Abstract
Polyketide natural products act as a broad range of therapeutics, including antibiotics, immunosuppressants and anti-cancer agents. This therapeutic diversity stems from the structural diversity of these small molecules, many of which are produced in an assembly line manner by modular polyketide synthases. The acyltransferase (AT) domains of these megasynthases are responsible for selection and incorporation of simple monomeric building blocks, and are thus responsible for a large amount of the resulting polyketide structural diversity. The substrate specificity of these domains is often targeted for engineering in the generation of novel, therapeutically active natural products. This review outlines recent developments that can be used in the successful engineering of these domains, including AT sequence and structural data, mechanistic insights and the production of a diverse pool of extender units. It also provides an overview of previous AT domain engineering attempts, and concludes with proposed engineering approaches that take advantage of current knowledge. These approaches may lead to successful production of biologically active ‘unnatural’ natural products.
Keywords: polyketide, acyltransferase, antibiotics, enzyme engineering
1. Introduction
Natural products have historically been an invaluable resource in the discovery and development of therapeutically active small molecules. Micro-organism-derived natural products became more prevalent following the discovery of penicillin and remained significant until the 1990s, when nearly 80 per cent of drugs were either natural products or natural product analogues [1]. Despite a subsequent decline in investments in natural product research by the pharmaceutical industry, the field is now better-equipped than ever to address critical therapeutic needs. Advancements in sequencing technologies and structural analysis techniques, combined with a better understanding of biosynthetic pathways and a wealth of unexplored source organisms, create an exciting environment for future drug discovery [1–3].
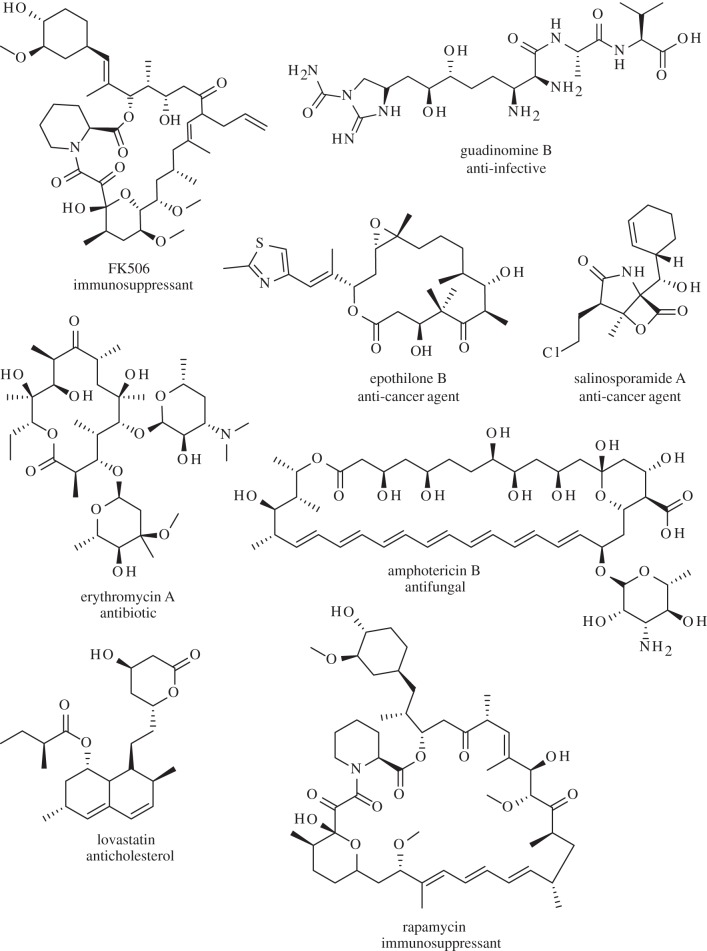
The polyketide class of natural products has seen huge success in the commercial drug arena, with a ‘hit rate’ that is several orders of magnitude above synthetic compound libraries [1]. Polyketides act as a wide range of therapeutics, including antibiotics (erythromycin A), immunosuppressants (FK506, rapamycin), anti-cancer agents (salinosporamide A, epothilone B) and others (figure 1). Many polyketides are produced by assembly line polyketide synthases (PKSs), which build polyketide chains by successive condensation of simple coenzyme A (CoA)-derived subunits. The structural and functional modularity of these enzyme complexes has placed them at the forefront of engineering endeavours in the pursuit of biologically active ‘unnatural’ natural products. A large amount of polyketide diversity stems from differences in building block selection by acyltransferase (AT) domains, and thus many engineering efforts have focused on altering the catalytic activity of these domains.
Figure 1.
The structural and therapeutic diversity of polyketide natural products. This diversity largely stems from differences in extender unit selection by the AT domain (for details, see text).
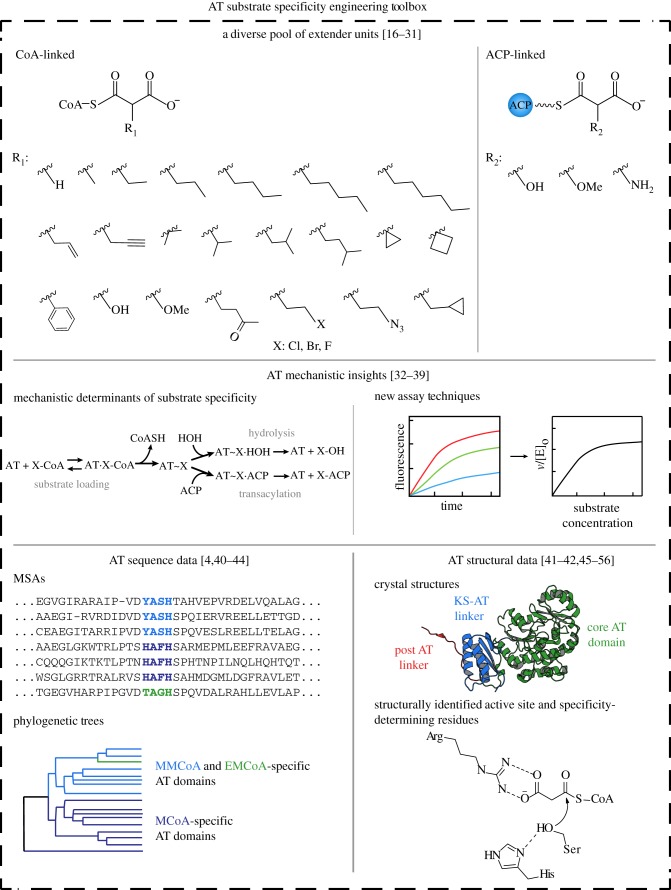
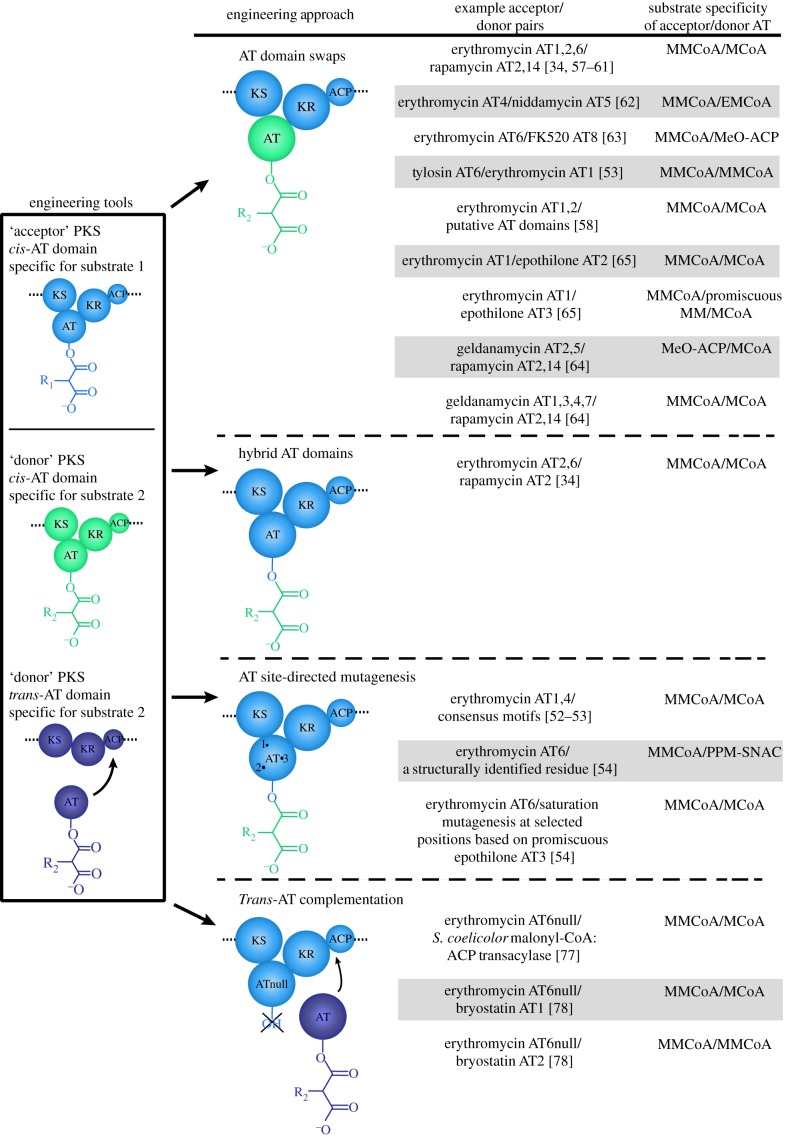
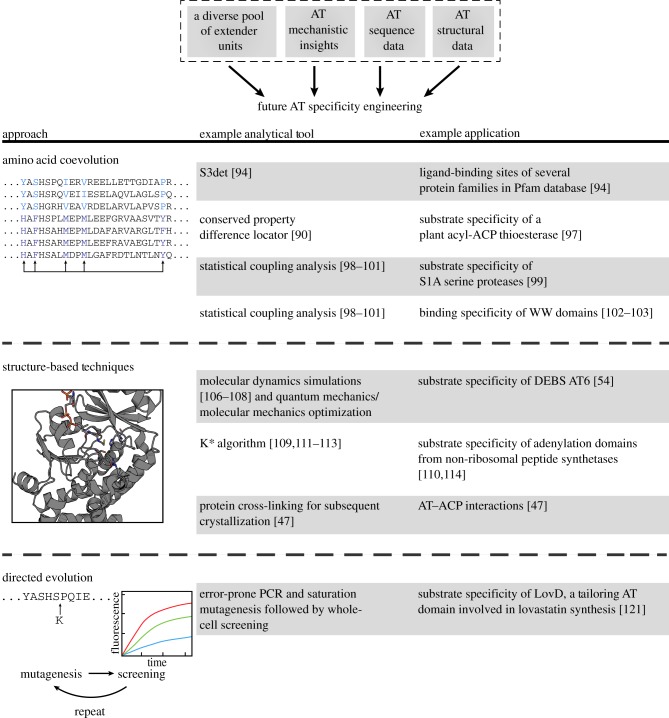
While the environment is right for major developments in polyketide engineering, these pursuits must be informed by previous efforts and new knowledge. This review begins by examining the characteristics of modular PKSs that originally placed them at the forefront of engineering efforts, and outlines recent developments that can be further used in the successful engineering of these enzymatic assembly lines (figure 2). We examine previous attempts to engineer PKS substrate selection by AT domain engineering, such as AT domain swapping, AT knockout and complementation by a domain that normally functions in trans, and AT site-directed mutagenesis (figure 3). We conclude by proposing future engineering approaches that we believe will be most successful in the quest for novel polyketides. These structure and sequence-based approaches take advantage of the aforementioned information and build upon successful engineering of similar protein families. This will set the stage for productive drug development in the near future (figure 4).
Figure 2.
Tools for engineering the substrate specificity of modular AT domains. A diverse pool of substrates, mechanistic insights and sequence and structural information combine to allow for productive AT substrate specificity engineering using a wide variety of techniques (for details, see text). (Online version in colour.)
Figure 3.
Previous attempts to engineer AT domain substrate specificity. The ‘acceptor’ AT domain is defined as the starting engineering scaffold, whereas the ‘donor’ AT domain possesses the final desired substrate specificity and is used to either fully replace or provide a template for mutation of the acceptor domain. MMCoA, methylmalonyl-CoA; MCoA, malonyl-CoA; EMCoA, ethylmalonyl-CoA; MeO-ACP, methoxymalonyl-ACP; PPM-SNAC, 2-propargylmalonyl-N-acetylcysteamine. (Online version in colour.)
Figure 4.
The future of AT domain engineering. Given the existing tools for engineering AT domain substrate specificity, several different techniques are feasible. The identified techniques have previously been used to engineer the substrate specificity of other protein families. (Online version in colour.)
2. The acyltransferase domain as an engineering target
2.1. Polyketide diversity: the importance of the acyltransferase domain
Modular type I PKSs are found primarily in Gram-negative and -positive bacteria, and are especially prevalent in actinomycetes [4]. These megasynthases are comprised of several catalytic modules that are each responsible for one round of polyketide chain elongation. Each module is composed of several different enzymatic domains, each of which plays a distinct role in the extension and modification of the growing polyketide. AT domains select the appropriate α-carboxyacyl-CoA building block and catalyse its loading onto the phosphopantetheine arm of the acyl carrier protein (ACP). Ketosynthase (KS) domains accept the polyketide chain from the upstream ACP and catalyse a Claisen-like condensation between it and the ACP-bound α-carboxyacyl-CoA extender unit. Together, the KS, AT and ACP comprise the minimal module that is necessary and sufficient for one round of chain elongation. Accessory domains such as dehydratase (DH), enoyl reductase (ER) and keto-reductase (KR) domains may also be present in a module and control the level of oxidoreductive modification of the β-keto group following chain elongation. The thioesterase (TE) domain is responsible for release of the final polyketide product, either by cyclization or by attack of an external nucleophile.
Modular PKS genes and their corresponding proteins are co-linear [5,6]—the organization of the assembly line (and thus the structure of the polyketide product) can often be predicted from the gene sequence. This combined modularity has made these catalysts attractive targets in the engineering of novel products via combinatorial biosynthesis [7,8]. Most domains in the assembly line receive their substrates from previous catalytic steps and not by diffusive loading; these enzymes therefore may not have been evolutionarily driven to develop strict substrate specificities and could be inherently robust to rearrangement [9]. Indeed, loading AT domains, KS, ACP, KR, DH and ER domains from the 6-deoxyerythronolide B synthase (DEBS) have been shown to accept substrates that vary considerably from their native substrates [10–15]. Alternatively, extender AT domains receive their substrates diffusively and usually exhibit strict specificity towards a single α-carboxyacyl-CoA building block [10,14]. In acting as the primary gatekeepers to building block incorporation, these domains play an important role in polyketide diversification and are often the primary target of engineering efforts. They are also the focus of this review.
2.2. Extender unit diversity
Extender AT domains in modular PKSs are naturally specific for several different α-carboxyacyl-CoA substrates, but malonyl-CoA and methylmalonyl-CoA are most commonly used in the biosynthetic pathways characterized to date [16]. AT domains have also been identified that are capable of recognizing ACP-bound substrates, such as those involved in the biosynthesis of geldanamycin [17], zwittermicin A [16,18–20] and guadinomine [21]. In vivo and in vitro studies aimed at interrogating AT substrate specificity have previously been limited by the cost and availability of these extender units [22–23], but methods for producing diverse pools of extender units enzymatically have emerged in recent years.
The discovery of crotonyl-CoA carboxylase/reductase (CCR) homologues in polyketide biosynthetic pathways more than doubled the number of CoA-linked extender units known to be incorporated into polyketides. These enzymes catalyse the reductive carboxylation of α,β-unsaturated acyl-CoA precursors (for review, see Wilson & Moore [24]). Originally discovered in the context of ethylmalonyl-CoA [25], CCR functionality quickly expanded to include biosynthesis of alkylmalonyl-CoA and both straight and branched chain malonyl-CoA derivatives. The AT domains associated with CCR-generated extender units typically show relaxed specificity, and several polyketide analogues have been made that take advantage of this fact, but more ambitious endeavours will benefit from rational engineering [24].
Malonyl-CoA can be produced by the direct activation of malonate by an ATP-dependent malonyl-CoA synthetase (MatB; [26,27]). The malonyl-CoA synthetases from Rhizobium trifolii [28] and Streptomyces coelicolor [22] were shown to have broad substrate tolerance, activating several different 1,3-dicarboxylate substrates with measurable efficiency. The S. coelicolor MatB was also shown to be promiscuous towards thiol acceptors, and is capable of producing N-acetylcysteamine- (SNAC-) and pantetheine-linked extender units [22]. AT domains have been shown to accept SNAC and pantetheine analogues, and these substrates are readily taken up by cells from exogenous media [22,29–35]. The activity of the R. trifolii MatB towards several natural and unnatural malonate derivatives was enhanced using structure-guided saturation mutagenesis [36,37]. Interestingly, the trans-AT domain from the kirromycin PKS was shown to be more promiscuous than originally anticipated when examined for transacylation activity with MatB-generated extender units [37].
A diverse pool of CoA-linked extender units provides a valuable toolbox for the polyketide engineer. The ability to regiospecifically incorporate many of the possible MatB or CCR-derived CoA-linked extender units into DEBS, for example, has the potential to yield over one million novel products alone. Availability of these extender units for in vivo and in vitro studies allows for interrogation of the structural and mechanistic determinants of substrate specificity. The emergence of AT domains capable of selecting substrates not present in the native host also provides new starting scaffolds in the pursuit of AT domains with altered specificity.
2.3. Acyltransferase mechanistic insights
AT-catalysed incorporation of CoA-derived extender units occurs by a ping–pong bi–bi mechanism [38,39] involving an acyl-AT intermediate that is subject to nucleophilic attack by the thiol residue found on the phosphopantetheine arm of the ACP. Early studies revealed that modular AT domains likely do not possess epimerase activity, and all domains in DEBS exclusively incorporate (2S)-methylmalonyl-CoA [40]. The rapid rate at which AT domains catalyse transacylation has made kinetic characterization of their activity apart from the rest of the assembly line difficult. Recently, the AT domain from module 3 of DEBS was characterized using a coupled enzymatic assay that allowed for continuous monitoring of AT catalytic activity and thus characterization of α-carboxyacyl-CoA and ACP specificity. This study revealed that the (2S)-methylmalonyl-CoA specificity of this enzyme lies in the first half reaction of the ping–pong mechanism (formation of the methylmalonyl-AT intermediate; [39]). Hydrolytic cleavage of incorrect substrates has been hypothesized to be a specificity-determining mechanism used by AT domains, but the reported importance of this mechanism has varied [39,41].
Several studies have established the importance of proper AT–ACP interactions during transacylation. Examination of domains from modules 3 and 6 of DEBS revealed that cognate AT–ACP pairs have at least 10-fold greater specificity for each other than for heterologous proteins [39,42]. Polyketide chain translocation [43] and elongation [15] are also dependent on proper protein–protein interactions within a module, and specific portions of the AT domain have been identified that contribute to these interactions. For example, the cleft formed by the KS domain, KS to AT linker, and AT domain was shown to be a possible docking site for the ACP during chain transfer and elongation [44,45]. Conservation of the protein–protein interactions necessary for all activities in the assembly line will be important when engineering AT domains for novel substrate incorporation.
2.4. Acyltransferase domain sequence and structure
The development of high-throughput sequencing technologies and a rise in whole-genome sequencing has led to a rapid increase in the number of PKS cluster sequences available [46]. Hundreds of AT domains have been sequenced and their clusters biochemically characterized [47], providing a useful dataset in the engineering of these domains using statistical, sequence-based methods. Many AT domains share high sequence homology and can be identified computationally based strictly on their sequence similarity to a template AT domain [48]. Interestingly, divergence occurs on the basis of extender unit specificity and not species of origin or composition of the rest of the PKS module [49]. Phylogenetic analyses of AT domains annotated for substrate specificity produce two major clades based on methylmalonyl- or malonyl-CoA specificity [48–50], and this separation is improved when considering only active site and highly conserved residues that vary with methylmalonyl- or malonyl-CoA substrate specificity [48]. Residues important in the prediction of ethylmalonyl- and methoxymalonyl-CoA specificity have also been identified [4], and the prediction of less common specificities will likely become more accurate as more sequenced AT domains are functionally characterized.
The X-ray crystal structures of two modular AT domains from DEBS have been solved (Protein Data Bank (PDB) entries 2QO3 and 2HG4) [51,52], as well as a trans-acting AT domain from the disorazole synthase (PDB entry 3RGI) [53]. Along with the multi-purpose FabD AT domains from Escherichia coli (PBD entry 1MLA) [54] and S. coelicolor (PDB entry 1NM2) [55], these crystal structures have facilitated the identification of residues that may play a role in determining substrate specificity. The E. coli FabD protein was crystallized with malonyl-CoA (PDB entry 2G2Z), allowing for identification of conserved oxyanion hole residues (Gln11, Leu93; 2G2Z numbering), and a conserved arginine residue (Arg117; 2G2Z numbering) that interacts with the carboxylate of the CoA-linked extender unit [56]. Crystal structures have also been useful in conjunction with multiple sequence alignments to identify residues in and around the active site that show strong correlations with substrate specificity, including four residues that have been identified repeatedly in different contexts (Leu93, Ser197, Val198, Ser200; 2G2Z numbering) [48,57–60]. The structure-based sequence analysis of polyketide synthases server allows for three-dimensional modelling of AT domains that have not been crystallized, and also allows for sequence-based comparisons and prediction of inter-subunit contacts [47]. Additionally, an AT domain with atypical specificity for long-chain acyl-CoA substrates from the mycobacterial PKS Pks13 (PDB entry 3TZX) was also crystallized recently, revealing a dedicated channel for the atypical long chain and providing an interesting comparison for previously crystallized methylmalonyl- and malonyl-CoA AT domains [61]. The structural advances made in elucidating AT domain architecture, as well as progress in describing the three-dimensional conformation of the rest of the modular PKS [62] position the protein engineer to pursue many different structure-based engineering approaches.
3. Previous attempts to engineer acyltransferase substrate specificity
3.1. Acyltransferase domain swaps
Despite the identification of specific residues that probably play a role in determining substrate specificity, the most common approach for altering AT catalytic activity to date is the swapping of an entire AT domain for a homologue with different specificity. Several desmethyl erythromycin, desmethyl-6-deoxyerythronolide B (desmethyl-6-dEB) or desmethyl triketide lactone analogues have been successfully produced by swapping methylmalonyl-CoA specific domains in DEBS for malonyl-CoA specific AT domains from the rapamycin PKS [40,63–67]. A 6-desmethyl-6-ethylerythromycin A analogue was produced by swapping AT4 of DEBS with an ethylmalonyl-CoA specific AT domain from the niddamycin PKS. When expressed in Saccharopolyspora erythraea, the hybrid PKS produced only erythromycin A unless the culture media was supplemented with precursors to ethylmalonate. The 6-desmethyl-6-ethylerythromycin A analogue was the primary macrolide produced when the ccr gene from Streptomyces collinus was also expressed in this strain [68]. A methoxymalonate building block was also incorporated into the 6-dEB scaffold to form 2-desmethyl-2-methoxy-6-dEB by swapping AT6 of DEBS with AT8 of the FK520 PKS [69]. AT domains in the geldanamycin PKS specific for methylmalonyl- or methoxymalonyl-CoA were successfully replaced with malonyl-CoA specific domains from the rapamycin PKS. These domain swaps led to the identification of a geldanamycin analogue with fourfold higher affinity towards Hsp90, a protein drug target [70]. Domain swaps have been successfully implemented with other PKSs as well [59,64,71].
Despite the apparent success of AT domain swaps in producing novel polyketide products, these successes often come at the cost of decreased product titres [72], and some AT domains appear to be especially difficult to replace [58]. While the causes of such shortcomings are not well understood, disruption of proper protein–protein interactions or an inability of downstream modules to handle altered substrates may be to blame [73]. Titres are decreased to varying degrees depending on the location of the swap in the PKS assembly line [40,64], indicating variability in the ability of downstream modules to process non-native substrates. The extensive interdomain interface shared by AT domains with their KS neighbours [51,52] could also be disrupted by domain swapping, as could the interactions of the ACP with the KS–AT didomain during chain elongation [44,74].
3.2. Generation of hybrid acyltransferase domains
A less common approach to altering AT catalytic activity involves creation of hybrid AT domains, in which possible specificity-determining regions are replaced by the analogous cassettes from an AT domain with a different α-carboxyacyl-CoA specificity. Hybrid AT domains created using methylmalonyl-CoA specific domains from DEBS and the malonyl-CoA specific AT domain from module two of the rapamycin PKS suggested that a short C-terminal ‘hypervariable region’ was the primary determinant of AT specificity [40]. The importance of this region in the context of other AT domains is not clear, but it is important to note given the success of hybrid domains in altering the reactivity of other modular PKS domains [44,74].
3.3. Acyltransferase site-directed mutagenesis
As mentioned above, examination of multiple sequence alignments and crystal structures has led to the identification of several residues in and adjacent to the AT active site that diverge based on α-carboxyacyl-CoA substrate specificity [48,57–60]. Site-directed mutagenesis of important residues has the potential to be minimally invasive, allowing the AT domain to remain in its native environment and causing minimal perturbation to important protein–protein interactions. A motif approximately 100 residues C-terminal of the active-site serine is commonly targeted for mutagenesis; mutation from the methylmalonyl-CoA specific YASH motif to the malonyl-CoA specific HAFH motif in AT1 [59], AT4 [58] and AT6 [60] of DEBS consistently led to promiscuous AT domains capable of incorporating both extender units. Mutations outside of this motif have also led to promiscuity, but have allowed incorporation of the non-native extender unit to a lesser extent [58,60]. Molecular dynamics (MD) simulations followed by Quantum Mechanics/Molecular Mechanics (QM/MM) optimizations on DEBS AT6 allowed for the identification of a residue in the active-site pocket that appeared to constrain the size of the extender unit side chain. Mutation of this residue from valine to alanine allowed for incorporation of the non-natural substrate 2-propargylmalonyl-SNAC, although at limited quantities [60].
It is important to note that alterations in AT substrate specificity are often quantified in vivo, and thus the availability of different α-carboxyacyl-CoA substrates cannot be readily controlled. The relative kinetic parameters describing the specificity of a mutated AT domain cannot be determined using in vivo techniques, and until recently the kinetic effects of such mutations were not known. In vitro analysis of a YASH to HAFH DEBS AT3 mutant, along with another point mutant previously examined in vivo [58,60] revealed that the activity of these mutants is drastically attenuated, implying that incorporation of non-natural extender units likely occurs as a result of diminished AT catalytic activity and not because of increased specificity towards the non-native substrate [39]. The engineering of AT domains by site-directed mutagenesis therefore has vast room for improvement and will benefit greatly from the use of advanced sequence- and structure-based techniques.
3.4. Trans-acyltransferase complementation
Several modular PKSs have been identified that lack canonical cis-AT domains and instead use freestanding domains that act in trans, similar to freestanding type II fatty acid synthase AT domains [75]. These trans-AT PKSs include those responsible for production of the disorazoles [76,77], kirromycin [78–80] and several others [75,81,82]. A potential engineering strategy involves the selective inactivation of a cis-AT domain and complementation using a trans-AT with differing specificity [9]. This approach has the potential to impose minimal conformational strain on the synthase. The majority of trans-AT domains are thought to be specific for malonyl-CoA, but domains with varying specificities have been proposed recently [80]. The ethylmalonyl-CoA specific kirromycin trans-AT domain was shown to be promiscuous, capable of catalysing the transacylation of several α-carboxyacyl-CoA substrates not found in its native host [37]. Further, several trans-AT domains have shown promiscuity towards their ACP substrates as well, with activities towards heterologous ACP domains that meet or surpass the activities of native cis-AT domains [42,83]. Several examples exist of trans-AT complementation in the context of an AT-null PKS module. The malonyl-CoA:ACP transacylase from S. coelicolor was used with an AT-null DEBS module 6 in the production of 2-desmethyl-6-dEB [83] and the trans-AT domains from the bryostatin PKS also complemented an AT-null DEBS module 6 [84].
In engineering cis- and trans-AT domains, it is important to note that these domains differ in their evolutionary origins. While cis domains are found mostly in actinomycetes, myxobacteria and cyanobacteria, trans domains stem largely from proteobacteria, myxobacteria and bacilli [75]. These domains appear to have evolved independently; cis-AT domains appear to have evolved largely by gene duplication and diversification, whereas trans domains evolved largely by horizontal gene transfer [49,75,81,85]. Trans-AT PKSs exhibit diverse modular architectures and deviate from normal co-linearity rules [81], and the KS domains of these PKSs have been used to predict product structure. Phylogenetic separation of these domains is largely based on substrate structure, and PKS module function and overall product structure can be deduced from the predicted substrate specificity of each KS domain [75,86,87].
4. The future of acyltransferase domain engineering
AT domain engineering by a variety of computational and experimental techniques is now possible due to the availability of high-quality sequence and structural data and advancements in in vitro assay techniques. Specifically, approaches in both directed evolution and rational design—hallmarks of protein engineering in the past several decades—can be pursued given the advancements in AT domain architecture and function outlined above. Future engineering of AT domain specificity will require consideration of protein–protein interactions and the effects of alterations on each step of the catalytic mechanism. We outline below several techniques that may be especially relevant for engineering AT domain specificity.
4.1. Amino acid coevolution
Given the extensive sequence data available for modular AT domains, techniques using multiple sequence alignments are particularly useful. Previous utilization of AT alignments has been limited to identification of partially conserved residues based on substrate specificity [48,57–60]. These residues were identified using phylogenetic trees in combination with multiple sequence alignments in order to identify patterns of sequence separation. Owing to the limited success of targeted mutagenesis attempts based on these analyses, it is apparent that a fundamental driving force for specificity is not being captured using conservation alone. The natural next step is incorporation of coevolutionary information, or the correlated mutation of residues in response to an evolutionary pressure. We speculate that, beyond the highly conserved motifs already identified in AT domains, there are other residues that have evolved in a compensatory manner. Several methods have been developed for analysing amino acid coevolution in protein sequences (see reviews [88–90]). We focus here on a few selected techniques that have been used successfully to alter or interrogate the substrate specificity of other protein families.
Residues that are differentially conserved based on subfamily, such as the residues that vary based on methylmalonyl- versus malonyl-CoA specificity, are often referred to as ‘specificity-determining positions’. It is assumed that these residues must coevolve because they show correlated mutational patterns [88]. Several computational techniques can be used to identify groups of coevolving residues in large multiple sequence alignments [91–101]. The JDet software package was developed recently for identification and manipulation of coevolving residues from an input alignment using several different methods [102]. One method, referred to as S3det, makes use of multiple correspondence analysis, which allows for extraction of independent sources of variation in the alignment. These variations can be linked to corresponding clusters of proteins in order to identify subfamilies. S3det was used to successfully identify ligand-binding sites in several protein families from the Pfam database [100]. As another example, the substrate specificity of a plant acyl-ACP TE was engineered using specificity-determining positions identified via a conserved property difference locator, which examines differential conservation as well as such consensus properties as size and hydrophobicity [96,103]. The use of computational techniques to identify specificity-determining positions could improve AT domain engineering, which may have previously been limited by sequence sample size and often relied on manual inspection or examination of crystal structures to identify specificity-determining residues.
Statistical coupling analysis (SCA) has been used on several occasions to identify groups of residues that have coevolved for the determination of substrate specificity. SCA goes a step beyond previously described methods by explicitly accounting for amino acid covariation. It uses a ‘SCA matrix’, or a conservation-weighted position correlation matrix that describes mathematically the conservation at individual amino acid positions and their inter-residue covariance [104,105]. Recent implementations of SCA identify statistically significant correlations among groups of residues, or ‘sectors’, by spectral decomposition of the SCA matrix followed by principal component analysis. This approach also allows for examination of the relationships between sequence divergence and amino acid covariance using a conservation-weighted sequence correlation matrix [106,107]. One of the first implementations of SCA successfully identified residues important for the substrate specificity of S1A serine proteases, and mutation of sector residues clearly perturbed the relative specificity constants (kcat/Km) for different substrates [105]. SCA was also used in the design of unnatural WW domains, small peptide recognition motifs that are classified based on the nature of their proline-containing target motif [108,109]. A group of eight residues that showed a high degree of coevolution was proposed to encode the binding specificity of these domains. These eight coevolving sector residues showed patterns of conservation that were strongly correlated with motif specificity [109]. SCA can identify residues that are not immediately obvious using conservation or structure examination alone and thus has the ability to expand and improve upon the cohort of previously identified AT specificity-determining residues.
The functional importance of residues identified using coevolution techniques depends on the constraints driving the evolution of the protein family of interest. The clear phylogenetic separation of AT domains into clades based on α-carboxyacyl-CoA substrate specificity provides evidence that evolutionary pressure likely exists for this function. Coevolution analyses on modular AT domains will therefore likely lead to the identification of previously elusive residues that, when modified, will lead to alterations in specificity. Further, coevolution analyses have been used to detect protein interaction surfaces (see reviews [88–90]), and thus may be helpful in identifying residues necessary for the protein–protein interactions that allow for proper functioning of the assembly line. Knowledge of the interaction surfaces between AT domains and other proteins in the synthase could enable engineering of specificity that properly maintains these interactions, and could also shed light on the proper junctions for engineering based on AT domain replacement.
4.2. Structure-based techniques
The availability of several X-ray crystal structures of AT domains provides the opportunity for utilization of structure-based engineering. These crystal structures have provided guidance in previous AT engineering attempts [48,57–60], but have mostly been used to pinpoint residues that might make contact with the substrate. More global approaches could allow for more successful engineering of AT specificity.
Understanding of the molecular basis for AT substrate specificity would be enhanced by MD simulations, which have largely been missing from modular PKS studies until recently [60,62,110,111]. MD simulations rely on Newtonian physics to approximate molecular interaction and conformation energetics and to model atomic movement [112–114]. A structural model of DEBS AT6 with (2S)-methylmalonyl-CoA docked into the active site was examined using a 30 ns MD simulation followed by QM/MM optimization. This simulation confirmed the role of proposed active-site residues and pinpointed residues important in maintaining the proper chemical environment for polar and non-polar regions of the substrate. MD simulations on mutated systems with the native methylmalonyl-CoA and non-native malonyl-CoA helped provide rationale for the observed in vivo substrate specificity of these mutant AT6 domains. Further, the MD simulations helped pinpoint a residue that appeared to prevent the incorporation of substrates with larger α-substituents. Mutation of this residue allowed for detectable utilization of a 2-propargylmalonyl-SNAC building block [60]. The utility of this initial MD simulation on a modular AT domain provides motivation for the future implementation of this technique. Allowing MD simulations to play a more active role in the design of experiments, as opposed to using them primarily to explain observed phenomena, will likely aid in the identification of residues outside the active site that indirectly affect active-site architecture. Examination of long-time dynamics in the presence of native and non-native substrates will be a likely strategy in the pursuit of this goal.
Other QM-based techniques have also been used to examine enzymatic substrate specificity. Adenylation domains from non-ribosomal peptide synthetases, which are analogous to AT domains in PKSs, have been engineered to take up different amino acid substrates using a structure-based algorithm known as K*. K* is an ensemble-based computational algorithm that models protein–ligand binding using approximate partition functions and energy minimization [115]. The phenylalanine-specific adenylation domain from the gramicidin synthase was first engineered for improved specificity towards leucine residues [116], and after several improvements to the K* algorithm [117–119], was engineered for a complete specificity swap to leucine and improved specificity towards several other amino acids [120].
Finally, the engineering of AT domains using structural techniques would be further improved by the structural elucidation of AT domains with other α-carboxyacyl-CoA specificity. With the exception of the AT domain from the mycobacterial Pks13 [61], all modular AT domains crystallized to date are specific for methylmalonyl- or malonyl-CoA [51–55]. Examination of the active-site architecture and global structural characteristics of domains with more unique specificities, along with simple structural alignments with currently elucidated structures, could shed light on the structural determinants of building block specificity. Further, the ACP specificity of AT domains and the role this specificity plays in allowing for productive transacylation could be elucidated via the crystallization of an AT–ACP complex. Structural characterization of these complexes has proven difficult, likely due to the naturally dynamic nature of the ACP domain. Several methods exist for the cross-linking of ACP and KS domains based on modification of the prosthetic phosphopantetheine group on the ACP [121,122], but no such methods exist for AT–ACP cross-linking. A method was devised for cross-linking and subsequent purification of the trans-AT from the disorazole PKS with its cognate ACP from module 1 with the intention of using this adduct as a crystallography tool. This method used an active-site serine to cysteine AT mutant, which was cross-linked to the phosphopantetheine arm of the ACP using dibromopropanone. Purification of the complex from free AT and ACP proteins was accomplished using a two-step affinity chromatography procedure [53]. Crystallization of this complex and others like it would be useful in understanding more fully the protein–protein interactions between AT and ACP domains, thus allowing for preservation of these domain interactions in engineered systems.
4.3. Directed evolution
Advancements in analytical techniques capable of rapidly analysing AT catalysis [39] make directed evolution of the AT domain an experimental possibility, although testing of a very large number of variants is still not technically feasible. Directed evolution techniques (for reviews, see [123–125]) rely on the creation of a mutant library by such methods as error-prone polymerase chain reaction (PCR), DNA shuffling or saturation mutagenesis. This library is then screened for improved function, and the identified mutants are subject to another round of mutagenesis and selection in a process that mimics natural evolution. This process is usually repeated several times until the desired function is achieved. Screening of all possible mutants is not feasible or desirable, and several in silico techniques have been developed to help design the most optimal mutant library [126]. While not necessary, computational optimization is helpful when an extremely high-throughput assay does not exist.
The substrate specificity of a tailoring enzyme involved in converting monacolin J acid (MJA) into the cholesterol-lowering drug lovastatin was engineered using directed evolution techniques. LovD, an AT that naturally transfers α-S-methylbutyrate to MJA was engineered for improved activity towards α-dimethylbutyrate, a substrate that when added to MJA creates the blockbuster cholesterol-lowering drug simvastatin. Mutant libraries were created by either error-prone PCR or saturation mutagenesis and screened using a whole-cell growth inhibition assay. Seven rounds of mutagenesis and screening produced a variant with six point mutations that exhibited approximately 11-fold higher whole-cell activity towards simvastatin synthesis and increased thermal stability. Crystallization revealed that mutated residues were scattered on the tertiary structure, distant from the active site and in both solvent-exposed and buried regions. These residues, like most residues identified using directed evolution, would have been difficult to identify using the crystal structure alone [127].
5. Conclusions and future outlook
The potential for engineering PKSs to produce novel bioactive molecules is essentially limitless. These molecular assembly lines lend themselves well to engineering, as the co-linearity of genes and gene products along with the predictability of the corresponding polyketide allow for rational manipulation with an intended product in mind. AT domains act as gatekeepers to polyketide extender unit incorporation and thus control a large source of the structural diversity of these natural products. The field is better-equipped than ever for AT domain engineering, with mechanistic, sequence, and structural knowledge on this domain growing at a fast pace in recent years. Many engineering techniques are now open to the AT domain engineer. It is likely that no single method will be entirely effective in altering the specificity of AT domains, and success will come with combinations of these techniques. Bridging of in vitro results to in vivo production environments will be important for the successful production of novel therapeutics. Further functional and structural characterization of sequenced AT domains, along with better tools for manipulating, expressing and analysing both individual domains and the overall PKS will continue to aid in making the production of novel polyketides a reality. The engineering of AT domain catalysis requires an in-depth knowledge of how such alterations affect the entire polyketide biosynthetic cycle. Thus, understanding of the fundamental catalytic mechanisms and protein–protein interactions necessary for polyketide biosynthesis are necessary for successful AT domain specificity engineering. This knowledge, coupled with the advancements outlined in this review, position the AT domain engineer to make huge strides in polyketide diversification in the very near future.
Acknowledgements
This research was supported by a grant from the National Institutes of Health (no. R01 GM 087934) to C.K.
References
- 1.Li JW, Vederas JC. 2009. Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–165 10.1126/science.1168243 (doi:10.1126/science.1168243) [DOI] [PubMed] [Google Scholar]
- 2.Butler MS. 2004. The role of natural product chemistry in drug discovery. J. Nat. Prod. 67, 2141–2153 10.1021/np040106y (doi:10.1021/np040106y) [DOI] [PubMed] [Google Scholar]
- 3.Wright GD. 2012. Antibiotics: a new hope. Chem. Biol. 19, 3–10 10.1016/j.chembiol.2011.10.019 (doi:10.1016/j.chembiol.2011.10.019) [DOI] [PubMed] [Google Scholar]
- 4.Minowa Y, Araki M, Kanehisa M. 2007. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 368, 1500–1517 10.1016/j.jmb.2007.02.099 (doi:10.1016/j.jmb.2007.02.099) [DOI] [PubMed] [Google Scholar]
- 5.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. 1991. Modular organization of genes required for complex polyketide biosynthesis. Science 252, 675–679 10.1126/science.2024119 (doi:10.1126/science.2024119) [DOI] [PubMed] [Google Scholar]
- 6.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. 1990. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature 348, 176–178 10.1038/348176a0 (doi:10.1038/348176a0) [DOI] [PubMed] [Google Scholar]
- 7.Tsoi CJ, Khosla C. 1995. Combinatorial biosynthesis of ‘unnatural’ natural products: the polyketide example. Chem. Biol. 2, 355–362 10.1016/1074-5521(95)90214-7 (doi:10.1016/1074-5521(95)90214-7) [DOI] [PubMed] [Google Scholar]
- 8.Khosla C, Zawada RJ. 1996. Generation of polyketide libraries via combinatorial biosynthesis. Trends Biotechnol. 14, 335–341 10.1016/0167-7799(96)10046-9 (doi:10.1016/0167-7799(96)10046-9) [DOI] [PubMed] [Google Scholar]
- 9.Weissman KJ, Leadlay PF. 2005. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 3, 925–936 10.1038/nrmicro1287 (doi:10.1038/nrmicro1287) [DOI] [PubMed] [Google Scholar]
- 10.Khosla C, Gokhale RS, Jacobsen JR, Cane DE. 1999. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68, 219–253 10.1146/annurev.biochem.68.1.219 (doi:10.1146/annurev.biochem.68.1.219) [DOI] [PubMed] [Google Scholar]
- 11.Wu N, Tsuji SY, Cane DE, Khosla C. 2001. Assessing the balance between protein–protein interactions and enzyme–substrate interactions in the channeling of intermediates between polyketide synthase modules. J. Am. Chem. Soc. 123, 6465–6474 10.1021/ja010219t (doi:10.1021/ja010219t) [DOI] [PubMed] [Google Scholar]
- 12.Watanabe K, Wang CC, Boddy CN, Cane DE, Khosla C. 2003. Understanding substrate specificity of polyketide synthase modules by generating hybrid multimodular synthases. J. Biol. Chem. 278, 42 020–42 026 10.1074/jbc.M305339200 (doi:10.1074/jbc.M305339200) [DOI] [PubMed] [Google Scholar]
- 13.Lau J, Cane DE, Khosla C. 2000. Substrate specificity of the loading didomain of the erythromycin polyketide synthase. Biochemistry 39, 10 514–10 520 10.1021/bi000602v (doi:10.1021/bi000602v) [DOI] [PubMed] [Google Scholar]
- 14.Liou GF, Lau J, Cane DE, Khosla C. 2003. Quantitative analysis of loading and extender acyltransferases of modular polyketide synthases. Biochemistry 42, 200–207 10.1021/bi0268100 (doi:10.1021/bi0268100) [DOI] [PubMed] [Google Scholar]
- 15.Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. 2006. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc. 128, 3067–3074 10.1021/ja058093d (doi:10.1021/ja058093d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan YA, Podevels AM, Kevany BM, Thomas MG. 2009. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 26, 90–114 10.1039/b801658p (doi:10.1039/b801658p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rascher A, Hu Z, Viswanathan N, Schirmer A, Reid R, Nierman WC, Lewis M, Hutchinson CR. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 218, 223–230 10.1016/S0378-1097(02)01148-5 (doi:10.1016/S0378-1097(02)01148-5) [DOI] [PubMed] [Google Scholar]
- 18.Chan YA, Thomas MG. 2010. Recognition of (2S)-aminomalonyl-acyl carrier protein (ACP) and (2R)-hydroxymalonyl-ACP by acyltransferases in zwittermicin A biosynthesis. Biochemistry 49, 3667–3677 10.1021/bi100141n (doi:10.1021/bi100141n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan YA, Boyne MT, 2nd, Podevels AM, Klimowicz AK, Handelsman J, Kelleher NL, Thomas MG. 2006. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc. Natl Acad. Sci. USA 103, 14 349–14 354 10.1073/pnas.0603748103 (doi:10.1073/pnas.0603748103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emmert EA, Klimowicz AK, Thomas MG, Handelsman J. 2004. Genetics of zwittermicin A production by Bacillus cereus. Appl. Environ. Microbiol. 70, 104–113 10.1128/AEM.70.1.104-113.2004 (doi:10.1128/AEM.70.1.104-113.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes TC, et al. 2012. Molecular insights into the biosynthesis of guadinomine: a type III secretion system inhibitor. J. Am. Chem. Soc. 134, 17 797–17 806 10.1021/ja308622d (doi:10.1021/ja308622d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes AJ, Keatinge-Clay A. 2011. Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB. Chem. Biol. 18, 165–176 10.1016/j.chembiol.2010.12.014 (doi:10.1016/j.chembiol.2010.12.014) [DOI] [PubMed] [Google Scholar]
- 23.Lombo F, Pfeifer B, Leaf T, Ou S, Kim YS, Cane DE, Licari P, Khosla C. 2001. Enhancing the atom economy of polyketide biosynthetic processes through metabolic engineering. Biotechnol. Prog. 17, 612–617 10.1021/bp010045j (doi:10.1021/bp010045j) [DOI] [PubMed] [Google Scholar]
- 24.Wilson MC, Moore BS. 2012. Beyond ethylmalonyl-CoA: the functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat. Prod. Rep. 29, 72–86 10.1039/c1np00082a (doi:10.1039/c1np00082a) [DOI] [PubMed] [Google Scholar]
- 25.O'Hagan D, Rogers SV, Duffin GR, Reynolds KA. 1995. The biosynthesis of monensin-A: thymine, beta-aminoisobutyrate and methacrylate metabolism in Streptomyces cinnamonensis. J. Antibiot. (Tokyo) 48, 1280–1287 10.7164/antibiotics.48.1280 (doi:10.7164/antibiotics.48.1280) [DOI] [PubMed] [Google Scholar]
- 26.Kim YS, Kang SW. 1994. Steady-state kinetics of malonyl-CoA synthetase from Bradyrhizobium japonicum and evidence for malonyl-AMP formation in the reaction. Biochem. J. 297, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YS, Chae HZ. 1991. Purification and properties of malonyl-CoA synthetase from Rhizobium japonicum. Biochem J. 273, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohl NL, Hans M, Lee HY, Kim YS, Cane DE, Khosla C. 2001. Remarkably broad substrate tolerance of malonyl-CoA synthetase, an enzyme capable of intracellular synthesis of polyketide precursors. J. Am. Chem. Soc. 123, 5822–5823 10.1021/ja0028368 (doi:10.1021/ja0028368) [DOI] [PubMed] [Google Scholar]
- 29.Cane DE, Yang CC. 1987. Macrolide biosynthesis. IV. Intact incorporation of a chain-elongation intermediate into erythromycin. J. Am. Chem. Soc. 109, 1255–1257 10.1021/ja00238a051 (doi:10.1021/ja00238a051) [DOI] [Google Scholar]
- 30.Pohl NL, Gokhale RS, Cane DE, Khosla C. 1998. Synthesis and incorporation of an N-acetylcysteamine analogue of methylmalonyl-CoA by a modular polyketide synthase. J. Am. Chem. Soc. 120, 11 206–11 207 10.1021/ja9830290 (doi:10.1021/ja9830290) [DOI] [Google Scholar]
- 31.Yue S, Duncan JS, Yamamoto Y, Hutchinson CR. 1987. Macrolide biosynthesis. Tylactone formation involves the processive addition of three carbon units. J. Am. Chem. Soc. 109, 1253–1255 10.1021/ja00238a050 (doi:10.1021/ja00238a050) [DOI] [Google Scholar]
- 32.Xie X, Watanabe K, Wojcicki WA, Wang CC, Tang Y. 2006. Biosynthesis of lovastatin analogs with a broadly specific acyltransferase. Chem. Biol. 13, 1161–1169 10.1016/j.chembiol.2006.09.008 (doi:10.1016/j.chembiol.2006.09.008) [DOI] [PubMed] [Google Scholar]
- 33.Mo S, et al. 2011. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J. Am. Chem. Soc. 133, 976–985 10.1021/ja108399b (doi:10.1021/ja108399b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klopries S, Sundermann U, Schulz F. 2013. Quantification of N-acetylcysteamine activated methylmalonate incorporation into polyketide biosynthesis. Beilstein J. Org. Chem. 9, 664–674 10.3762/bjoc.9.75 (doi:10.3762/bjoc.9.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier JL, Burkart MD. 2009. Chapter 9. Synthetic probes for polyketide and nonribosomal peptide biosynthetic enzymes. Methods Enzymol. 458, 219–254 10.1016/S0076-6879(09)04809-5 (doi:10.1016/S0076-6879(09)04809-5) [DOI] [PubMed] [Google Scholar]
- 36.Koryakina I, Williams GJ. 2011. Mutant malonyl-CoA synthetases with altered specificity for polyketide synthase extender unit generation. Chembiochem 12, 2289–2293 10.1002/cbic.201100383 (doi:10.1002/cbic.201100383) [DOI] [PubMed] [Google Scholar]
- 37.Koryakina I, McArthur J, Randall S, Draelos MM, Musiol EM, Muddiman DC, Weber T, Williams GJ. 2013. Poly specific trans-acyltransferase machinery revealed via engineered acyl-CoA synthetases. ACS Chem. Biol. 8, 200–208 10.1021/cb3003489 (doi:10.1021/cb3003489) [DOI] [PubMed] [Google Scholar]
- 38.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. 2007. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 76, 195–221 10.1146/annurev.biochem.76.053105.093515 (doi:10.1146/annurev.biochem.76.053105.093515) [DOI] [PubMed] [Google Scholar]
- 39.Dunn BJ, Cane DE, Khosla C. 2013. Mechanism and specificity of an acyltransferase domain from a modular polyketide synthase. Biochemistry 52, 1839–1841 10.1021/bi400185v (doi:10.1021/bi400185v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau J, Fu H, Cane DE, Khosla C. 1999. Dissecting the role of acyltransferase domains of modular polyketide synthases in the choice and stereochemical fate of extender units. Biochemistry 38, 1643–1651 10.1021/bi9820311 (doi:10.1021/bi9820311) [DOI] [PubMed] [Google Scholar]
- 41.Bonnett SA, Rath CM, Shareef AR, Joels JR, Chemler JA, Hakansson K, Reynolds K, Sherman DH. 2011. Acyl-CoA subunit selectivity in the pikromycin polyketide synthase PikAIV: steady-state kinetics and active-site occupancy analysis by FTICR-MS. Chem. Biol. 18, 1075–1081 10.1016/j.chembiol.2011.07.016 (doi:10.1016/j.chembiol.2011.07.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong FT, Chen AY, Cane DE, Khosla C. 2010. Protein–protein recognition between acyltransferases and acyl carrier proteins in multimodular polyketide synthases. Biochemistry 49, 95–102 10.1021/bi901826g (doi:10.1021/bi901826g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu N, Cane DE, Khosla C. 2002. Quantitative analysis of the relative contributions of donor acyl carrier proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry 41, 5056–5066 10.1021/bi012086u (doi:10.1021/bi012086u) [DOI] [PubMed] [Google Scholar]
- 44.Kapur S, Chen AY, Cane DE, Khosla C. 2010. Molecular recognition between ketosynthase and acyl carrier protein domains of the 6-deoxyerythronolide B synthase. Proc. Natl Acad. Sci. USA 107, 22 066–22 071 10.1073/pnas.1014081107 (doi:10.1073/pnas.1014081107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charkoudian LK, Liu CW, Capone S, Kapur S, Cane DE, Togni A, Seebach D, Khosla C. 2011. Probing the interactions of an acyl carrier protein domain from the 6-deoxyerythronolide B synthase. Protein Sci. 20, 1244–1255 10.1002/pro.652 (doi:10.1002/pro.652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong FT, Khosla C. 2012. Combinatorial biosynthesis of polyketides—a perspective. Curr. Opin. Chem. Biol. 16, 117–123 10.1016/j.cbpa.2012.01.018 (doi:10.1016/j.cbpa.2012.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anand S, Prasad MV, Yadav G, Kumar N, Shehara J, Ansari MZ, Mohanty D. 2010. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 38, W487–496 10.1093/nar/gkq340 (doi:10.1093/nar/gkq340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yadav G, Gokhale RS, Mohanty D. 2003. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328, 335–363 10.1016/S0022-2836(03)00232-8 (doi:10.1016/S0022-2836(03)00232-8) [DOI] [PubMed] [Google Scholar]
- 49.Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. 2005. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22, 2027–2039 10.1093/molbev/msi193 (doi:10.1093/molbev/msi193) [DOI] [PubMed] [Google Scholar]
- 50.Ridley CP, Lee HY, Khosla C. 2008. Evolution of polyketide synthases in bacteria. Proc. Natl Acad. Sci. USA 105, 4595–4600 10.1073/pnas.0710107105 (doi:10.1073/pnas.0710107105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. 2006. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl Acad. Sci. USA 103, 11 124–11 129 10.1073/pnas.0601924103 (doi:10.1073/pnas.0601924103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. 2007. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem. Biol. 14, 931–943 10.1016/j.chembiol.2007.07.012 (doi:10.1016/j.chembiol.2007.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong FT, Jin X, Mathews II, Cane DE, Khosla C. 2011. Structure and mechanism of the trans-acting acyltransferase from the disorazole synthase. Biochemistry 50, 6539–6548 10.1021/bi200632j (doi:10.1021/bi200632j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serre L, Verbree EC, Dauter Z, Stuitje AR, Derewenda ZS. 1995. The Escherichia coli malonyl-CoA:acyl carrier protein transacylase at 1.5-Å resolution. Crystal structure of a fatty acid synthase component. J. Biol. Chem. 270, 12 961–12 964 10.1074/jbc.270.22.12961 (doi:10.1074/jbc.270.22.12961) [DOI] [PubMed] [Google Scholar]
- 55.Keatinge-Clay AT, Shelat AA, Savage DF, Tsai SC, Miercke LJ, O'Connell JD, 3rd, Khosla C, Stroud RM. 2003. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure 11, 147–154 10.1016/S0969-2126(03)00004-2 (doi:10.1016/S0969-2126(03)00004-2) [DOI] [PubMed] [Google Scholar]
- 56.Oefner C, Schulz H, D'Arcy A, Dale GE. 2006. Mapping the active site of Escherichia coli malonyl-CoA–acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 62, 613–618 10.1107/S0907444906009474 (doi:10.1107/S0907444906009474) [DOI] [PubMed] [Google Scholar]
- 57.Haydock SF, et al. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA : acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374, 246–248 10.1016/0014-5793(95)01119-Y (doi:10.1016/0014-5793(95)01119-Y) [DOI] [PubMed] [Google Scholar]
- 58.Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, McDaniel R. 2001. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry 40, 15 464–15 470 10.1021/bi015864r (doi:10.1021/bi015864r) [DOI] [PubMed] [Google Scholar]
- 59.Del Vecchio F, et al. 2003. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biot. 30, 489–494 [DOI] [PubMed] [Google Scholar]
- 60.Sundermann U, Bravo-Rodriguez K, Klopries S, Kushnir S, Gomez H, Sanchez-Garcia E, Schulz F. 2012. Enzyme-directed mutasynthesis: a combined experimental and theoretical approach to substrate recognition of a polyketide synthase. ACS Chem. Biol. 8, 443–450 10.1021/cb300505w (doi:10.1021/cb300505w) [DOI] [PubMed] [Google Scholar]
- 61.Bergeret F, et al. 2012. Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase Pks13. J. Biol. Chem. 287, 33 675–33 690 10.1074/jbc.M111.325639 (doi:10.1074/jbc.M111.325639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keatinge-Clay AT. 2012. The structures of type I polyketide synthases. Nat. Prod. Rep. 29, 1050–1073 10.1039/c2np20019h (doi:10.1039/c2np20019h) [DOI] [PubMed] [Google Scholar]
- 63.Oliynyk M, Brown MJ, Cortes J, Staunton J, Leadlay PF. 1996. A hybrid modular polyketide synthase obtained by domain swapping. Chem. Biol. 3, 833–839 10.1016/S1074-5521(96)90069-1 (doi:10.1016/S1074-5521(96)90069-1) [DOI] [PubMed] [Google Scholar]
- 64.Ruan X, et al. 1997. Acyltransferase domain substitutions in erythromycin polyketide synthase yield novel erythromycin derivatives. J. Bacteriol. 179, 6416–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Ashley G. 1999. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel ‘unnatural’ natural products. Proc. Natl Acad. Sci. USA 96, 1846–1851 10.1073/pnas.96.5.1846 (doi:10.1073/pnas.96.5.1846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu L, Thamchaipenet A, Fu H, Betlach M, Ashley G. 1997. Biosynthesis of 2-nor-6-deoxyerythronolide B by rationally designed domain substitution. J. Am. Chem. Soc. 119, 10 553–10 554 10.1021/ja972451y (doi:10.1021/ja972451y) [DOI] [Google Scholar]
- 67.Ranganathan A, et al. 1999. Knowledge-based design of bimodular and trimodular polyketide synthases based on domain and module swaps: a route to simple statin analogues. Chem. Biol. 6, 731–741 10.1016/S1074-5521(00)80020-4 (doi:10.1016/S1074-5521(00)80020-4) [DOI] [PubMed] [Google Scholar]
- 68.Stassi DL, et al. 1998. Ethyl-substituted erythromycin derivatives produced by directed metabolic engineering. Proc. Natl Acad. Sci. USA 95, 7305–7309 10.1073/pnas.95.13.7305 (doi:10.1073/pnas.95.13.7305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato Y, Bai L, Xue Q, Revill WP, Yu TW, Floss HG. 2002. Functional expression of genes involved in the biosynthesis of the novel polyketide chain extension unit, methoxymalonyl-acyl carrier protein, and engineered biosynthesis of 2-desmethyl-2-methoxy-6-deoxyerythronolide B. J. Am. Chem. Soc. 124, 5268–5269 10.1021/ja0127483 (doi:10.1021/ja0127483) [DOI] [PubMed] [Google Scholar]
- 70.Patel K, Piagentini M, Rascher A, Tian ZQ, Buchanan GO, Regentin R, Hu Z, Hutchinson CR, McDaniel R. 2004. Engineered biosynthesis of geldanamycin analogs for Hsp90 inhibition. Chem. Biol. 11, 1625–1633 10.1016/j.chembiol.2004.09.012 (doi:10.1016/j.chembiol.2004.09.012) [DOI] [PubMed] [Google Scholar]
- 71.Petkovic H, et al. 2008. Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase. Org. Biomol. Chem. 6, 500–506 10.1039/b714804f (doi:10.1039/b714804f) [DOI] [PubMed] [Google Scholar]
- 72.Liou GF, Khosla C. 2003. Building-block selectivity of polyketide synthases. Curr. Opin. Chem. Biol. 7, 279–284 10.1016/S1367-5931(03)00016-4 (doi:10.1016/S1367-5931(03)00016-4) [DOI] [PubMed] [Google Scholar]
- 73.Hans M, Hornung A, Dziarnowski A, Cane DE, Khosla C. 2003. Mechanistic analysis of acyl transferase domain exchange in polyketide synthase modules. J. Am. Chem. Soc. 125, 5366–5374 10.1021/ja029539i (doi:10.1021/ja029539i) [DOI] [PubMed] [Google Scholar]
- 74.Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. 2012. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation. Proc. Natl Acad. Sci. USA 109, 4110–4115 10.1073/pnas.1118734109 (doi:10.1073/pnas.1118734109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen T, et al. 2008. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 26, 225–233 10.1038/nbt1379 (doi:10.1038/nbt1379) [DOI] [PubMed] [Google Scholar]
- 76.Carvalho R, Reid R, Viswanathan N, Gramajo H, Julien B. 2005. The biosynthetic genes for disorazoles, potent cytotoxic compounds that disrupt microtubule formation. Gene 359, 91–98 10.1016/j.gene.2005.06.003 (doi:10.1016/j.gene.2005.06.003) [DOI] [PubMed] [Google Scholar]
- 77.Kopp M, Irschik H, Pradella S, Muller R. 2005. Production of the tubulin destabilizer disorazol in Sorangium cellulosum: biosynthetic machinery and regulatory genes. Chembiochem 6, 1277–1286 10.1002/cbic.200400459 (doi:10.1002/cbic.200400459) [DOI] [PubMed] [Google Scholar]
- 78.Weber T, Laiple KJ, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W. 2008. Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chem. Biol. 15, 175–188 10.1016/j.chembiol.2007.12.009 (doi:10.1016/j.chembiol.2007.12.009) [DOI] [PubMed] [Google Scholar]
- 79.Laiple KJ, Hartner T, Fiedler HP, Wohlleben W, Weber T. 2009. The kirromycin gene cluster of Streptomyces collinus Tu 365 codes for an aspartate-alpha-decarboxylase, KirD, which is involved in the biosynthesis of the precursor beta-alanine. J. Antibiot. (Tokyo). 62, 465–468 10.1038/ja.2009.67 (doi:10.1038/ja.2009.67) [DOI] [PubMed] [Google Scholar]
- 80.Musiol EM, Hartner T, Kulik A, Moldenhauer J, Piel J, Wohlleben W, Weber T. 2011. Supramolecular templating in kirromycin biosynthesis: the acyltransferase KirCII loads ethylmalonyl-CoA extender onto a specific ACP of the trans-AT PKS. Chem. Biol. 18, 438–444 10.1016/j.chembiol.2011.02.007 (doi:10.1016/j.chembiol.2011.02.007) [DOI] [PubMed] [Google Scholar]
- 81.Piel J. 2010. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27, 996–1047 10.1039/b816430b (doi:10.1039/b816430b) [DOI] [PubMed] [Google Scholar]
- 82.Musiol EM, Weber T. 2012. Discrete acyltransferases involved in polyketide biosynthesis. Med. Chem. Comm. 3, 871–886 10.1039/c2md20048a (doi:10.1039/c2md20048a) [DOI] [Google Scholar]
- 83.Kumar P, Koppisch AT, Cane DE, Khosla C. 2003. Enhancing the modularity of the modular polyketide synthases: transacylation in modular polyketide synthases catalyzed by malonyl-CoA:ACP transacylase. J. Am. Chem. Soc. 125, 14 307–14 312 10.1021/ja037429l (doi:10.1021/ja037429l) [DOI] [PubMed] [Google Scholar]
- 84.Lopanik NB, Shields JA, Buchholz TJ, Rath CM, Hothersall J, Haygood MG, Hakansson K, Thomas CM, Sherman DH. 2008. In vivo and in vitro trans-acylation by BryP, the putative bryostatin pathway acyltransferase derived from an uncultured marine symbiont. Chem. Biol. 15, 1175–1186 10.1016/j.chembiol.2008.09.013 (doi:10.1016/j.chembiol.2008.09.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piel J, Hui D, Fusetani N, Matsunaga S. 2004. Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ. Microbiol. 6, 921–927 10.1111/j.1462-2920.2004.00531.x (doi:10.1111/j.1462-2920.2004.00531.x) [DOI] [PubMed] [Google Scholar]
- 86.Teta R, Gurgui M, Helfrich EJ, Kunne S, Schneider A, Van Echten-Deckert G, Mangoni A, Piel J. 2010. Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial phylum Bacteroidetes. Chembiochem 11, 2506–2512 10.1002/cbic.201000542 (doi:10.1002/cbic.201000542) [DOI] [PubMed] [Google Scholar]
- 87.Jenner M, Frank S, Kampa A, Kohlhaas C, Poplau P, Briggs GS, Piel J, Oldham NJ. 2013. Substrate Specificity in ketosynthase domains from trans-AT polyketide synthases. Angew. Chem. Int. Ed. Engl. 52, 1143–1147 10.1002/anie.201207690 (doi:10.1002/anie.201207690) [DOI] [PubMed] [Google Scholar]
- 88.de Juan D, Pazos F, Valencia A. 2013. Emerging methods in protein co-evolution. Nat. Rev. Genet. 14, 249–261 10.1038/nrg3414 (doi:10.1038/nrg3414) [DOI] [PubMed] [Google Scholar]
- 89.Sandler I, Abu-Qarn M, Aharoni A. 2013. Protein co-evolution: how do we combine bioinformatics and experimental approaches? Mol. Biosyst. 9, 175–181 10.1039/c2mb25317h (doi:10.1039/c2mb25317h) [DOI] [PubMed] [Google Scholar]
- 90.Codoner FM, Fares MA. 2008. Why should we care about molecular coevolution? Evol. Bioinform. Online 4, 29–38 [PMC free article] [PubMed] [Google Scholar]
- 91.del Sol Mesa A, Pazos F, Valencia A. 2003. Automatic methods for predicting functionally important residues. J. Mol. Biol. 326, 1289–1302 10.1016/S0022-2836(02)01451-1 (doi:10.1016/S0022-2836(02)01451-1) [DOI] [PubMed] [Google Scholar]
- 92.Casari G, Sander C, Valencia A. 1995. A method to predict functional residues in proteins. Nat. Struct. Biol. 2, 171–178 10.1038/nsb0295-171 (doi:10.1038/nsb0295-171) [DOI] [PubMed] [Google Scholar]
- 93.Lichtarge O, Bourne HR, Cohen FE. 1996. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 257, 342–358 10.1006/jmbi.1996.0167 (doi:10.1006/jmbi.1996.0167) [DOI] [PubMed] [Google Scholar]
- 94.Mirny LA, Gelfand MS. 2002. Using orthologous and paralogous proteins to identify specificity determining residues. J. Mol. Biol. 321, 7–20 10.1016/S0022-2836(02)00587-9 (doi:10.1016/S0022-2836(02)00587-9). [DOI] [PubMed] [Google Scholar]
- 95.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. 2002. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 18(Suppl. 1), S71–S77 10.1093/bioinformatics/18.suppl_1.S71 (doi:10.1093/bioinformatics/18.suppl_1.S71) [DOI] [PubMed] [Google Scholar]
- 96.Mayer KM, McCorkle SR, Shanklin J. 2005. Linking enzyme sequence to function using conserved property difference locator to identify and annotate positions likely to control specific functionality. BMC Bioinformatics 6, 284. 10.1186/1471-2105-6-284 (doi:10.1186/1471-2105-6-284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalinina OV, Rassel RB, Rakhmaninova AB, Gel'fand MS. 2007. Computational method for prediction of protein functional sites using specificity determinants. Mol. Biol. (Mosk) 41, 151–162 10.1134/S0026893307010189 (doi:10.1134/S0026893307010189) [DOI] [PubMed] [Google Scholar]
- 98.Bonella S, Rocchia W, Amat P, Nifosí R, Tozzini V. 2009. SDPhound, a mutual information-based method to investigate specificity-determining positions. Algorithms 2, 764–789 10.3390/a2020764 (doi:10.3390/a2020764) [DOI] [Google Scholar]
- 99.Mazin PV, Gelfand MS, Mironov AA, Rakhmaninova AB, Rubinov AR, Russell RB, Kalinina OV. 2010. An automated stochastic approach to the identification of the protein specificity determinants and functional subfamilies. Algorithms Mol. Biol. 5, 29. 10.1186/1748-7188-5-29 (doi:10.1186/1748-7188-5-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rausell A, Juan D, Pazos F, Valencia A. 2010. Protein interactions and ligand binding: from protein subfamilies to functional specificity. Proc. Natl Acad. Sci. USA 107, 1995–2000 10.1073/pnas.0908044107 (doi:10.1073/pnas.0908044107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ye K, Feenstra KA, Heringa J, Ijzerman AP, Marchiori E. 2008. Multi-RELIEF: a method to recognize specificity determining residues from multiple sequence alignments using a machine-learning approach for feature weighting. Bioinformatics 24, 18–25 10.1093/bioinformatics/btm537 (doi:10.1093/bioinformatics/btm537) [DOI] [PubMed] [Google Scholar]
- 102.Muth T, Garcia-Martin JA, Rausell A, Juan D, Valencia A, Pazos F. 2012. JDet: interactive calculation and visualization of function-related conservation patterns in multiple sequence alignments and structures. Bioinformatics 28, 584–586 10.1093/bioinformatics/btr688 (doi:10.1093/bioinformatics/btr688) [DOI] [PubMed] [Google Scholar]
- 103.Mayer KM, Shanklin J. 2007. Identification of amino acid residues involved in substrate specificity of plant acyl-ACP thioesterases using a bioinformatics-guided approach. BMC Plant Biol. 7, 1. 10.1186/1471-2229-7-1 (doi:10.1186/1471-2229-7-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lockless SW, Ranganathan R. 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 10.1126/science.286.5438.295 (doi:10.1126/science.286.5438.295) [DOI] [PubMed] [Google Scholar]
- 105.Halabi N, Rivoire O, Leibler S, Ranganathan R. 2009. Protein sectors: evolutionary units of three-dimensional structure. Cell 138, 774–786 10.1016/j.cell.2009.07.038 (doi:10.1016/j.cell.2009.07.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smock RG, Rivoire O, Russ WP, Swain JF, Leibler S, Ranganathan R, Gierasch LM. 2010. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol. Syst. Biol. 6, 414. 10.1038/msb.2010.65 (doi:10.1038/msb.2010.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reynolds KA, McLaughlin RN, Ranganathan R. 2011. Hot spots for allosteric regulation on protein surfaces. Cell 147, 1564–1575 10.1016/j.cell.2011.10.049 (doi:10.1016/j.cell.2011.10.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. 2005. Evolutionary information for specifying a protein fold. Nature 437, 512–518 10.1038/nature03991 (doi:10.1038/nature03991) [DOI] [PubMed] [Google Scholar]
- 109.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. 2005. Natural-like function in artificial WW domains. Nature 437, 579–583 10.1038/nature03990 (doi:10.1038/nature03990) [DOI] [PubMed] [Google Scholar]
- 110.Anand S, Mohanty D. 2012. Modeling holo-ACP:DH and holo-ACP:KR complexes of modular polyketide synthases: a docking and molecular dynamics study. BMC Struct. Biol. 12, 10. 10.1186/1472-6807-12-10 (doi:10.1186/1472-6807-12-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anand S, Mohanty D. 2012. Inter-domain movements in polyketide synthases: a molecular dynamics study. Mol. Biosyst. 8, 1157–1171 10.1039/c2mb05425f (doi:10.1039/c2mb05425f) [DOI] [PubMed] [Google Scholar]
- 112.Durrant JD, McCammon JA. 2011. Molecular dynamics simulations and drug discovery. BMC Biol. 9, 71. 10.1186/1741-7007-9-71 (doi:10.1186/1741-7007-9-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cornell WD, et al. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 10.1021/ja00124a002 (doi:10.1021/ja00124a002) [DOI] [Google Scholar]
- 114.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. 2004. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 10.1002/jcc.20035 (doi:10.1002/jcc.20035) [DOI] [PubMed] [Google Scholar]
- 115.Lilien RH, Stevens BW, Anderson AC, Donald BR. 2005. A novel ensemble-based scoring and search algorithm for protein redesign and its application to modify the substrate specificity of the gramicidin synthetase a phenylalanine adenylation enzyme. J. Comput. Biol. 12, 740–761 10.1089/cmb.2005.12.740 (doi:10.1089/cmb.2005.12.740) [DOI] [PubMed] [Google Scholar]
- 116.Stevens BW, Joska TM, Anderson AC. 2005. Progress toward re-engineering non-ribosomal peptide synthetase proteins: a potential new source of pharmacological agents. Drug Dev. Res. 66, 9–18 10.1002/ddr.20041 (doi:10.1002/ddr.20041) [DOI] [Google Scholar]
- 117.Georgiev I, Lilien RH, Donald BR. 2006. Improved pruning algorithms and divide-and-conquer strategies for dead-end elimination, with application to protein design. Bioinformatics 22, e174–e183 10.1093/bioinformatics/btl220 (doi:10.1093/bioinformatics/btl220) [DOI] [PubMed] [Google Scholar]
- 118.Georgiev I, Donald BR. 2007. Dead-end elimination with backbone flexibility. Bioinformatics 23, i185–i194 10.1093/bioinformatics/btm197 (doi:10.1093/bioinformatics/btm197) [DOI] [PubMed] [Google Scholar]
- 119.Georgiev I, Lilien RH, Donald BR. 2008. The minimized dead-end elimination criterion and its application to protein redesign in a hybrid scoring and search algorithm for computing partition functions over molecular ensembles. J. Comput. Chem. 29, 1527–1542 10.1002/jcc.20909 (doi:10.1002/jcc.20909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen CY, Georgiev I, Anderson AC, Donald BR. 2009. Computational structure-based redesign of enzyme activity. Proc. Natl Acad. Sci. USA 106, 3764–3769 10.1073/pnas.0900266106 (doi:10.1073/pnas.0900266106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. 2006. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chem. Biol. 1, 687–691 10.1021/cb6003965 (doi:10.1021/cb6003965) [DOI] [PubMed] [Google Scholar]
- 122.Kapur S, Worthington A, Tang Y, Cane DE, Burkart MD, Khosla C. 2008. Mechanism based protein crosslinking of domains from the 6-deoxyerythronolide B synthase. Bioorg. Med. Chem. Lett. 18, 3034–3038 10.1016/j.bmcl.2008.01.073 (doi:10.1016/j.bmcl.2008.01.073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jackel C, Kast P, Hilvert D. 2008. Protein design by directed evolution. Annu. Rev. Biophys. 37, 153–173 10.1146/annurev.biophys.37.032807.125832 (doi:10.1146/annurev.biophys.37.032807.125832) [DOI] [PubMed] [Google Scholar]
- 124.Tracewell CA, Arnold FH. 2009. Directed enzyme evolution: climbing fitness peaks one amino acid at a time. Curr. Opin. Chem. Biol. 13, 3–9 10.1016/j.cbpa.2009.01.017 (doi:10.1016/j.cbpa.2009.01.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dalby PA. 2011. Strategy and success for the directed evolution of enzymes. Curr. Opin. Struct. Biol. 21, 473–480 10.1016/j.sbi.2011.05.003 (doi:10.1016/j.sbi.2011.05.003) [DOI] [PubMed] [Google Scholar]
- 126.Damborsky J, Brezovsky J. 2009. Computational tools for designing and engineering biocatalysts. Curr. Opin. Chem. Biol. 13, 26–34 10.1016/j.cbpa.2009.02.021 (doi:10.1016/j.cbpa.2009.02.021) [DOI] [PubMed] [Google Scholar]
- 127.Gao X, Xie X, Pashkov I, Sawaya MR, Laidman J, Zhang W, Cacho R, Yeates TO, Tang Y. 2009. Directed evolution and structural characterization of a simvastatin synthase. Chem. Biol. 16, 1064–1074 10.1016/j.chembiol.2009.09.017 (doi:10.1016/j.chembiol.2009.09.017) [DOI] [PMC free article] [PubMed] [Google Scholar]