Abstract
We isolated mouse embryo fibroblasts (MEFs) from N-acetylglucosaminyltransferase Va (GnT-Va) knockout mice and studied the effects of loss of expression of GnT-Va on asparagine- linked glycans (N-glycan) synthesis and the gene expression of groups of glycosyltransferases and galectins. Loss of GnT-Va expression caused aberrant expression of several N-glycan structures, including N-linked β(1,6) branching, poly-N-lactosamine, bisecting N-acetylglucosamine (GlcNAc) and sialic acid. Using quantitative reverse transcriptase-PCR (qRT-PCR), altered gene expression of several groups of glycosyltransferases and galectins was observed in GnT-Va null MEFs, supporting the observed changes in N-glycan structures. These results suggest that genetic disruption of GnT-Va ultimately resulted in altered MEFs gene expression and decreased tumor progression associated with loss of GnT-Va observed may result in part from a combination of effects from these altered gene expressions.
Keywords: Mouse embryo fibroblasts (MEFs), GnT-Va (Mgat5), Knockout, Glycosyltransferase, Galectin, qRT-PCR
1. Introduction
Many studies show that alterations in N-linked oligosaccharides of tumor cells are associated with tumorigenesis, progression and metastasis. N-linked β(1,6)-N-acetylglucosamine (GlcNAc) synthesized by N-acetylglucosaminyltransferase Va (GnT-Va or Mgat5) is one of the more common glycans up-regulated during malignant transformation [1]. Studies have proposed a close relationship between increased GnT-Va activity and its glycan products with enhanced cell invasiveness, and in some cases metastatic potential [2–4]. In GnT-Va deficient mice, the progression of mammary tumors induced by polyoma middle T oncoprotein (PyMT) expression was significantly suppressed [5].
It is well-documented that modification of cell surface adhesion molecules and growth factor receptors by GnT-Va is implicated in GnT-Va mediated, invasiveness-related phenotypes [2,6–8]. A recent report showed that aberrant expression of GnT-Va caused altered expression of surface sialyl Lewis X (Slex), a sialylated fucose-containing antigen related to tumor metastasis, by changing the expression of Slex biosynthesis-related glycosyltransferases [9]. We found that deletion of GnT-Va in mouse embryo fibroblasts (MEFs) causes up-regulation of β1 integrin transcripts, which consequently increased fibronectin- induced cell adhesion and reduced cell motility [10]. These results suggest that aberrant glycosylation caused by GnT-Va inhibition might regulate the gene expression of other proteins.
To further elucidate the molecular mechanisms whereby GnT-Va-modified asparagine-linked glycans (N-glycans) may affect tumor invasiveness-related phenotypes in MEFs and mammary tumor cells [7,10], we explored the effect of genetic disruption of GnT-Va on the expression of other glycosyltransferase and galectin transcripts. We report here, for the first time, that the genetically targeted deletion of GnT-Va causes the altered gene expression of several glycosyltransferases and galectins in MEFs using a newly developed qRT-PCR transcript analysis platform [11]. We speculate that some of the inhibited invasiveness-related phenotypes in MEFs caused by deletion of GnT-Va may result, at least in part, from the combination of altered expression of these glycosyltansferases and/or galectins.
2. Materials and methods
2.1. Antibodies and chemicals
Antibodies against galectin-1 and galectin-3, ERK1/2, and HRP-labeled anti-rabbit IgG and anti-mouse IgG were from Santa Cruz Biotechnology. Monoclonal antibody against β1 integrin was a product of BD Biosciences. OL.28 anti-polysialic acid antibody was kind gift from Dr. Karen Colley. Streptavidin–HRP and streptavidin–agarose were obtained from Rockland. WGA–agarose and biotinylated lectins were products of Vector Laboratories.
2.2. Isolation of MEFs
Mouse embryo fibroblasts (MEFs) were isolated from E13.5 embryos from heterozygous GnT-Va knock-out mice mating, and the genotype of MEFs was identified by PCR analysis using genomic DNA as described earlier [6].
2.3. GnT-V activity assay
The assay of GnT-V activity was performed using a synthetic trisaccharide acceptor as previously described [2].
2.4. Assay of cell growth and survival
MEFs were harvested, re-suspended in DMEM growth medium and seeded into six-well plates with 2–3 × 105 cells/well. Assay of cell growth and survival was performed as previously described [12].
2.5. N-Glycan analysis by HPLC
About 100 mg liver tissue were homogenized and N-glycans were released from glycoproteins by digestion of the sample with 10 U PN-Gase- F at 37 °C overnight. Released glycans were fluorescently labeled with 2-aminopyridine (2-AP) as previously reported [13]. Excess 2- AP was removed by gel filtration using a Sephadex G-15 column (1 × 50 cm) and the 2-AP labeled glycans were fractionated on a 4.6 × 250 mm TSK Amide-80 column as described previously [14].
2.6. Immuno- and lectin-blotting and lectin pull-down
Cells were harvested, lysed, and 20 μg of protein was used for immuno- and lectin-blotting [10]. For some experiments, cell lysates were boiled in denaturing buffer containing 0.1% SDS and 1 mM DTT for 5 min and treated with PNGase-F at 37 °C overnight before they were subjected to lectin-blotting. For lectin pull-down experiments, 500 μg of protein was incubated with 5 μg of a biotinylated lectin or lectin–agarose at 4 °C overnight, followed by addition of agarose–streptavidin.
2.7. Quantitative RT-PCR analysis
Primer pairs for investigated genes and control genes were designed within a single exon using conditions described in Nairn et al. [11] and listed in Table 1. Primers were validated with respect to primer efficiency and single product detection [11].
Table 1.
Sequences of primers used in qRT-PCR
| Gene | Abbreviation | Accession number | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|---|---|
| Mannoside acetylglucosaminyltransferase 1 | Mgat1 | NM_010794 | CCCTTCACCCAGTTGGACCTG | GCACCATAGACCTGGGCGAG |
| Mannoside acetylglucosaminyltransferase 2 | Mgat2 | NM_146035 | TGCTGGAGACTGTGGTATGC | ACTCAATTTGGGCACTCTGG |
| Mannoside acetylglucosaminyltransferase | Mgat3 | NM_010795 | GCGTGATGGTGTGCTGTTCC | ACAGGGACTTCCGCATGTGG |
| Mannoside acetylglucosaminyltransferase 4, isoenzyme A | Mgat4a | NM_173870 | GCGACAGACAGAAGGCAAACC | CCGACAGAGACGAGTGTAGGC |
| Mannoside acetylglucosaminyltransferase 4, isoenzyme B | Mgat4b | NM_145926 | AGGTGACGTGGTGGACATTT | GCTTCAGGCTCTCTTGCTCA |
| Mannoside N-acetylglucosaminyltransferase Va | Mgat5a | NM_145128 | CCCTGGAAGTTGTCCTCTCA | TCCTCTGCCAGTGCCTTAAT |
| Mannoside acetylglucosaminyltransferase 5, isoenzyme B | Mgat5b | NM_172948 | TGCCCTGTGACAGCACTGAG | GTAGGTAGCACTCTTGGCCGG |
| Glucosaminyl (N-acetyl) transferase 1, core2 | Gcnt1 | NM_173442 | CGAAGGCCATGTTTCCAACGG | TCCGAAGACGCACACAGAGC |
| Glucosaminyltransferase, I-branching enzyme | Gcnt2 | NM_008105 | CGGTTGATTTGCTCCATTG | TATTGAGCGTCACCCAGAAAT |
| Glucosaminyl (N-acetyl) transferase 3, mucin type | Gcnt3 | NM_028087 | ACCCAGGCTCTGCTGAATAA | AGGTAGTCGGCCTCTGTGAA |
| Beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) | B3gat1 | NM_029792 | ACCGTGACATCGTGGAAGTGG | GGATGGTGGGCAGCGTGTC |
| Beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) | B3gat2 | NM_172124 | CACACTCGGACAGAGAAGGTC | CCTCAATGTTCACTGTGTCCA |
| UDP-Gal:betaGlcNAc beta-1,4-galactosyltransferase, polypeptide 1 | B4galt1 | NM_022305 | GGCGTCACCCTCGTCTATTA | GCCCTGCAGTGTAGAGGAGA |
| UDP-Gal:betaGlcNAc beta-1,4-galactosyltransferase, polypeptide 2 | B4galt2 | NM_017377 | GCCGCTATTCTCCACCCGAC | GGTGCTCCCGGTGTCTAAAGG |
| UDP-Gal:betaGlcNAc beta-1,4-galactosyltransferase, polypeptide 3 | B4galt3 | NM_020579 | TGAAGAATGGGACTGCTTGTT | TCGCACACATACAGGTTATGG |
| UDP-Gal:betaGlcNAc beta-1,4-galactosyltransferase, polypeptide 4 | B4galt4 | NM_019804 | GACGTGGACCTGGTGCCTG | GTTCCGGCCCACCACCAAG |
| UDP-Gal:betaGlcNAc beta-1,4-galactosyltransferase, polypeptide 5 | B4galt5 | NM_019835 | CGGTCCTCCTGAGACACCT | CACATAGAAGGCGAACTGCA |
| UDP-Gal:betaGlcNAc beta-1,4-galactosyltransferase, polypeptide 6 | B4galt6 | NM_019737 | CGGCACACAACCATTCAAT | ATCACACAGTCCCAGGCTCT |
| Xylosylprotein beta1,4-galactosyltransferase, polypeptide 7 | B4galt7 | NM_146045 | CGGGCAGCACTCATCAATGTG | GAGGGAGCAGGTCCACATCG |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 | B3gnt1 | NM_175383 | GGAGAGCTTGCTGAGACCTG | CCTTTCCTCCAGCCACATAA |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 | B3gnt2 | NM_016888 | TTAGGTGGGTGAGCACTTCC | GATGGGTGTTCACAAACACG |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 | B3gnt3 | NM_028189 | TCTTCCTTGTGGGCTCAGAC | GTATTTCTGCGCTTCCAGCT |
| Beta-1,3-N-acetylglucosaminyltransferase-4 | B3gnt4 | NM_198611 | CCAGCCGTCATCGCCTCTTC | CACACTCCGAAGGCTCCAGC |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 | B3gnt5 | NM_052993 | GCAAGGCATTCAGATGACAA | ATGTTGGCTGGAATCTGCAT |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 | B3gnt7 | NM_145222 | GGGCCACGAATGGGAGTCAG | TCCTCTTGCTTGGAGGCTGTG |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 | B3gnt8 | NM_146184 | CGGCGCTATGGTGACCTACTG | TCAGCAGCAGCAGGTCCTTG |
| Sialyltransferase 4A (beta-galactosidase alpha-2,3-sialytransferase) | St3gal1 | NM_009177 | TCCTACAACTGCACAGCGTCG | TGTTTCGCCTGGTGCCTGG |
| Sialyltransferase 5 (N-acetyllacosaminide alpha 2,3-sialyltransferase) | St3gal2 | NM_178048 | GCTCTCTTCGGGTGTGGTTCC | ATGCTGTGGTGCGAGTAGGTG |
| Sialyltransferase 6 (N-acetyllacosaminide alpha 2,3-sialyltransferase) | St3gal3 | NM_009176 | CAGCAAGAAACCCAGACCAT | ATGAATGGCTCCGTCCATAG |
| Sialyltransferase 4C (beta-galactosidase alpha-2,3-sialytransferase) | St3gal4 | NM_009178 | GCTCCTGTGGCTGGCTACG | GGGTCAAAGTGGGCCGACTC |
| Sialyltransferase 9 (CMP-NeuAc:lactosylceramide alpha-2,3-sialyltransferase) | St3gal5 | NM_011375 | AGCCTCTTGGATATGCTGCCC | CGTTCCCAACAACCACACAGC |
| Sialyltransferase 10 (alpha-2,3-sialyltransferase VI) | St3gal6 | NM_018784 | ATGGTGGCATTCCCGTAGTA | AAGTGCACCTCGCTGGTTT |
| Sialyltransferase 1 (beta-galactoside alpha-2,6-sialyltransferase) | St6gal1 | NM_145933 | TGCGTGTGGAAGAAAGGGAGC | CTCCTGGCTCTTCGGCATCTG |
| Beta-galactoside alpha-2,6-sialyltransferase II (ST6GalII-pending) | St6gal2 | NM_172829 | AGCAATCCTGCGGCACCTATG | CCGCTGCTTGCCCTGTAGAG |
| Sialyltransferase 8A (alpha-2,8-sialytransferase) | St8sia1 | NM_011374 | TTTGGTGAGTGCAGCCTTGGG | TGCATGTTCACCGAGAAGGGC |
| Sialyltransferase 8B (alpha-2,8-sialytransferase) | St8sia2, STX | NM_009181 | CGGCCTCAATGGGAGCATCC | AGCATTGACCCACTCCACACG |
| Sialyltransferase 8C (alpha-2,8-sialytransferase) | St8sia3 | NM_009182 | TCCAACCACAGCACAAACAT | AATAACTTCCGGTCCCTGCT |
| Sialyltransferase 8D (alpha-2,8-sialytransferase) | St8sia4, PST | NM_009183 | CCTAGCACAGGTCTCCTCATG | GGAAATGGCCAGAATCCATA |
| ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 | St8sia5 | NM_153124 | CCTGGTATCCTTCCGAGTCA | TGCGGGTGGAAGAAGTAGAC |
| Alpha-2,8-sialyltransferase VI | St8sia6 | NM_145838 | GTTGATGGAGGGAAGCAGAG | GGGCTCCAGAGTGTCTTCAG |
| Lectin, galactose binding, soluble 1 | Lgals1 | NM_008495 | CAGCAACAACCTGTGCCTAC | ACAATGGTGTTGGCGTCTC |
| Lectin, galactose binding, soluble 2 | Lgals2 | NM_025622 | AAGAAACCCTCAACCTGCATT | GGCCACCTTCACTGGTGTTA |
| Lectin, galactose binding, soluble 3 | Lgals3 | NM_010705 | CCTTGCCTGGAGGAGTCAT | TGTTTGCGTTGGGTTTCAC |
| Lectin, galactose binding, soluble 4 | Lgals4 | NM_010706 | TCGTCCGGAGACATAGCTTT | CATAAAGCTGTTGCGAACCA |
| Lectin, galactose binding, soluble 6 | Lgals6 | NM_010707 | CAACTTCTCCCATCGGTTCC | TGAGGTCACCATTGATCTCCA |
| Lectin, galactose binding, soluble 7 | Lgals7 | NM_008496 | CTATGCGGTGAGGAGCAAG | ACAACCTCGGAAGTGTCCAG |
| Lectin, galactose binding, soluble 8 | Lgals8 | NM_018886 | GACTTTCAGCTGGGCAACA | TGAACCGAGGGTTAAAGTGG |
| Lectin, galactose binding, soluble 9 | Lgals9 | NM_010708 | CCGTTTCAATGAGAATGCTG | AGCAGACTTCGCTCTTCCTG |
| Lectin, galactose binding, soluble 12 | Lgals12 | NM_019516 | TCAGCCAGATGTTGCCTTC | GTGGGTGTTGCAGATGACAT |
| Ribosomal Protein L4a | RPL4 | NM_024212 | GACAGCCCTATGCCGTCAGTG | GCCACAGCTCTGCCAGTACC |
Normalization control gene.
The RNeasy kit (Qiagen) was used to isolate total RNA from MEFs. One microgram of total RNA was used for a 20 μl cDNA synthesis reaction using the SuperScript III First-Strand Synthesis System (Invitrogen). Reactions were assembled as described [11]. The control gene, Ribosomal Protein L4 (RPL4, NM_024212) was included on each plate to control for run variation and to normalize individual gene expression.
Triplicate cycle threshold (Ct) values for each gene were averaged and the standard deviation was calculated. Samples that resulted in a standard deviation of >0.5 Ct units were re-run until values with standard deviations within an acceptable range were acquired. The logarithmic average Ct value for each gene and the control gene was converted to a linear value using the conversion: 2−Ct. Converted values were normalized to RPL4 by dividing the individual gene value by the control gene value. Normalized values were scaled so that genes that were below the level of detection were given a value of 1 × 10−6 and this value was used as the lower limit on histograms. Following normalization and scaling, expression data is reported as the relative transcript abundance for each gene assayed. Error bars represent one standard deviation from the mean value.
3. Results
3.1. Characterization of MEFs from embryos
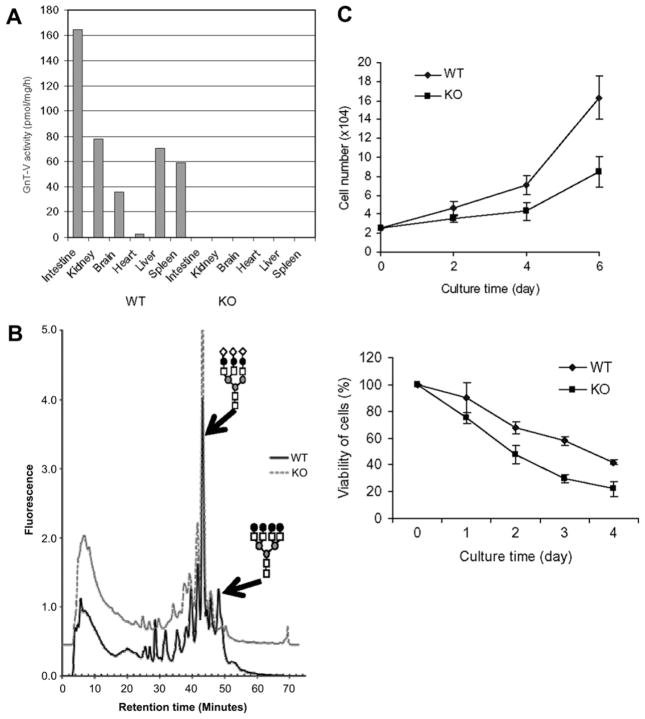
Compared with wild-type, GnT-Va activity was absent in major tissues from GnT-Va null mice (Fig. 1A). Analysis of 2-AP labeled N-linked glycans by HPLC indicated a complete disappearance of the peak representing the asialo tetra-antennary N-glycan in liver (Fig. 1B), indicating the disappearance of N-linked β(1,6) branching caused by the absence of GnT-Va. We have previously observed that GnT-Va null MEFs displayed undetectable GnT-Va activity and lost the typical morphology of a fibroblast when compared to wild-type cells [10]. GnT-Va deficient (KO) and wild-type (WT) MEFs were isolated from 13.5 days embryos. Cell growth in serum-containing medium (Fig. 1C, top) and cell viability in serum-free medium (Fig. 1C, bottom) were measured. GnT-Va null MEFs showed significant decreases in both cell growth and cell survival indicating that the rate of proliferation of MEFs was affected by deletion of GnT-Va.
Fig. 1.
Isolation of mouse embryonic fibroblasts (MEFs) from GnT-Va null mice. (A) Mouse tissues were collected, homogenized, and GnT-Va activities determined. (B) N-Glycans were released from liver tissue and fluorescently labeled with 2-PA, N-glycan structures were characterized by HPLC. N-glycan structural symbol: tri-antennary N-glycan (top) and tetra-antennary N-glycan (bottom). (C) (top) growth curve of MEFs cultured in serum-containing medium; (bottom) viability of cells cultured in serum-free medium. Each point represents the mean value of cell number from three independent experiments. WB: Western blotting.
3.2. Aberrant N-glycan expressions in GnT-Va deficient MEFs
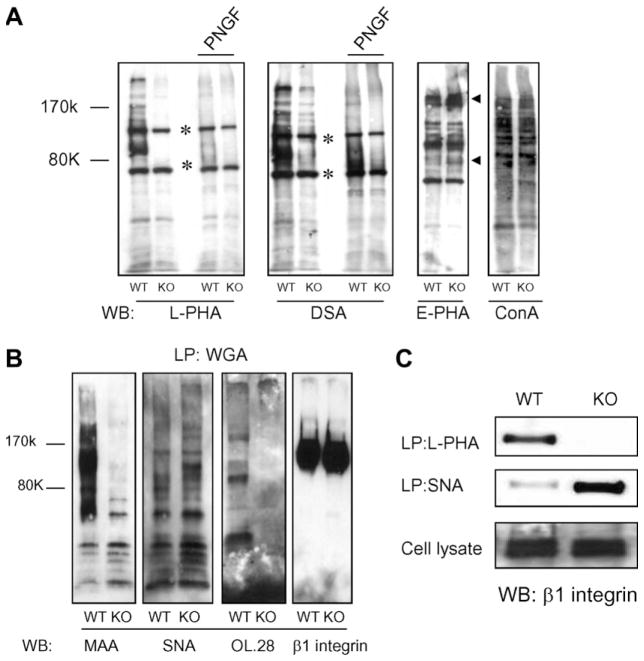
To further study N-glycans changes caused by deletion of GnT-Va in MEFs, lectin-blotting and -precipitation experiments were performed. The binding of not only leucoagglutinating phytohemagglutinin (L-PHA), but also DSA was extensively suppressed in GnT-Va null MEFs (Fig. 2A), indicating the suppression of both β(1,6) branched and poly-N-acetyllactosamine structures, respectively. This result confirmed our observation that poly-N-acetyllactosamine is synthesized preferentially on N-glycans expressing the β(1,6) branch [6]. After treatment of cell lysates with PNGase-F, most binding of L-PHA and DSA were abolished, confirming these bound N-linked glycans. The expression of either high mannose or biantennary N-linked oligosaccharides was not significantly affected by deletion of GnT-Va, as detected by ConA binding (Fig. 2A). Increased E-PHA binding was observed for a few, but not all proteins from GnT-Va knockout cells (indicated as arrows, Fig. 2A), indicating selective increased expression of bisecting N-linked GlcNAc. α(1,3) fucosylation did not appear to be affected, as detected by Lotus agglutinin (LTA) binding (data not shown). To study the effect of GnT-Va knockout on sialylation, we used WGA–agarose first to pull-down sialic acid-containing glycoproteins, followed by blotting with MAA, SNA and anti-polysialic acid antibody (OL.28), respectively. Surprisingly, α(2,6)-sialylation detected by Sambucus nigra agglutinin (SNA) was increased, while α(2,3)-sialylation and α(2,8)-polysialylation, detected by Maakia ameurinsin agglutinin (MAA) binding and OL.28, respectively, were dramatically decreased in GnT-Va null MEFs (Fig. 2B). Consistent with these findings, β1 integrin, an abundant surface proteins on MEFs, showed undetectable expression of β(1,6) branching in GnT-Va null MEFs, but a significant increase of α(2,6)-sialylation, compared to wild-type cells (Fig. 2C). These results indicate that deletion of GnT-Va caused significant changes in not only β(1,6) branching, but also other N-glycan expression in MEFs.
Fig. 2.
Deletion of GnT-Va caused altered N-glycan expression. (A) N-Glycan structures were detected using cell lysates (with or without PNGase-F pre-treatment) by lectin-blotting. *Non-specific binding. The small arrows indicated increased binding proteins. L-PHA, leucoagglutinating phytohemagglutinin; DSA, D. stramonium agglutinin; E-PHA, erythroagglutinating phytohemagglutinin; ConA, concanavalin A; PNGF, PNGase-F; WB:Western blotting. (B) Glycoproteins were lectin precipitated (LP) from cell lysates using WGA–agarose, followed by blotting with lectins and antibodies. Anti-β1 integrin was used for loading control. WGA, wheat germ agglutinin; MAA, M. amurensis agglutinin; SNA, S. nigra agglutinin; OL.28, anti-polysialic acid antibody. (C) Cell lysates were subjected to L-PHA and SNA precipitation (LP), respectively, followed by SDS–PAGE, blotting, and detection using antibody against β1 integrin (top two panels). Immunoblots of cell lysates with anti-β1 integrin were performed as control to confirm amount of precipitated β1 integrin in both cell lines (bottom panel).
3.3. Altered gene expression of glycosyltransferses in GnT-Va null MEFs
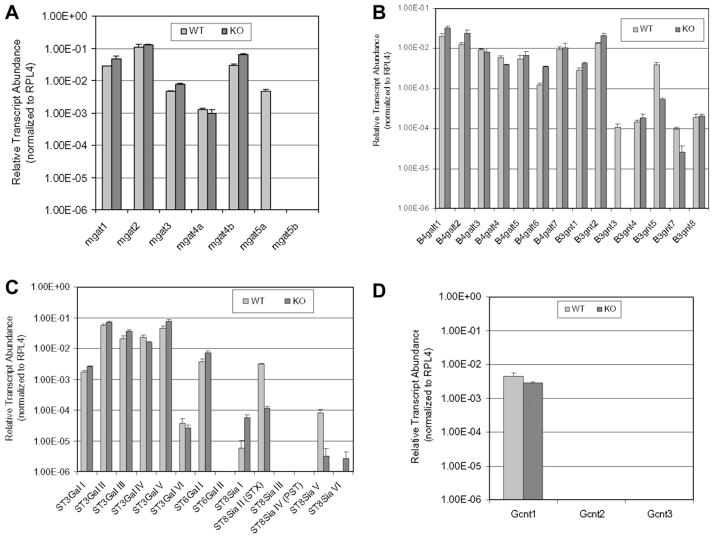
To begin a study of the mechanisms by which the different N-glycan structures were changed in GnT-Va deficient MEFs, transcript levels of relevant groups of glycosyltransferases, listed in Table 2, were detected by qRT-PCR using total RNA isolated from MEFs. As shown in Fig. 3 and Table 2, altered gene expression patterns were observed for several groups of glycosyltransferases after deletion of GnT-Va. The expression of 7N-acetylglucosaminyltransferase (GnT) transcripts was determined (Fig. 3A and Table 2). The expression of GnT-I, GnT-III and GnT-IVb was significantly increased in GnT-Va null cells. GnT-Va was detected in wild-type but not in GnT-Va null cells. GnT-Vb was not detected in either wild-type or GnT-Va null cells. Two other groups of glycosyltransferases that regulate the initiation and extension of N-acetyllactosamine chains on glycoproteins and glycolipids, β(1,3)-N-acetylglucosaminyltransferases (β3GnT) and β(1,4)galactosyltransferases (β4GalT), were affected by either up- or down-regulation after deletion of GnT-Va. Among them, β3GnT-III, V and VII were extremely reduced in GnT-Va null MEFs, while β3GnT-I and II and β4GalT-I, II, VI, were increased (Fig. 3B and Table 2).
Table 2.
Altered gene expression profile of glycosyltransferases in GnT-Va null MEFs
| Gene | WT (mean)a | KO (mean)a | Ratio (WT/KO) |
|---|---|---|---|
| Mgat1 | 0.027673 | 0.046369 | 0.596791 |
| Mgat2 | 0.108606 | 0.124820 | 0.870100 |
| Mgat3 | 0.004548 | 0.007684 | 0.591858 |
| Mgat4a | 0.001230 | 0.000953 | 1.290909 |
| Mgat4b | 0.029364 | 0.063232 | 0.464377 |
| Mgat5a | 0.004605 | 0.000001 | 4605.313126 |
| Mgat5b | 0.000001 | 0.000001 | 1.000000 |
| Gcnt1 | 0.004553 | 0.002759 | 1.650467 |
| Gcnt2 | 0.000001 | 0.000001 | 1.000000 |
| Gcnt3 | 0.000001 | 0.000001 | 1.000000 |
| B4galt1 | 0.019953 | 0.032838 | 0.607618 |
| B4galt2 | 0.012594 | 0.024487 | 0.514288 |
| B4galt3 | 0.009568 | 0.008144 | 1.174870 |
| B4galt4 | 0.005826 | 0.003895 | 1.495915 |
| B4galt5 | 0.005516 | 0.006892 | 0.800403 |
| B4galt6 | 0.001240 | 0.003591 | 0.345244 |
| B4galt7 | 0.009342 | 0.010353 | 0.902375 |
| B3gnt1 | 0.002855 | 0.004278 | 0.667305 |
| B3gnt2 | 0.013662 | 0.020811 | 0.656500 |
| B3gnt3 | 0.000110 | 0.000001 | 110.138458 |
| B3gnt4 | 0.000144 | 0.000184 | 0.781529 |
| B3gnt5 | 0.003916 | 0.000536 | 7.311063 |
| B3gnt7 | 0.000098 | 0.000027 | 3.681326 |
| B3gnt8 | 0.000195 | 0.000205 | 0.948217 |
| St3gal1 | 0.001698 | 0.002569 | 0.660952 |
| St3gal2 | 0.057057 | 0.070288 | 0.811768 |
| St3gal3 | 0.021313 | 0.036801 | 0.579139 |
| St3gal4 | 0.023461 | 0.015987 | 1.467521 |
| St3gal5 | 0.044011 | 0.077479 | 0.568035 |
| St3gal6 | 0.000038 | 0.000026 | 1.446651 |
| St6gal1 | 0.003774 | 0.007324 | 0.515373 |
| St6gal2 | 0.000001 | 0.000001 | 1.000000 |
| St8sia1 | 0.000006 | 0.000056 | 0.103812 |
| St8sia2, STX | 0.003064 | 0.000116 | 26.359330 |
| St8sia3 | 0.000001 | 0.000001 | 1.000000 |
| St8sia4, PST | 0.000001 | 0.000001 | 1.000000 |
| St8sia5 | 0.000078 | 0.000003 | 23.826403 |
| St8sia6 | 0.000001 | 0.000003 | 0.376690 |
0.000001 = below the level of detection.
The ratio is a simple division of the data for WT and KO for a specific gene.
Relative transcript abundance (as described in Section 2).
Fig. 3.
Deletion of GnT-Va resulted in altered gene expression of glycosyltransferases. (A) Expression of several N-acetylglucsoaminyltransferases (gnt). (B) Relative transcript levels of β4-galactosyltransferases (B4galt 1–7) and β3-N-acetylglucosaminyltransferases (B3gnt 1–8). (C) Relative transcript levels of α2,3-sialyltransferases (ST3Gal I–VI), α2,6-sialyltransferases (ST6Gal I–II) and α2,8-sialyltransferases (ST8Sia I–VI). (D) Relative transcript levels of core2N-acetylglucosaminyltransferase (Gcnt1), I-branching N-acetylglucosaminyltransferase (Gcnt2), mucin-type N-acetylglucosaminyltransferase (Gcnt3). Data expressed as “relative transcript abundance” are plotted on a log scale for each gene assayed. Error bars represent one standard deviation from the mean of triplicate values.
An expression pattern of either up- or down-regulation was observed within another group of glycosyltransferases, the sialyltransferases (ST), which consist of three specific subfamilies based on their substrate specificity and type of sialyl linkages of the products (α2,3, α2,6 and α2,8). The α(2,3)sialyltransferases (ST3Gals) catalyze the formation of α(2,3) sialylated structures and play an essential role in performing the sialylation of LeX structure (sLex). As shown in Fig. 3C, the expression of ST3Gal-I, III and V were increased, while ST3Gal-IV and VI were reduced after deletion of GnT-Va. Decreased expression of ST3Gal-IV and VI could result in the observed significant inhibition of the biosynthesis of α(2,3) sialylated structures (Fig. 2B). The α(2,6) sialyltransferases (ST6Gals) are another group of the sialyltransferase family and generate α(2,6)-linked sialylated structures. A significant increase in ST6Gal-I transcripts was found in GnT-Va null cells, which was consistent with N-glycan changes detected by SNA binding (Fig. 2B). A terminal linear homopolymer of α(2,8) linked sialic acids is another important form of sialylation catalyzed by α(2,8) sialyltransferase (ST8sia), which participates in neural development [15]. Two major ST8sia which are responsible for the biosynthesis of polysialylation (PSA) are ST8sia-II/STX and ST8sia-IV/PST [16,17]. Interestingly, the expression of ST8sia-II/STX was extremely decreased in GnT-Va null MEFs, which was consistent with reduced polysialylation determined by anti-polysialylation antibody binding, whereas the level of ST8sia-IV/PST was undetectable (Fig. 3C). A large decrease of ST8sia-V was also observed in null MEFs, although its expression level was, however, very low.
In addition, deletion of GnT-Va also caused changes in some glycosyltransferase transcripts responsible for the biosynthesis of O-glycans. The expression of core2GnT-I, which catalyzes the formation of the branched core2 and core4 O-glycans [18] was reduced (Fig. 3D), while core2GnT-II and III expression were very low and undetectable, respectively, in MEFs.
3.4. Altered gene expression of galectins in GnT-Va null MEFs
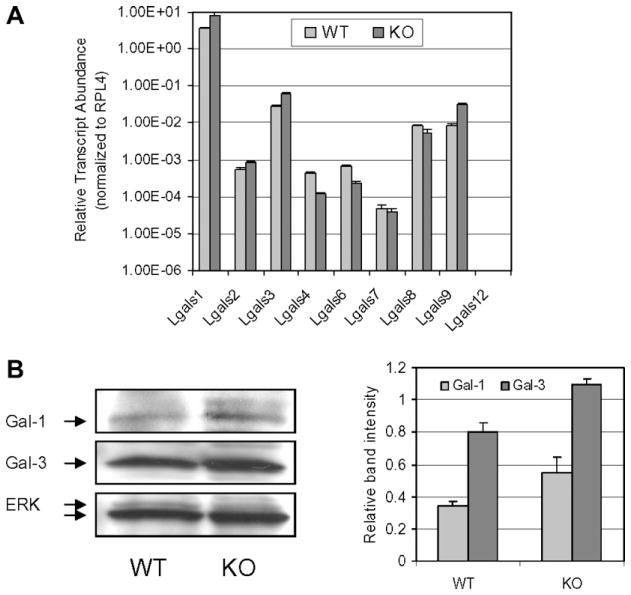
Galectins (Gal) are a family of carbohydrate-binding proteins with an affinity for β-galactosides [19]. To date, 15 mammalian galectins have been identified. To investigate if deletion of GnT-Va causes altered expression of galectins in MEFs, the transcript expression pattern of galectins was studied. The transcript abundance of Gal-1, Gal-2, Gal-3 and Gal-9 was increased to different degrees, while transcript levels of Gal-4, Gal-6 and Gal-8 were reduced in GnT-Va null MEFs (Fig. 4A and Table 3). Consistent with the transcript expression, Western blotting showed increased protein expression of both Gal-1 and Gal-3 (Fig. 4B), the two most abundant galectins in mouse, in GnT-Va null MEFs.
Fig. 4.
Deletion of GnT-Va resulted in altered galectins gene expression. (A) Relative transcript levels for galectins: galectin 1 (Lgals1), galectin 2 (Lgals2), galectin 3 (Lgals3), galectin 4 (Lgals4), galectin 6 (Lgals6), galectin 7 (Lgals7), galectin 8 (Lgals8), galectin 9 (Lgals9) and galectin 12 (Lgals12). (B) The expression levels of galectins-1 and -3 and ERK in MEFs were detected by immunoblotting using the respective antibodies (left panel). Relative levels of gal-1 and -3 were calculated by band intensity of gal-1 or gal-3/intensity of ERK, and data represent the mean (±S.D.) of three independent experiments (right panel).
Table 3.
Altered gene expression profile of galectins in GnT-Va null MEFs
| Gene | WTa | KOa | Ratio (WT/KO) |
|---|---|---|---|
| Lgals1 | 3.569301 | 7.843389 | 0.455071 |
| Lgals2 | 0.000560 | 0.000838 | 0.668573 |
| Lgals3 | 0.027261 | 0.062521 | 0.436029 |
| Lgals4 | 0.000433 | 0.000118 | 3.661759 |
| Lgals6 | 0.000673 | 0.000227 | 2.963587 |
| Lgals7 | 0.000048 | 0.000038 | 1.282823 |
| Lgals8 | 0.008210 | 0.005495 | 1.494213 |
| Lgals9 | 0.008108 | 0.031355 | 0.258580 |
| Lgals12 | 0.000001 | 0.000001 | 1.000000 |
0.000001 = below the level of detection.
The ratio is a simple division of the data for WT and KO for a specific gene.
Relative transcript abundance (as described in Section 2).
4. Discussion
In the present study, we isolated MEFs from GnT-Va knockout mice and explored the effects of genetic disruption of one glycosyltransferase, GnT-Va, on the gene expressions of other glycosyltransferases and galectins. We showed that deletion of GnT-Va had marked effects on MEFs growth on plastic and cell survival. Supporting these observation, the phosphorylation of both ERK and PKB was significantly inhibited in GnT-Va null mammary tumor cells and MEFs [5,7], suggesting a possible role of GnT-Va in regulating cell growth and apoptosis.
Consistent with undetectable activity of GnT-Va in the null MEFs, we found that the expression of β(1,6) branched oligosaccharides and poly-N-acetyllactosamine, determined by L-PHA and DSA binding, respectively, were almost entirely inhibited after deletion of GnT-Va in MEFs. Interestingly, “bisected” N-linked glycans produced by GnT-III, and recognized by E-PHA binding, were increased in GnT-Va null MEFs, consistent with the possible competition between GnT-III and GnT-Va for biantennary substrates during glycoprotein biosynthesis in the Golgi [20]. Of note, significant decreases in α(2,3) sialylation and α(2,8) polysialylation were observed after deletion of GnT-Va, reflecting changes in enzymatic activity of both α(2,3) sialyltransferases and α(2,8) polysialyltransferases. A significant increase in α(2,6) sialylation, detected by SNA binding was observed in GnT-Va null cells, indicating increased activity of ST6Gal resulting from deletion of GnT-Va. This result was consistent with our recent finding that knockdown of GnT-Va by siRNA led to increased α(2,6) sialylation expression on cell surface EGFR in human mammary tumor cells [8]. Supporting these findings, a recent report [21] demonstrated that targeted deletion of Mgat4a also caused absence of N-linked β(1,6) branching on Glut-2 of pancreatic islet β cells. These results suggested that genetic disruption of one glycosyltransferase could affect others changes in glycan expression.
To further investigate if altered N-glycan structures resulted from changes in gene expression of different glycosyltransferases, transcript levels of different families of glycosyltransferases were evaluated using qRT-PCR. In the family of GnTs, several transcripts were up-regulated due to the deletion of GnT-Va. Increased gene expression of GnT-III may account for the enhanced E-PHA binding in GnT-Va null MEFs, indicating for the first time that the opposing expression pattern of GnT-Va and GnT-III glycan products may be due to regulation of these enzymes at the level of transcription. This result is further supported by our recent observation that increased gene expression of GnT-Va was accompanied by reduced GnT-III gene expression in Her-2/neu-induced mouse mammary tumor tissue (unpublished data), indicating that these two genes might be regulated by an opposing manner.
Poly-N-acetyllactosamine is synthesized preferentially on N-glycans expressing the β(1,6) branch. In our study, we found that poly-N-acetyllactosamine showed a concomitant decreased expression when N-linked β(1,6) branches were eliminated by knockout of GnT-Va. Although some of the transcript levels of glycosyltransferases which are involved in the biosynthesis of poly-N-acetyllactosamine were extremely down-regulated, enhanced gene expression within this family was also observed. Reduced expression of poly-N-acetyllactosamine, detected by DSA binding likely resulted, at least in part, from altered gene expression of relevant glycosyltransferases.
Sialylation is another important modification of glycoproteins and aberrant sialylation has been reported to relate to the progression of several types of human tumors [22,23]. In mammalian cells, six α(2,3) sialyltransferases (ST3Gal I–VI) have been identified. Down-regulation of ST3Gal IV and VI was observed in GnT-Va knockout MEFs, which likely accounts for the significant decrease in expression of α(2,3) sialylation detected by MAA binding.
The transcripts for ST6GalTs, forming α(2,6) sialylation structures, were significantly up-regulated in GnT-Va deficient MEFs, which is consistent with the observed increased expression of α(2,6) sialylation, detected by SNA binding, suggesting that elimination of GnT-Va expression negatively regulate the formation of α(2,6) sialylation. This hypothesis was further supported by our recent observations that knockdown of GnT-Va caused increased α(2,6) sialylation on EGFR [8], and enhanced gene expression of GnT-Va in Her-2/neu induced mammary tumor tissue was accompanied by down-regulation of ST6GalT (unpublished data). Indeed, α(2,6) sialylation of cell surface N-glycans was reported to inhibit tumor formation in vivo [24] and attenuate the neoplastic phenotype of human colon cancer cells [22], indicating an opposing role of increased ST6GalT, compared to increased GnT-Va, in mediating tumorigenesis.
Polysialic acid (PSA) is another post-translational modification consisting of a homopolymer of α(2,8) linked sialic acid, which plays a critical role in regulating neural development and tumor progression [15]. Dramatically, concomitant with a marked reduction of polysialylation detected by anti-poly-sialylation antibody, we found that the transcript levels of ST8Sia II/STX were significantly reduced after deletion of GnT-Va, while the level of ST8Sia IV/PST was very low. N-CAM is a major carrier of PSA [25], and the presence of PSA attenuates the adhesive property of N-CAM and increases cell motility. Decreased expression of PSA caused by down-regulation of ST8Sia II/STX transcripts after deletion of GnT-Va may, therefore, lead to a phenotype with increased cell adhesive properties and decreased cell motility. This hypothesis is consistent with inhibited cell growth and supportive of previous findings that GnT-Va null MEFs and tumor cells showed decreased cell motility [7,10].
Intriguingly, deletion of GnT-Va also affected the transcript expression level of core2GnT-I, a Golgi enzyme that catalyzes the formation of β(1,6) branched structure attached to O-Gal-NAc [18]. This enzyme was found to be expressed in the developing mouse embryo and was closely correlated with vessel invasion and depth of invasion of several human cancers.
Among 15 identified mammalian galectins, Gal-1 and -3 have been intensively studied for their roles in regulating cell proliferation, apoptosis and tumor progression [26,27]. In our study, we found that knockout of GnT-Va also resulted in increased transcript levels of Gal-1 and Gal-3, accompanied by increased protein expression detected by Western blotting. Gal-9 was originally identified as a tumor antigen and is associated with suppression of cell growth and induces apoptosis. Increased Gal-1, -3 and -9 may be related to the inhibited cell growth and survival of GnT-Va null MEFs observed in our study. Of note, gene expression of Gal-1 was inhibited in Her-2/neu-induced mouse mammary tumor tissue and cells [28], suggesting that up-regulated gene expression of Gal-1 after deletion of GnT-Va may function in GnT-Va-mediated mammary tumor progression[5]. Among the down-regulated galectins, Gal-8 showed relatively higher expression in GnT-Va null MEFs. This galectin has been reported to be a matrix protein that can positively or negatively modulate cell adhesion [29] and its down-regulation may, therefore, contribute to some of the altered cell adhesion properties caused by deletion of GnT-Va in MEFs [10].
The altered gene expression of several groups of glycosyltransferases and galectins caused by genetic disruption of one specific glycosyltransferase has not as yet been reported. Studies have shown that knockout or knockdown of GnT-Va caused aberrant N-glycosylation of surface growth factor receptors and are associated with altered signaling pathway mediated by these receptors [7,8,10,30]. These aberrant signals might up- or down-regulate gene expression patterns of some glycosyltransferases and glycan-recognition proteins. We did find that knockout of GnT-Va in MEFs caused increased gene expression of β1 integrin [10]. Furthermore, increased gene expression of some integrins was also observed in SH-SY5Y cells after GnT-Vb knockdown by siRNA [31]. These findings suggest that the expression level of GnT-Va indeed played a role in regulating expression of other proteins. In addition, it is believed that the galectin binding increases if galactose is attached to an N-acetylglucosamine of glycoproteins on the cell surface forming N-acetyllactosamine [32]. A recent paper shows that galectins cross-link glycoproteins on the cell surface with avidities dependent on N-glycan structure and number [30]. Knockout of GnT-Va caused reduced expression of poly-N-acetyllactosamine structures, preferred ligands of the galectins, which could possibly render feedback regulation on the expression of some galectins, causing increased gene expression of Gal-1 and Gal-3. In conclusion, our results suggest possible coordination between GnT-Va expression and the expression of several glycosyltransferases and lectins, although the mechanisms that result in this coordination are, as yet, unknown.
Acknowledgments
We thank Dr. Karen Colley for providing with anti-polysialylation antibody (OL.28). This research was supported by NIH Grant RR018502 (to K.M. and M.P.).
Abbreviations
- MEFs
mouse embryo fibroblasts
- GnT-Va
N-acetylglucosaminyltransferase Va
- GlcNAc
N-acetylglucosamine
- N-glycan
asparagine-linked glycans
- L-PHA
leucoagglutinating phytohemagglutinin
- qRT-PCR
quantitative reverse transcriptase-PCR
References
- 1.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci USA. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo HB, Lee I, Kamar M, Akiyama SK, Pierce M. Aberrant N-glycosylation of beta1 integrin causes reduced alpha5beta1 integrin clustering and stimulates cell migration. Cancer Res. 2002;62:6837–6845. [PubMed] [Google Scholar]
- 3.Yamamoto H, Swoger J, Greene S, Saito T, Hurh J, Sweeley C, Leestma J, Mkrdichian E, Cerullo L, Nishikawa A, Ihara Y, Taniguchi N, Moskal JR. Beta-1,6-N-acetylglucosamine- bearing N-glycans in human gliomas: implications for a role in regulating invasivity. Cancer Res. 2000;60:134–142. [PubMed] [Google Scholar]
- 4.Demetriou M, Nabi IR, Coppolino M, Dedhar S, Dennis JW. Reduced contact-inhibition and substratum adhesion in epithelial cells expressing GlcNAc-transferase V. J Cell Biol. 1995;130:383–392. doi: 10.1083/jcb.130.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 6.Guo HB, Lee I, Kamar M, Pierce M. N-acetylglucosaminyltransferase Va expression levels regulate cadherin-associated homotypic cell–cell adhesion and intracellular signaling pathways. J Biol Chem. 2003;278:52412–52424. doi: 10.1074/jbc.M308837200. [DOI] [PubMed] [Google Scholar]
- 7.Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 8.Guo HB, Randolph M, Pierce M. Inhibition of a specific N-glycosylation activity results in attenuation of breast carcinoma cell invasiveness-related phenotypes: inhibition of epidermal growth factor-induced dephosphorylation of focal adhesion kinase. J Biol Chem. 2007;282:22150–22162. doi: 10.1074/jbc.M611518200. [DOI] [PubMed] [Google Scholar]
- 9.Guo P, Zhang Y, Shen ZH, Zhang XY, Chen HL. Effect of N-acetylglucosaminyltransferase Va on the expressions of other glycosyltransferases. FEBS Lett. 2004;562:93–98. doi: 10.1016/S0014-5793(04)00188-7. [DOI] [PubMed] [Google Scholar]
- 10.Guo HB, Lee I, Bryan BT, Pierce M. Deletion of mouse embryo fibroblast N-acetylglucosaminyltransferase Va stimulates alpha5beta1 integrin expression mediated by the protein kinase C signaling pathway. J Biol Chem. 2005;280:8332– 8342. doi: 10.1074/jbc.M413532200. [DOI] [PubMed] [Google Scholar]
- 11.Nairn AV, Kinoshita-Toyoda A, Toyoda H, Xie J, Harris K, Dalton S, Kulik M, Pierce JM, Toida T, Moremen KW, Linhardt RJ. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res. 2007;6:4374–4387. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo HB, Liu F, Chen HL. Increased susceptibility to apoptosis of human hepatocarcinoma cells transfected with antisense N-acetylglucosaminyltransferase Va cDNA. Biochem Biophys Res Commun. 1999;264:509–517. doi: 10.1006/bbrc.1999.1303. [DOI] [PubMed] [Google Scholar]
- 13.Hase S. High-performance liquid chromatography of pyridylaminated saccharides. Method Enzymol. 1994;230:225–237. doi: 10.1016/0076-6879(94)30015-1. [DOI] [PubMed] [Google Scholar]
- 14.Kamar M, Alvarez-Manilla G, Abney T, Azadi P, Kumar Kolli VS, Orlando R, Pierce M. Analysis of the site-specific N-glycosylation of beta1,6 N-acetylglucosaminyltransferase Va. Glycobiology. 2004;14:583–592. doi: 10.1093/glycob/cwh062. [DOI] [PubMed] [Google Scholar]
- 15.Finne J, Finne U, Deagostini-Bazin H, Goridis C. Occurrence of alpha 2–8 linked polysialosyl units in a neural cell adhesion molecule. Biochem Biophys Res Commun. 1983;112:482–487. doi: 10.1016/0006-291x(83)91490-0. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama J, Fukuda MN, Fredette B, Ranscht B, Fukuda M. Expression cloning of a human polysialyl-transferase that forms the polysialylated neural cell adhesion molecule present in embryonic brain. Proc Natl Acad Sci USA. 1995;92:7031–7035. doi: 10.1073/pnas.92.15.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckhardt M, Muhlenhoff M, Bethe A, Koopman J, Frosch M, Gerardy-Schahn R. Molecular characterization of eukaryotic polysialyltransferase-1. Nature. 1995;373:715–718. doi: 10.1038/373715a0. [DOI] [PubMed] [Google Scholar]
- 18.Lowe JB, Marth JD. A genetic approach to mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 19.Barondes SH, Castronovo V, Cooper DN, Cummings RD, Drickamer K, Feizi T, Gitt MA, Hirabayashi J, Hughes C, Kasai K, et al. Galectins: a family of animal beta-galactoside- binding lectins. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 20.Sasai K, Ikeda Y, Eguchi H, Tsuda T, Honke K, Taniguchi N. The action of N-acetylglucosaminyltransferase- V is prevented by the bisecting GlcNAc residue at the catalytic step. FEBS Lett. 2002;522:151–155. doi: 10.1016/s0014-5793(02)02916-2. [DOI] [PubMed] [Google Scholar]
- 21.Ohtsubo K, Takamatsu S, Minowa MT, Yoshida A, Takeuchi M, Marth JD. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123:1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 22.Chiricolo M, Malagolini N, Bonfiglioli S, Dall’Olio F. Phenotypic changes induced by expression of beta-galactoside alpha2,6 sialyltransferase I in the human colon cancer cell line SW948. Glycobiology. 2006;16:146–154. doi: 10.1093/glycob/cwj045. [DOI] [PubMed] [Google Scholar]
- 23.Cheung IY, Vickers A, Cheung NK. Sialyltransferase STX (ST8SiaII): a novel molecular marker of metastatic neuroblastoma. Int J Cancer. 2006;119:152–156. doi: 10.1002/ijc.21789. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto H, Oviedo A, Sweeley C, Saito T, Moskal JR. Alpha-2,6-sialylation of cell-surface N-glycans inhibits glioma formation in vivo. Cancer Res. 2001;61:6822–6829. [PubMed] [Google Scholar]
- 25.Fukuda M. Possible roles of tumor-associated carbohydrate antigens. Cancer Res. 1996;56:2237–2244. [PubMed] [Google Scholar]
- 26.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 27.Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim Biophys Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Astolfi A, Landuzzi L, Nicoletti G, De Giovanni C, Croci S, Palladini A, Ferrini S, Iezzi M, Musiani P, Cavallo F, Forni G, Nanni P, Lollini PL. Gene expression analysis of immune-mediated arrest of tumorigenesis in a transgenic mouse model of Her-2/neu-positive basal-like mammary carcinoma. Am J Pathol. 2005;166:1205–1216. doi: 10.1016/S0002-9440(10)62339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy Y, Arbel-Goren R, Hadari YR, Eshhar S, Ronen D, Elhanany E, Geiger B, Zick Y. Galectin-8 functions as a matricellular modulator of cell adhesion. J Biol Chem. 2001;276:31285–31295. doi: 10.1074/jbc.M100340200. [DOI] [PubMed] [Google Scholar]
- 30.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 31.Abbott KL, Troupe K, Lee I, Pierce M. Integrindependent neuroblastoma cell adhesion and migration on laminin is regulated by expression levels of two enzymes in the O-mannosyl-linked glycosylation pathway, PomGnT1 and GnT-Vb. Exp Cell Res. 2006;312:2837–2850. doi: 10.1016/j.yexcr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Hirabayashi J, Hashidate T, Arata Y, Nishi N, Nakamura T, Hirashima M, Urashima T, Oka T, Futai M, Muller WE, Yagi F, Kasai K. Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim Biophys Acta. 2002;1572:232–254. doi: 10.1016/s0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]