for Japanese isolates. Further analysis of chloroplast genomes from various ecotypes, particularly focusing on chlB, chlL, and chlN, may shed light on their adaptational strategies to various light environments. In particular, drought-tolerant bryophytes could survive in sunny locations, where DPOR requirement would be less, suggesting the possibility that chlB, chlL, and chlN genes on their chloroplast genomes may initiate pseudogenization if day length is the main selection pressure for this gene disposition. The molecular evolutionary analysis of genes for DPOR subunits on their chloroplast genomes may prove our hypothesis presented in this study.
In contrast, it is likely that land plants, including angiosperms, several gymnosperms, and some Pteridophytes have established additional and more elaborate regulatory systems for Chl biosynthesis to inhibit the aberrant Pchlide accumulation that have led to the complete elimination of DPOR from their chloroplast genomes. In case of A. thaliana, the regulation of Chl biosynthesis has been attributed to metabolic feedback control of glutamyl-tRNA reductase, the first enzyme committed to tetrapyrrole synthesis, by heme and another negative regulator, FLU protein. The FLU protein contains a coiled-coil and tetratricopeptide repeat (TPR) domains, both implicated in protein–protein interactions. The TPR domain of FLU is required for interaction with the C-terminus of GluTR. Given that GluTR inhibition by heme requires the N-terminal region, heme and FLU seem to act independently on the same target to regulate Chl biosynthesis (fig. 4A) (Meskauskiene et al. 2001; op den Camp et al. 2003; Kauss et al. 2012). In addition to FLU, LPOR expression (PORA and PORB paralogs in A. thaliana) is positively regulated in the dark by DELLA, which is considered to be a transcriptional factor having unclear DNA-binding domain and plant-specific GRAS domain, and LPOR proteins bound to Pchlide to form a prolamellar body in chloroplasts, circumventing the photodamage caused by free Pchlide accumulation (Cheminant et al. 2011).
In M. polymorpha, our genome analysis revealed the presence of homologs for the genes involved in the control of Chl biosynthesis (table 2), suggesting the possibility that control systems analogous to those of A. thaliana are present and functional. However, we hypothesize that this condition is unlikely, given that pale green phenotype and Pchlide accumulation could not be prevented in the chlB mutant of M. polymorpha under short-day conditions (fig. 3A: table 1). Another reason that would support this speculation is the simplicity of the M. polymorpha genome. Only one homologous gene has been observed for each of the gene families, LPOR, DELLA, and HEMA (the gene encoding GluTR). Gene duplication must have played a major role in the evolution of elaborate mechanisms for controlling Chl biosynthesis in land plants (table 2). In A. thaliana, genes encoding LPOR have been duplicated, and the light-response expression among the duplicated genes has diversified. PORA and PORB expressions are primarily induced in darkness. Their expression in the dark can alleviate photodamage during the dark-to-light transition, with complex formation containing Pchlide (fig. 4A). On the other hand, PORC expression is strongly induced in light. Given that the ethylene signaling pathway involves PORC expression, duplicated LPOR genes are subjected to different hormone signaling pathways (Runge et al. 1996; Sperling et al. 1997; Bae and Choi 2008; Kauss et al. 2012). HEMA is also duplicated in angiosperms. They show different tissue expression patterns and distinct interaction ability with FLU among them (references cited in Tanaka R and Tanaka A 2007).
Table 2.
Homologous Genes Involved in Regulation of Chl Biosynthesis in the Dark in Marchantia polymorpha
| Gene Name in Arabidopsis thaliana | Gene ID in A. thaliana | Gene ID of Homologs in M. polymorpha |
|---|---|---|
| CHL27 | At3g56940 | AB889743 |
| FLU | At3g14110 | AB889744 |
| HEMA | At1g58290 (HEMA1) | AB889745 |
| At1g09940 (HEMA2) | ||
| LPOR | At5g54190 (PORA) | AB889746 |
| At4g27440 (PORB) | ||
| At1g03630(PORC) | ||
| DELLA | At2g01570 (RGA) | AB889747 |
| At1g14920 (GAI) | ||
| At1g66350 (RGL1) | ||
| At3g03450 (RGL2) | ||
| At5g17490 (RGL3) |
Note.—Gene IDs of LPOR and DELLA in A. thaliana refer to Cheminant et al. (2011).
The genome organization of M. polymorpha is much simpler than that of A. thaliana and shows lower gene redundancy, resulting in that gene functions tend to be in the primitive stage of subfunctionalization in M. polymorpha (Ueda et al. 2013). In the case of A. thaliana, LPOR accumulation with complex formation containing Pchlide in darkness can alleviate photodamage during the dark-to-light transition. The induction of MpLPOR expression in the chlB mutants should be beneficial to protect against photodamage because DPOR deficiency increases Pchlide accumulation. However, qRT-PCR analysis revealed that MpLPOR expression in the chlB mutants was not induced in darkness (fig. 4C), which suggests that the response of LPOR expression to Pchlide accumulation is absent or incomplete in M. polymorpha. Under this circumstance, it would be difficult to establish an elaborate regulatory system for Chl biosynthesis, which is comparable with that of A. thaliana. To investigate the actual function of the genes homologous to LPOR, HEMA (the gene encoding GluTR), and DELLA, it would be interesting to perform gene disruption experiments by exploiting the genetic tractability of M. polymorpha.
Development of a regulatory system for Chl biosynthesis and its harmonization to the land environment may eliminate genes for DPOR from the chloroplast genome. Our study provides insights for the detailed process, by which genes for DPOR have been eliminated from the chloroplast genome, and further analysis will shed light on the diversification between the nuclear and chloroplast gene expression networks during the evolution of land plants.
Supplementary Material
Supplementary tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Preliminary M. polymorpha sequence data were obtained from the Department of Energy Joint Genome Institute and Dr Takayuki Kohchi (Kyoto University). The authors thank Dr Yuichi Fujita (Nagoya University), Ms Junko Kishimoto (Hokkaido University), and Dr T. Kohchi for kindly providing the primary antibodies against ChlB, technical support for HPLC analysis, and critical reading of our manuscript, respectively. For experimental support, they thank Ms Nanami Kishida, Dr Nobuyoshi Mochizuki, Dr Akira Nagatani, and Ms Kanako Nariyama, Kyoto University. This work was supported by the Funding Program for Next Generation World-Leading Researchers (GS015, NEXT Program) from the Precursory Research for Embryonic Science and Technology (PRESTO), Japan Society of Promoting Science (JSPS) to Y.N. and the JSPS KAKENHI Grant number 25650126 to M.U.
Literature Cited
- Armstrong GA. Greening in the dark: light-independent chlorophyll biosynthesis from anoxygenic photosynthetic bacteria to gymnosperms. J Photochem Photobiol B Biol. 1998;43:87–100. [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Beale SI. Green genes gleaned. Trends Plant Sci. 2005;10:309–312. doi: 10.1016/j.tplants.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Cheminant S, et al. DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell. 2011;23:1849–1860. doi: 10.1105/tpc.111.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyoda S, Ishizaki K, Kataoka H, Yamato KT, Kohchi T. Direct transformation of the liverwort Marchantia polymorpha L. by particle bombardment using immature thalli developing from spores. Plant Cell Rep. 2008;27:1467–1473. doi: 10.1007/s00299-008-0570-5. [DOI] [PubMed] [Google Scholar]
- Chiyoda S, et al. Simple and efficient plastid transformation system for the liverwort Marchantia polymorpha L. suspension-culture cells. Transgenic Res. 2007;16:41–49. doi: 10.1007/s11248-006-9027-1. [DOI] [PubMed] [Google Scholar]
- Crandall-Stotler B, Stotler RE, Long DG. Morphology and classification of the Marchantiophyta. In: Shaw AJ, Goffinet B, editors. Bryophyte biology. Cambridge: Cambridge University Press; 2008. pp. 1–54. [Google Scholar]
- Fujita Y, Takagi H, Hase T. Identification of the chlB gene and the gene product essential for the light-independent chlorophyll biosynthesis in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1996;37:313–323. doi: 10.1093/oxfordjournals.pcp.a028948. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Takahashi Y, Shonai F, Ogura Y, Matsubara H. Cloning, nucleotide sequences and differential expression of the nifH and nifH-Like (frxC) genes from the filamentous nitrogen-fixing cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1991;32:1093–1106. [Google Scholar]
- Glime JM. Bryophyte ecology. In: Glime JM, editor. 2007. Light: the shade plants. Vol. 1. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. [cited 2014 Mar10]. Available from: http://www.bryoecol.mtu.edu/ [Google Scholar]
- Gray MW. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- Jagerbrand AK, Lindblad KEM, Bjork RG, Alatalo JM, Molau U. Bryophyte and lichen diversity under simulated environmental change compared with observed variation in unmanipulated alpine tundra. Biodivers Conserv. 2006;15:4453–4475. [Google Scholar]
- Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this path way. FEBS Lett. 2012;586:211–216. doi: 10.1016/j.febslet.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Apel K, Laloi C. No single way to understand singlet oxygen signalling in plants. EMBO Rep. 2008;9:435–439. doi: 10.1038/embor.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi J, Sato A, Tachida H SMBE Tri-National Young Investigators. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Relaxation of functional constraint on light-independent protochlorophyllide oxidoreductase in Thuja. Mol Biol Evol. 2006;23:941–948. doi: 10.1093/molbev/msj097. [DOI] [PubMed] [Google Scholar]
- Li J, Goldschmidt-Clermont M, Timko MP. Chloroplast-encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii. Plant Cell. 1993;5:1817–1829. doi: 10.1105/tpc.5.12.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Kowallik KV. Annotated english translation of Mereschkowsky's 1905 paper ‘Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Eur J Phycol. 1999;34:287–295. [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereschkowski C. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralbl. 1905;25:593–604. [Google Scholar]
- Meskauskiene R, et al. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Tanaka R, Satoh S, Tanaka A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell. 2005;17:233–240. doi: 10.1105/tpc.104.027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Tanaka R, Tanaka A. The major route for chlorophyll synthesis includes [3,8-divinyl]-chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:1803–1808. doi: 10.1093/pcp/pcm153. [DOI] [PubMed] [Google Scholar]
- Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. [Google Scholar]
- Ohyama K, et al. Gene content, organization and molecular evolution of plant organellar genomes and sex chromosomes: insights from the case of the liverwort Marchantia polymorpha. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:108–124. doi: 10.2183/pjab.85.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp RG, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, et al. Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 2010;15:614–624. doi: 10.1016/j.tplants.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Apel K, Lebedev N. Evolution of chlorophyll biosynthesis-the challenge to survive photooxidation. Cell. 1996;86:703–705. doi: 10.1016/s0092-8674(00)80144-0. [DOI] [PubMed] [Google Scholar]
- Runge S, Sperling U, Frick G, Apel K, Armstrong GA. Distinct roles for light-dependent NADPH:protochlorophyllide oxidoreductases (POR) A and B during greening in higher plants. Plant J. 1996;9:513–523. doi: 10.1046/j.1365-313x.1996.09040513.x. [DOI] [PubMed] [Google Scholar]
- Schoefs B, Franck F. Protochlorophyllide reduction: mechanisms and evolutions. Photochem Photobiol. 2003;78:543–557. doi: 10.1562/0031-8655(2003)078<0543:prmae>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Shikanai T, et al. Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci U S A. 1998;95:9705–9709. doi: 10.1073/pnas.95.16.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling U, van Cleve B, Frick G, Apel K, Armstrong GA. Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 1997;12:649–658. doi: 10.1046/j.1365-313x.1997.00649.x. [DOI] [PubMed] [Google Scholar]
- Suzuki JY, Bauer CE. Light-independent chlorophyll biosynthesis: involvement of the chloroplast gene chlL (frxC) Plant Cell. 1992;4:929–940. doi: 10.1105/tpc.4.8.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Takio S, Yamamoto I, Satoh T. Characterization of cDNA of the liverwort phytochrome gene, and phytochrome involvement in the light-dependent and light-independent protochlorophyllide oxidoreductase gene expression in Marchantia paleacea var. diptera. Plant Cell Physiol. 2001;42:576–582. doi: 10.1093/pcp/pce070. [DOI] [PubMed] [Google Scholar]
- Takenaka M, et al. Direct transformation and plant regeneration of the haploid liverwort Marchantia polymorpha L. Transgenic Res. 2000;9:179–185. doi: 10.1023/a:1008963410465. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- Ueda M, Kadowaki K. Gene content and gene transfer from mitochondria to the nucleus during evolution. In: Marechal-Drouard L, editor. Advances in botanical research: mitochondrial genome evolution. Oxford: Academic Press; 2012. pp. 21–40. [Google Scholar]
- Ueda M, et al. Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. Plant J. 2012;72:683–693. doi: 10.1111/j.1365-313X.2012.05115.x. [DOI] [PubMed] [Google Scholar]
- Ueda M, et al. Subfunctionalization of sigma factors during the evolution of land plants based on mutant analysis of liverwort (Marchantia polymorpha L.) MpSIG1. Genome Biol Evol. 2013;5:1836–1848. doi: 10.1093/gbe/evt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitt DH, Pakarinen P. The bryophyte vegetation, production, and organic components of Truelove Lowland. In: Bliss LC, editor. Truelove Lowland, Devon Island, Canada: a high arctic ecosystem. Alberta (Canada): University of Alberta Press; 1977. pp. 225–244. [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM, Huang YY. Chloroplast phylogenomics indicates that Ginkgo biloba is sister to cycads. Genome Biol Evol. 2013;5:243–254. doi: 10.1093/gbe/evt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.