Abstract
For symmetrically dividing cells, large variations in the cell cycle time are typical, even among clonal cells. The consequence of this variation is important in stem cell differentiation, tissue and organ size control, and cancer development, where cell division rates ultimately determine the cell population. We explore the connection between cell cycle time variation and population-level fluctuations using simple stochastic models. We find that standard population models with constant division and death rates fail to predict the level of population fluctuation. Instead, variations in the cell division time contribute to population fluctuations. An age-dependent birth and death model allows us to compute the mean squared fluctuation or the population dispersion as a function of time. This dispersion grows exponentially with time, but scales with the population. We also find a relationship between the dispersion and the cell cycle time distribution for synchronized cell populations. The model can easily be generalized to study populations involving cell differentiation and competitive growth situations.
Keywords: stochastic population dynamics, cell cycle variation, evolutionary models
1. Introduction
All cell types reproduce by repeated cycles of division and growth. If a cell divides symmetrically, then both daughter cells are expected to divide roughly at the same rate when environmental factors and cell population densities are kept constant. In a recent experiment where individual bacterial cell division events were monitored, it was shown that the time between cell divisions (or generation time) is a highly stochastic variable, even for clonal cells [1]. Furthermore, the generation time distribution is a non-monotonic functions of chronological cell age, a, defined as the time since its birth. Typical probability distributions of the time between successive cell divisions from experiments are shown in figure 1b,c. This behaviour is not restricted to prokaryotes, and is also true for eukaryotes. It is known that the probability for cell division depends both on age and on cell size [1,2]. For instance, for cells of the same size (mouse lymphoblasts L1210) the probability of division increases with cell age and for cells of the same age, the probability of division increases with cell size [2]. Cell division and cell growth cycles seem to be interrelated [3]. Other factors are also involved in the division decision. For instance, in Escherichia coli cells the specific growth rate of a population is found to linearly correlate with RNA/protein ratio and ribosome production [4]. The linear correlation holds for cell growth with doubling times varying from minutes to hours. These studies and others demonstrate that the cell cycle decision is an intrinsically noisy process where cell age and other factors are important, but biochemical noise also dominates.
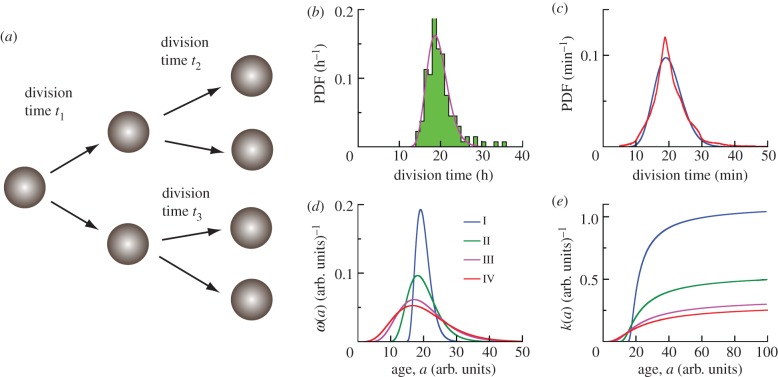
Figure 1.
Cell age and division time distributions. (a) For symmetrically dividing cells, the time between cell divisions is stochastic, and can be described by a probability distribution function (PDF). (b) Histogram of division time distribution for human dermal fibroblast cells studied in the present work (133 cells). The average division time is τ = 19.8 h. The solid line represents the fit by the shifted gamma distribution with parameters α = 12.5, β = 0.72 h and a0 = 10.4 h. (c) Division time distribution for E. coli, MG1655 strain (22 959 cells) with τ = 21.4 min and σ = 5.4 min from Wang et al. [1] shown by red line. Blue line is the best fit by the shifted gamma distribution with parameters α = 22.9, β = 0.87 min and a0 = 0 min with τfit = 20.0 min and σfit = 4.2 min. (d) Theoretical generation time distributions ω(a) studied in the present work are described by shifted gamma distributions with parameters α = 6 (I–IV), and β = 0.91, 1.82, 2.86, 3.33 arb. units, and a0 = 14.55, 9.09, 2.86, 0 arb. units for I, II, III, IV, respectively (see equation (2.17)). The average generation time for distributions I–IV is the same τ = 20 arb. units. (e) The function k(a) is the probability per unit time that a cell divides into two new cells for theoretical distributions of generation times I–IV (see legends in (d)). The asymptotic value of k(a) is 1/β (arb. units)–1.
Given the probabilistic nature of cell division events, we ask how cell division time variation ultimately translates to cell number fluctuations in a growing population. Understanding this connection is important for understanding tissue growth and homeostatsis, where not just the average population but also population fluctuations must be carefully controlled [5]. Quantitative models for this question must be stochastic in nature. The simplest model for studying stochastic population dynamics is a Markovian master equation (or a birth–death process) with constant division and death probabilities per unit time [6]. The average population and population fluctuations for this model can be solved exactly (see the electronic supplementary material, section A). At long times, the relative population fluctuation is
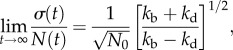 |
2.1 |
where N(t) is the average population and σ(t) is the average population fluctuation (dispersion). kb and kd are the cell birth and death rates, respectively. N0 is the initial population at t = 0. This scaling result is simple. However, since the model assumes constant division and death probabilities, it is in fact not applicable to typical cell division processes since the division probability per unit time is clearly not constant (figure 1). A constant division rate amounts to the assumption that a newly born cell has just as much likelihood to divide as a mature cell. But this is not the case, as shown in figure 1. In this paper, we develop a stochastic model where division and death probabilities are functions of cell age, and use experimentally measured division time distributions to predict population fluctuations. We consider a homogeneous cell population and with no correlation between interdivision times for cells in different generations. The age a of cells is considered explicitly as the variable determining the propensity of cell division. We examine growth dynamics when the mean division time is kept constant, but the spread of the division time distribution changes (figure 1d). We show that our model is equivalent to the solution of the von Foerster equation when considering the average population. However, we also obtain quantitative results regarding population fluctuations as a function of time. We find that there is a direct link between division time distribution, ω(a), and population fluctuations. In the case of synchronized populations, a simple relationship between ω(a) and σ(t) is found. We also examine more realistic models of cell population homeostasis where the death rate (or the net growth rate) is controlled by the overall population.
Note that in the long time limit, population growth described by the age-dependent model exhibits some properties of the age-independent (stationary) stochastic model, e.g. the average number of cells grows exponentially. However, the predicted exponential population growth rate and population size fluctuations (dispersion) around the mean are different in these models, even when the average cell cycle times are kept constant. These differences are highlighted in this paper. There have been other quantitative studies of cell growth with time-dependent division rates [5,7–10]. For instance, Horowitz et al. [7] consider a probabilistic model of microbial growth and mortality in which both cell division and cell death transition probabilities depend on time, e.g. in the form of logistic expressions. Two simulation algorithms are considered: one for tracking the fates of individual cells; another for simulation at the population level using a simplified model. Although the authors have succeeded in reproducing the experimental growth curves for a bacteria, the age of cells as an important characteristics of mitosis is not considered. Another approach is applied for stochastic modelling of population growth by Pin & Baranyi [8] based on the assignment of division times from the empirical generation time distributions.
Other theoretical studies have considered stochastic processes in cell division, synchronous growth curves and/or age distributions of exponential cultures within deterministic models [11–15]. For instance, Bremer [14] has reduced the cell cycle variability of E. coli to variability of times between the end of DNA replication and the next cell division. This is also a variation of the original idea by Smith & Martin [11] that there exists a highly variable period before DNA replication. Engelberg [16] has derived a simple model establishing the relationship between the loss of synchrony in cell divisions and the width of division time distribution in synchronized cell cultures. However, there is a lack of systematic studies analysing population size fluctuations of growing cell colonies. Our paper is aimed at providing such analysis and establishing the link between population size fluctuations and the stochastic nature of the cell division process at different growth conditions (both in the absence and in the presence of cell death, for conditions of restrictive growth, etc.).
Our age-dependent model is in principle applicable to any cell growth situation, e.g. in vitro in a reactor as well as in vivo where multiple cell types may influence each other. In conditions of saturating nutrient and low cell densities, cell division and cell death probabilities are known and the application of our model is straightforward. In conditions of limiting nutrient and/or space, competition between individuals cells results in death rate proportional to total number of cells. A simple example of this situation is also considered below. For multiple types of interacting cells, cell division and death probabilities will depend on signalling, cooperative as well as competitive interactions. Additional models and information are needed to quantify cell division and cell death probabilities. Our model is not applicable, however, to cell division probabilities that has memory of prior growth conditions. This is because our model is Markovian. In addition, our model currently does not consider spatial patterns of growing cell colonies, only the overall population in a well mixed situation.
2. Model
2.1. Cell population and division in the age-time representation
Before discussing the stochastic dynamics of cell population, it is useful to consider deterministic dynamics of the average population as a function of time. For this purpose, the von Foerster equation describes the cell number density as a function of cell age a and time t [17]. The age of a cell, a, is defined as the time elapsed since its birth. The ongoing time, t, can be chosen arbitrarily, but it is natural to measure this from the time of cell culture seeding. In the age-time representation the equation is written as
| 2.2 |
where n(a,t) da represents the number of cells in the age interval from a to a + da at time t and λ is a cell loss factor. The total number of cells at time t is therefore
| 2.3 |
The loss factor λ is assumed to be due to both cell division (mitosis) as well as cell death or disappearance. The von Foerster equation is a form of a conservation equation that describes the flow of population densities. From conservation of population, the number of cells n(a + Δa, t + Δt)Δa of the age a + Δa at time t + Δt equals the number of cells that have matured from an earlier age a at time t, i.e. n(a, t)Δa less the number of cells that have been lost. The number of lost cells is assumed to be proportional to the number of cells n(a, t)Δa and the time interval Δt. Cell loss due to mitosis should be understood as the mother cell of age a is replaced by two replicas of zero age (a = 0). If there are no other reasons for cell loss besides mitosis, then λ = k(a), where k(a) is an age-dependent probability per unit time that a cell undergoes mitosis. The solution and the properties of the von Foerster equation regardless the initial and boundary condition have been discussed by Trucco [18,19]. Rubinow [13] gave a solution for the specific initial and boundary conditions
| 2.4 |
and cell birth is described by the boundary condition
| 2.5 |
Thus, the age-dependent function k(a) accounts for the variability in observed generation times. Here, we note that the total number of cells N(t) at time t is obtained by integrating n(a,t) for all ages and is found as the infinite sum of terms (j = 1, 2,…), each represents the contribution of the jth generation (see the electronic supplementary material, section B).
The probability per unit time that a cell undergoes mitosis is related to probability density function ω(a) for cell division times:
| 2.6 |
This relation is derived as follows. Let us assume that we have  cells of zero age at time t = 0. In this case, time t and age a are synchronized and can be considered as a single variable, say a. By time a > 0, the number of cells that remain undivided is
cells of zero age at time t = 0. In this case, time t and age a are synchronized and can be considered as a single variable, say a. By time a > 0, the number of cells that remain undivided is 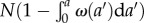 . The number of cells that undergo mitosis during a time interval from a to a + Δa is Nω(a)Δa. Thus, the conditional probability that a cell divides between a and a + Δa (provided it had not divided earlier) is given by the ratio of the number of cells divided during this time interval to the total (remaining) number of cells at time a, i.e.
. The number of cells that undergo mitosis during a time interval from a to a + Δa is Nω(a)Δa. Thus, the conditional probability that a cell divides between a and a + Δa (provided it had not divided earlier) is given by the ratio of the number of cells divided during this time interval to the total (remaining) number of cells at time a, i.e. 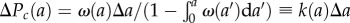 . The cell division probability per unit of time k(a) is obtained from ΔPc(a) above resulting in equation (2.6).
. The cell division probability per unit of time k(a) is obtained from ΔPc(a) above resulting in equation (2.6).
Following Powell [12], in the limit of long times, the solution of the von Foerster equation reduces to exponential growth: 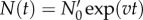 with the steady-state growth rate, v, determined by generation time distribution ω(a) only. The pre-factor
with the steady-state growth rate, v, determined by generation time distribution ω(a) only. The pre-factor 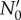 depends on the initial age distribution and is proportional to the initial number of live cells N0. The following ansatz satisfies the von Foerster equation at long times:
depends on the initial age distribution and is proportional to the initial number of live cells N0. The following ansatz satisfies the von Foerster equation at long times:
| 2.7 |
where r(a) is a function that only depends on the age a. Substitution equation (2.7) into equation (2.2) gives
| 2.8 |
and the solution is
| 2.9 |
Using equation (2.9), we re-write the boundary condition equation (2.5) in the form (after cancelling the non-negative term exp(vt)r(0) on both sides)
| 2.10 |
From equation (2.6), one may show that
| 2.11 |
This equation has a simple physical interpretation. If we consider mitosis as a one-step irreversible transition, the first term is the transition probability per unit of time, while the second term is the survival probability Ω(a). Thus, the distribution of cell division times can be expressed through the survival probability as ω(a) = −(dΩ(a))/(da). Equation (2.10) then becomes
| 2.12 |
This equation gives the fundamental relationship between division time distribution, ω(a), and the long time growth rate of the population, v.
Using the long time ansatz solution, we can also find the steady-state age distribution g(a), which is
| 2.13 |
Since 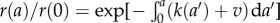 , and in light of equation (2.10), we have
, and in light of equation (2.10), we have
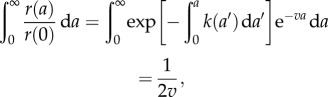 |
2.14 |
where we have used the fact that 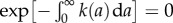 . Thus, for the age distribution, we obtain
. Thus, for the age distribution, we obtain
| 2.15 |
We now can examine the behaviour for different generation time distributions ω(a). When the generation time is not dispersed and all cells divide exactly at the same age, then ω(a) = δ(a − τ), where δ(a − τ) is the Dirac delta function satisfying the condition 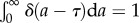 . The growth rate is v = ln 2/τ by equation (2.12) and the doubling time of the cell population equals the average generation time τ* ≡ ln 2/v = τ. The age distribution is
. The growth rate is v = ln 2/τ by equation (2.12) and the doubling time of the cell population equals the average generation time τ* ≡ ln 2/v = τ. The age distribution is
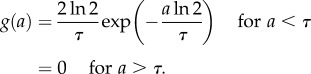 |
2.16 |
The age distribution described by equation (2.16) is an ideal age distribution assuming that the population has reached the steady state. In reality g(a) is smoothed-out. The average age of population with an ideal age distribution is obtained as 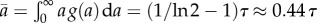 . Let us consider another hypothetical case when the rate of cell division is age-independent k(a) = 1/τ corresponding to exponential distribution for generation times ω(a) = 1/τ exp(−a/τ). In this case, the average growth rate is equal to v = 1/τ by equation (2.12) and the average doubling time is equal to τ* ≡ ln 2/v = τ ln 2. The age distribution is the exponential function g(a) = 2/τ exp(−2a/τ) with the average age of population equal to a half of the average generation time
. Let us consider another hypothetical case when the rate of cell division is age-independent k(a) = 1/τ corresponding to exponential distribution for generation times ω(a) = 1/τ exp(−a/τ). In this case, the average growth rate is equal to v = 1/τ by equation (2.12) and the average doubling time is equal to τ* ≡ ln 2/v = τ ln 2. The age distribution is the exponential function g(a) = 2/τ exp(−2a/τ) with the average age of population equal to a half of the average generation time 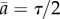 . Powell has pointed out that in general (see equation (2.12)) the average doubling time τ* is not equal to the average generation time τ but in practice this difference varies depending on the division time distribution.
. Powell has pointed out that in general (see equation (2.12)) the average doubling time τ* is not equal to the average generation time τ but in practice this difference varies depending on the division time distribution.
For most cells, experimental data show that the cell division time distribution is neither exponential nor a delta function. Some typical results are given in figure 1. The realistic generation time distribution is often best described by the a0-shifted gamma distribution [9,12,13,20–22]
| 2.17 |
where α > 1, β > 0 and a0 ≥ 0 are parameters of shape, scale and shift of generation time distribution, respectively. Typically, data show that ω(a) is not symmetric but skewed towards higher values (the skewness of gamma distribution is 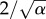 ). The gamma distribution is widely used in chemical kinetics for dwell-time distributions (e.g. in analysing enzyme kinetics) [23,24]. The probability distribution of first passage times for a multi-step irreversible process
). The gamma distribution is widely used in chemical kinetics for dwell-time distributions (e.g. in analysing enzyme kinetics) [23,24]. The probability distribution of first passage times for a multi-step irreversible process 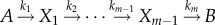 with m rate-limiting steps can be shown to correspond to equation (2.17). The average growth rate of the population is the solution of equation (2.12) which reads
with m rate-limiting steps can be shown to correspond to equation (2.17). The average growth rate of the population is the solution of equation (2.12) which reads
| 2.18 |
For simplicity, take a0 = 0. The average doubling time in this case is τ* = ln2/v = βln/(21/α − 1), whereas τ = αβ. For 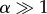 , the average doubling time,
, the average doubling time, 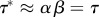 , equals the average generation time.
, equals the average generation time.
Interestingly, for E. coli the division time distribution (figure 1 and [1]) is not completely described by the shifted gamma distribution. Rather, it is better described by a combination of two exponential functions with a sharp peak. The mechanistic argument for this was discussed by Bremer [14].
In addition to cell birth, cell death can also contribute to cell loss. In this paper, we consider constant cell death probability per unit time kd. In this case, λ = k(a) + kd in equation (2.2) with the same boundary condition given by equation (2.5). If we are interested in the long time limit solution identical to equation (2.7), the net growth rate in the presence of cell death becomes v = v0 − kd, where is v0 is the growth rate in the absence of cell death found in equation (2.10). Of course, if kd depends on the cell age, this simple relation does not hold. The electronic supplementary material, figure S1 of SI shows simulation results of v as a function of constant kd for age-dependent cell division. Note that cell death with age-independent probability does not perturb steady-state age distribution since cells of different ages die with the same rate. Constant cell death rescales both numerator and denominator of equation (2.13) by a factor 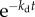 .
.
The von Foerster equation describes the average age-dependent population change. In this sense, it does not contain information about population fluctuations. In order to obtain fluctuation information, we must incorporate age dependence in a stochastic model of cell population growth. In §2.2, we describe a stochastic simulation of age-dependent population growth and analyse the results. In general, the mathematical quantity we have to consider is the probability distribution of cell number density as a function of age P[n(a),t]. This quantity is a functional and it is possible to write a stochastic equation to describe its evolution. The theoretical treatment of this model will be discussed elsewhere.
2.2. Stochastic simulations of age-dependent cell populations
Our stochastic algorithm takes explicitly into consideration the age-dependence of rates of ‘elementary’ events. Mitosis is considered in the present work as a one-step event with a probability per unit time k(a) that depends on the cell age a, i.e. the time elapsed since its birth. Our simple simulation model is a version of the kinetic Monte Carlo algorithm in which the acceptance probability of any event for each cell depends on the exponential factor exp[−(k(aj) + kd)Δt], where k(aj) and kd are cell division and cell death probabilities per unit time for the jth cell, respectively. Δt is a time step. A random number 0 < r1 < 1 is compared with exp[−(k(aj) + kd)Δt] to determine whether an event occurs. At each time step, t = mΔt, a particular cell remains alive but undivided if r1 < f1(aj) = exp[−(k(aj) + kd)Δt]. Otherwise this cell dies or divides. If an event occurs for the jth cell, the choice between mitosis and cell death is made by comparing another random number 0 < r2 < 1 with f2(aj) = k(aj)/(k(aj) + kd). Thus, cell division occurs if r1 > f1(aj) and r2 < f2(aj), giving rise to a pair of newborn cells with ages equal to 0. The jth cell dies if r1 > f1(aj) and r2 > f2(aj). Note, that if no event occurs with a particular cell at a given time step, the cell becomes older by a = a + Δt. As time is incremented by Δt, a small portion of the cells undergoes mitosis or death. Thus, ages of cells are tracked explicitly. This procedure generates a histogram of cells with different ages nh(a, t)Δa, from which we can obtain the number density of cells as a function of cell age at time t, where Δa is the bin size of the histogram. nh is the cell number density from the particular simulation trajectory. This algorithm amounts to a Markovian stochastic simulation of age-dependent cell division and growth.
Note that since the model is stochastic, for each simulation, the obtained cell number density is different. The proper interpretation is that the age distribution averaged over many simulations, 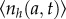 , will reproduce the von Foerster equation result. The average is over many simulations. From the average population density, we can also compute the total number of cells by integrating over age. We also compute the population fluctuation, which is the average variation of a particular simulation from the mean, defined below in equation (3.3).
, will reproduce the von Foerster equation result. The average is over many simulations. From the average population density, we can also compute the total number of cells by integrating over age. We also compute the population fluctuation, which is the average variation of a particular simulation from the mean, defined below in equation (3.3).
Figure 2 shows the average number of cells N(t) plus and minus standard deviation σ(t) of the mean obtained from our simulations. The population is grown from a single cell of age a = 0. Results of our simulations are in excellent agreement with analytical solution of the von Foerster equation shown also for comparison in figure 2. The contributions from different generations to N(t) are also shown. All contributions excluding the initial generation go through a maxima. In separate short runs, we analyse the distribution of waiting times to first cell division starting from a single cell of zero age. This histogram is also in the full agreement with theoretical generation time distribution (see the inset in figure 2). Thus, these two comparisons demonstrate the relevance and accuracy of our simulation algorithm.
Figure 2.
Growth of a clonal cell colony using age-dependent stochastic model. (a) The average number of cells, N(t), grown from a single cell of age a = 0 (synchronized initial condition) for division time distribution II is shown from 1000 simulations (thick green line). Magenta lines represent N(t) plus and minus the standard deviation σ(t). The red line is the analytical solution of the von Foerster equation for synchronized populations. The thin curves (with maxima) represent the contributions from different generations (see the electronic supplementary material, section B). The results of our simulations are in excellent agreement with analytic solution. The inset depicts the probability density function of division times obtained from simulations (106 runs) for the theoretical division time distribution II (see caption to figure 1d). Our simulation recovers the original division time distribution ω(a) (red line). (b) the computed average doubling times compared to the theoretical result of equation (2.12) for division time distributions (I–IV) and the exponential distribution (V). There is complete agreement between the theoretical prediction and our simulation.
3. Results
3.1. Average growth rates of cell colonies and doubling times
We first examine growing clonal populations with no cell death. In most of the simulations, cells are grown from a single clone with age a = 0. In some simulations, we start with more than one cell. We perform simulations for (a) initially synchronized and (b) non-synchronized cell populations. In the first case (a), the initial age of the cell (or cells) is taken to be zero. In the second case (b), the initial age of the cells is chosen randomly between 0 and T (typically, T = τ). We find that in all cases at long times the average number of cells N(t), defined
| 3.1 |
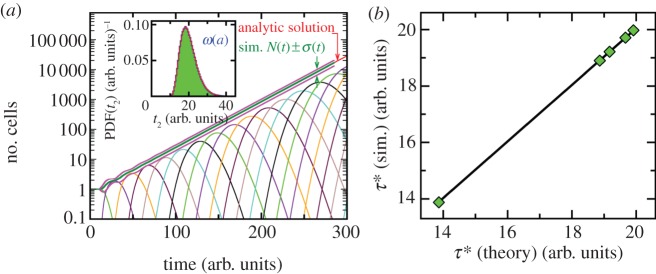
grows exponentially (see the electronic supplementary material, figures S2 and S3 of SI). For initially synchronized populations for the case of the distribution with the smallest spread (distribution I, see electronic supplementary material, figure S4), we observe the step-like pattern in the average growth trajectory (‘mitotic waves’) [25–27]. However, eventually these ‘oscillations’ are damped at long times. The loss of synchrony results from the variation in division times. The growth rates v are found from exponential fits to the average number of cells as a function of times. For comparison, for the exponential distribution of division times V, the cell division rate is constant, so synchronization does not matter. Note that for all these distributions I–V, the average generation times are the same (τ = 20 arb. units) but their spreads are different. We characterize the width of generation time distributions by the dimensionless coefficient of variation of τ defined as
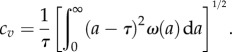 |
3.2 |
For the exponential division time distribution cv = 1. For division time distributions I–IV in figure 1, 0 < cv < 1. We run the simulation many times and obtain the average growth rate. The doubling time of the cell cultures is τ* = ln2/v. Figure 2b shows the comparison between theory and simulation results for the doubling time. Excellent agreement is observed. For generation time distribution with larger spread (larger cv), the difference between the average generation time τ and the average doubling time τ* is larger than for more narrow distribution of τ in full accord with theory.
The effect of initial age distribution on the average growth rates is negligible (see the electronic supplementary material, figure S4 of SI). For instance, for distribution I the doubling times for initially synchronized and non-synchronized cell populations are τ* = 19.972 ± 0.004 and τ* = 19.934 ± 0.001. For wider distribution II, these values are τ* = 19.713 ± 0.002 and τ* = 19.695 ± 0.001, respectively.
3.2. Steady-state age distribution in the growing colony
At long times, the age distribution of cells in the colony should not depend on time, as indicated from the long time ansatz of the von Foerster equation. Indeed, we observe stationary age distributions in our simulations, regardless of the initial age distribution (figure 3). We also observe oscillations in the average age of populations because of sequential cell divisions. However, these oscillations eventually disappear at long times. The rate at which these oscillations disappear depends on cv of the division time distribution (figure 4). The age distribution at long times in all cases has a maximum at zero age a = 0 and decreases monotonically for a > 0. For the narrowest distribution (case I with the smallest cv), the age distribution is closer to the ‘ideal’ age distribution with two distinct regions: exponential decrease of probability density g(a) ∼ exp(−va) for a < τ followed by much faster decay of g(a) for a > τ. Our simulation results for the steady-state age distributions are in excellent agreement with theoretical ones obtained by equation (2.15).
Figure 3.
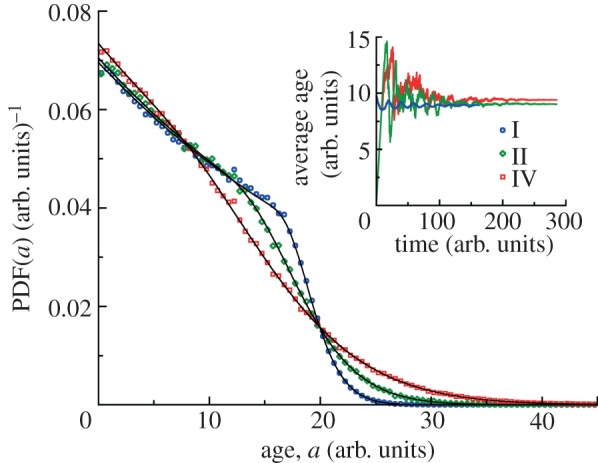
Steady-state age distributions for clonal colonies. Age distributions for division time distributions I, II and IV (τ = 20 arb. units, see caption to figure 1d) are obtained from simulations. Each distribution is obtained by analysing 25 growth trajectories with final ages recorded at the end of each run. For distributions II and IV the populations are grown from a single cell of a = 0 and the final ages are analysed at t = 285 arb. units. For the narrow distribution I, the initial number of cells is 200 for each run with uniform initial age distribution between 0 and a = 20 arb. units. The final ages are analysed at t = 160 arb. units. The solid black lines represent theoretical results of the long time ansatz in equation (2.15). The inset depicts the convergence of the average age of cells in the growing population to a constant value at long times. The average ages of cells for distributions I, II and IV are 8.94, 9.02 and 9.40 arb. units, respectively, are in agreement with theoretical values for the same distributions.
Figure 4.
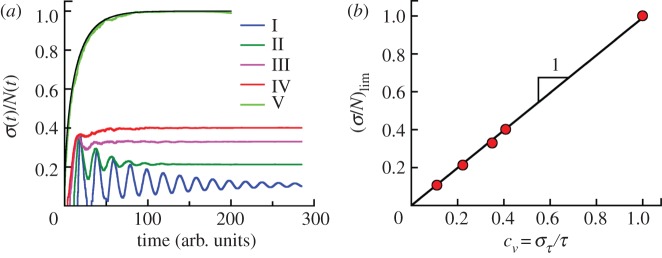
Population fluctuations in growing clonal colonies with initially synchronized population. (a) The average relative population fluctuations in clones grown from a single cell (N0 = 1) with no cell death. σ(t) is defined in equation (3.3). The fluctuations depend on the division time distribution. Curves I–IV are for ω(a) in figure 1d. Curve V corresponds to exponential distribution of division times (i.e. constant division rate). The relative fluctuations reach constant values for long times. (b) The asymptotic values of the relative population fluctuations of synchronized cell populations (σ/N)lim at long times for theoretical distributions I–V taken from figure 4a are shown as a function of coefficient of variation of generation times cv defined by equation (3.2). The solid line represents the linear fit (σ/N)lim = bσcv with bσ = 0.99 ± 0.01. The condition of synchronized populations at t = 0 is essential to observe this simple relation between (σ/N)lim and cv for age-dependent cell division probability per unit time (cases I–IV) but does not matter for constant cell division rate (case V). The coefficients of variation for theoretical distributions I–V by equation (3.2) are cv = 0.111, 0.222, 0.350, 0.408 and 1, respectively.
3.3. Population fluctuations in initially synchronized growing clones
Beyond the average clone population and the age distribution, we obtain results for population fluctuations defined as
| 3.3 |
Simulations show that σ(t) grows exponentially at long times with the same rates as the average numbers of cells N(t). Thus, relative standard deviations defined as σ(t)/N(t) converge to constant values at long times. The results of our simulations for different generation time distributions I–IV in the absence of the cell death are shown in figure 4. Distribution V is the exponential generation time distribution, which corresponds to a constant division probability per unit time (see above). Exact analytic results are available (see the electronic supplementary material, section A). Note that these populations have the same average division time. Thus, we see that the width (or the dispersion) of the generation time distribution has a dramatic effect on the population fluctuation; this dependence should have important consequences in real biological settings.
We will consider the asymptotic value of σ(t)/N(t) at t → ∞, which we denote as (σ/N)lim. However, if we consider the time dependence of the relative fluctuation, we will keep t in the notation. In general, the asymptotic value is less than 1. From our simulations, it appears that the following simple relation for (σ/N)lim for initially synchronized cell populations is valid:
| 3.4 |
cv being defined earlier by equation (3.2). It is easy to see that equation (3.4) is valid for delta function (cv = 0) and exponential (cv = 1) generation time distributions. However, the initial population must be synchronized to satisfy the simple relation 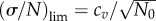 . Note that for the constant cell division rate (exponential distribution), the asymptotic value of standard deviation remains the same regardless of the initial age distribution. This conjecture seems to be reasonable because the more dispersed distribution of division times results in larger variation of the growth rates and eventually the larger relative fluctuations of sizes of cell populations around mean values. Note that only the dispersion but not the general shape of ω(a) matters. For other shapes such as the combination of two exponential functions proposed by Bremer [14], equation (3.4) is also valid.
. Note that for the constant cell division rate (exponential distribution), the asymptotic value of standard deviation remains the same regardless of the initial age distribution. This conjecture seems to be reasonable because the more dispersed distribution of division times results in larger variation of the growth rates and eventually the larger relative fluctuations of sizes of cell populations around mean values. Note that only the dispersion but not the general shape of ω(a) matters. For other shapes such as the combination of two exponential functions proposed by Bremer [14], equation (3.4) is also valid.
3.4. Effect of cell death and initial conditions on population fluctuations
We now examine other aspects of typical cell population that effect population fluctuations. First, we model cell death with a constant death rate kd that does not depend on the age of individuals. The results show that cell death has a significant effect on the average population N(t) and population fluctuations σ(t) (figure 5). The average growth curves for kd > 0 are shown in electronic supplementary material, figure S5 of SI. Both N(t) and σ(t) grow exponentially as 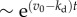 with the net growth rate v = v0 − kd, where v0 is the mean growth rate in absence of cell death. The relative standard deviation σ(t)/N(t) converges to an asymptotic value that is larger than in absence of cell death. Figure 5a shows that the relative fluctuations grow with increasing cell death rate kd. Apparently, a similar result is seen in models with constant (age-independent) cell division k(a) = kb and cell death kd rates. In this case, σ(t)/N(t) actually diverge when kd approaches kb, i.e. equation (2.1) (see electronic supplementary material, section A). For
with the net growth rate v = v0 − kd, where v0 is the mean growth rate in absence of cell death. The relative standard deviation σ(t)/N(t) converges to an asymptotic value that is larger than in absence of cell death. Figure 5a shows that the relative fluctuations grow with increasing cell death rate kd. Apparently, a similar result is seen in models with constant (age-independent) cell division k(a) = kb and cell death kd rates. In this case, σ(t)/N(t) actually diverge when kd approaches kb, i.e. equation (2.1) (see electronic supplementary material, section A). For 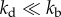 , we have
, we have
| 3.5 |
However, in the age-dependent cell division case, the relative fluctuation does not agree with equation (3.5). Here, analytical results are not available, and the quantitative dependence is an open question. We compare the relative population fluctuations as a function of constant cell death rate kd for age-dependent model with age-independent model in figure 5b. A significant difference between these models is seen. The effect of cell death on asymptotic relative fluctuations is stronger for the age-dependent model (cf. the initial slopes of dependencies shown in figure 5b). The dependence is not linear for small death rates. Since cell division processes are generally age-dependent, this suggests that a more sophisticated understanding of population fluctuations is needed.
Figure 5.
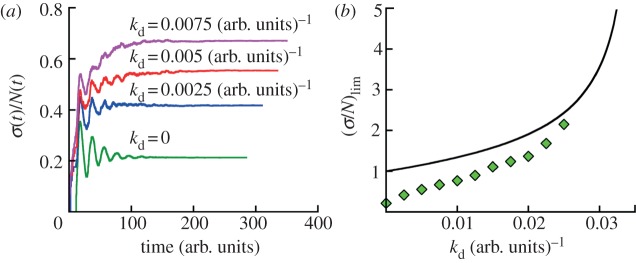
Effect of cell death on cell population fluctuations. (a) A constant cell death rate always increases the relative population fluctuation. The bottom line is the simulation result with no cell death taken from figure 4 (generation time distribution II, see caption to figure 1d). The other curves are for cell death rates kd = 0.0025, 0.005 and 0.0075 (arb. units.)−1, respectively. σ(t) and N(t) are obtained at least for 200 runs for all cases. The populations are grown from a single cell of the zero age a = 0. The respective growth curves from simulations for some kd are presented in electronic supplementary material, figure S5 of SI. (b) Asymptotic values of (σ/N)lim are shown as a function of cell death rate kd. The symbols are simulation data for age-dependent cell division k(a) (generation time distribution II) and constant cell death kd obtained by analysing at least 200 runs for each case. The black line represents theoretical dependence for both contant rates kb and kd given by equation (S9) in electronic supplementary material, section A.
In addition to cell death, the initial age distribution of the cell population also affects the long-term population fluctuation. In figure 6, we compare the relative population fluctuation for two different division time distributions. The initial age distribution is taken to be a delta function, or a uniform distribution between 0 and τ, where τ is the average division time. We see that (σ/N)lim is larger for non-synchronized populations when compared with synchronized cell cultures. The same trend is also true for the absolute standard deviations σ(t). Note that the effect of initial age distribution is more pronounced for the distribution of division times with the smaller spread. Again, for constant division probability, the effect of initial age distribution on σ(t) and σ(t)/N(t) is absent.
Figure 6.
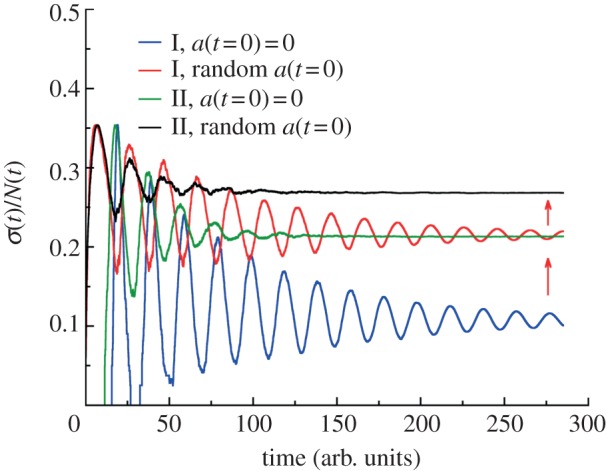
Effect of initial age distribution on population fluctuations. The growth of cell populations is simulated from a single cell for generation time distributions I and II (see legends and caption to figure 1d). For synchronized populations the initial age of a starting cell is taken to be zero. For asynchronous growth the initial age of a cell is chosen randomly from 0 to τ. The red arrows show increasing the asymptotic values of relative dispersions for asynchronous growth compared with synchronized condition. The effect of initial age distribution on (σ/N)lim is greater for less dispersed generation time distribution (I verus II) and is absent for constant cell division rate.
The initial total number of cells, N0, also affects population fluctuation. However, the dependence is trivial since σ(t) generally scales as 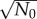 for independent cells. Therefore, σ(t)/N(t) scales as
for independent cells. Therefore, σ(t)/N(t) scales as 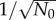 . This is true regardless of whether the model incorporates age-dependent cell division or constant cell division rates (see the electronic supplementary material, figures S6 and S7 of SI). For asynchronous populations, the scaling of (σ/N)lim with N0 also remains the same (see the electronic supplementary material, figure S8).
. This is true regardless of whether the model incorporates age-dependent cell division or constant cell division rates (see the electronic supplementary material, figures S6 and S7 of SI). For asynchronous populations, the scaling of (σ/N)lim with N0 also remains the same (see the electronic supplementary material, figure S8).
3.5. Growth of competitive populations
In organized and biologically realistic cell colonies, cell division and cell death are carefully controlled. The control strategy has a strong effect on the total cellular population as well as population fluctuations. There are many possible models to describe these situations. Here, we examine a competitive population model where in addition to natural cell division k(a) and cell death rates kd the competition gives rise to an additional death rate γ = γ0(n − 1) proportional to the number of other cells N in the colony at time t. For the age-independent case, k(a) = kb, and constant kd and γ0, this model is well known as the Malthus–Verhulst problem [28]. The equation for the average number of cells is
| 3.6 |
which has a stable steady-state population Nss:
| 3.7 |
In our model, we consider the age-dependent cell division rate k(a) and all other rates are taken the same as in the Malthus–Verhulst problem, i.e. independent of the age of cells. We analyse the results for the growth of a competitive population from a single cell of the zero age a = 0 from our simulations. The ‘natural’ cell death rate is taken to be zero for simplicity kd = 0, while γ0 > 0. Indeed, we observe the steady state at long times (figure 7a) corresponding to a population of the macroscopic size equal to
| 3.8 |
where v is the growth rate of a population in the absence of any cell death events. Recall that this growth rate appears as the solution of the long time ansatz of the von Foerster equation, see equation (2.12). Thus, for cells with the same average division time, but different shapes ω(a), we expect a different steady-state population.
Figure 7.
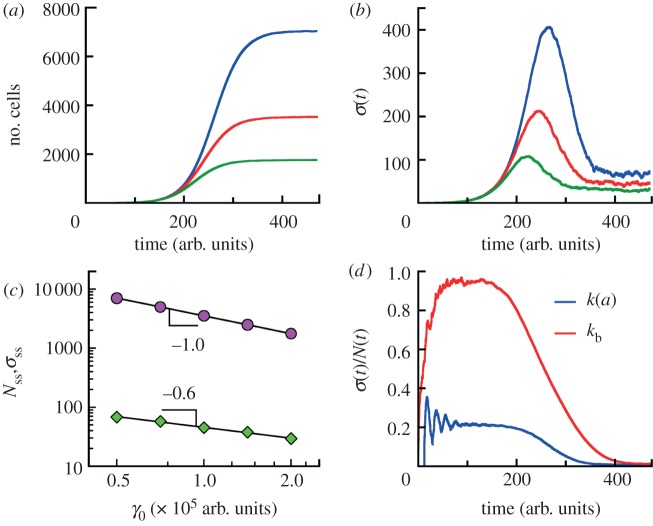
Competitive growth. (a) The average numbers of cells as functions of time for competitive populations with age-dependent cell division rate k(a) (distribution II, see caption to figure 1d) and death rates proportional to the number of other individuals γ = γ0(n − 1). The populations are grown from a single cell of the zero age a = 0. The numbers of cells reach the steady state Nss at long times. The data for each γ0 are obtained from simulations for 120 growth trajectories. (b) The standard deviations σ(t) of the mean sizes of competitive populations from data in (a) are non-monotonic functions of time. At long times, σ(t) approach the limiting values σss in contrast to exponentially growing populations. The population size distributions at steady state are stationary Gaussian around mean sizes Nss (see the electronic supplementary material, figure S9 of SI). (c) The average numbers of cell Nss (circles) and its standard deviations σss (diamonds) at steady state are shown as a function of ‘competitive’ cell death rate γ0. The data are analysed in the time interval from t = 400 arb. units to t = 474 arb. units for all values of γ0. The mean population is inversely proportional to γ0. (d) Comparison of the population fluctuation for age-dependent (blue) and age-independent (constant division rate kb) (red) models. γ0 for these models are identical. Parameters are chosen so that the average growth rates in the absence of cell death are identical. The age-dependent model shows a significantly different behaviour.
At long times, fluctuations around the mean population approach a steady value as in the Malthus–Verhulst problem. Interestingly, the standard deviation σ(t) of the mean is a non-monotonic function of time (see figure 7b). At early times when the cell number is small and death events are rare, σ(t) grows exponentially. In the crossover regime, σ(t) goes through the maximum and at last reaches its steady-state value. The steady-state value of σ(t), however, does depend on γ0 and whether the model considers cell age. Figure 7d shows the comparison between the age-independent model (see the electronic supplementary material, section C) and the age-dependent model. The population fluctuation at steady-state scales as 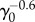 for the age-dependent model. For the age-independent model, σss scales as
for the age-dependent model. For the age-independent model, σss scales as 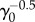 (see the electronic supplementary material, section C). In addition, the behaviours of σ(t) before reaching steady state are also quite different (figure 7d).
(see the electronic supplementary material, section C). In addition, the behaviours of σ(t) before reaching steady state are also quite different (figure 7d).
In spite of the fact that the average cell death rate depends on time, the age distribution in this case is the same as in absence of cell death. We have analysed age distributions for the case of growth of competitive population at two times. One (t = 252 arb. units) is chosen around the maximum of σ(t), and the other time (t = 450 arb. units) is within the steady-state regime. For these times, age distributions turned out to be the same (the results are not shown) and identical to steady-state age distribution in absence of cell death (figure 3). Since the cell death rate is age-independent, cells of different ages have the same probability to die, so the age distribution remains intact.
4. Discussion
In this paper, we consider the relationship between cell division time variability and the fluctuations in cell population using a stochastic modelling approach. We examine the simple case of symmetrical cell division, and explicitly consider the time-dependent probability of cell division. The average population in this model exactly coincides with the deterministic von Foerster equation for age-dependent cell populations. Indeed, our simulations confirm this, and accurately reproduce the growth curves of cell populations. As expected, the width of the division time distribution influences the overall cell colony growth rate, even when the average cell division time is identical. This is in contrast with models with constant cell division and death rates, which does not reproduce the correct population growth.
The main focus of our study, unlike the deterministic von Foerster model, is to delineate the underlying cause of cell population fluctuations. We find that the division time distribution, ω(a), has a strong influence on the population-level fluctuation, even for long times. For initially synchronized cells, we find a simple relation between the asymptotic values of relative fluctuations (σ/N)lim, and the width of the division time distributions cv. We also find that the initial age distribution affects the long time population fluctuation. Asynchronous population has a larger population fluctuation. We also incorporate cell death in the model and compute the population fluctuation as a function of an age-independent cell death rate, kd. Results show that cell death generally increases cell population fluctuation, but the scaling with respect to kd does not follow equation (2.1). Finally, since in most cell colonies both cell division and death are affected by the overall population, we study a simple model of a competitive growth where the death rate increases with the overall population. Once again, the age-dependent model predicts a different fluctuation behaviour. We expect that in general, age-dependent cell growth models will have qualitatively different results than the constant birth–death models. This fundamental result suggests a revision of the usual theoretical approach to model growth and evolution of cell populations.
To predict time evolution of cell populations in biologically realistic situations, it is important to obtain good knowledge of the cell division and death rates as functions of cell age. In our studies, we have incorporated realistic cell division rates but assumed that the death rate is a constant. Note that our approach is Markovian, i.e. there is no memory between successive division events. This assumption is justified by our data and others [1] which showed a lack of correlation between successive divisions. However, cell division and death rates are regulated by other factors such as population density and nutrient levels. Cells may also transform into a different state where cell division is inhibited. These factors suggest that rich and interesting population dynamics exist, even in the simplest colonies. For the living tissue, it is known that several cell types must coexist, and transformations from stem or progenitor cells will influence the final population. Our work here suggests that an age-dependent approach may be appropriate to understand those situations as well.
Equation (3.4) suggests that the distributions of cell colony sizes broaden with increasing spread of the generation time distribution. This result is for exponentially growing colonies, but similar effects may be present for homeostatic populations. During organ growth, for example, the final size of the organ and the variation in organ size may be related to cell population fluctuations. This fluctuation may be influenced by the age-dependence of cell events. Age-dependent dynamics are also expected to be important in colonies seeded by a few cells where population fluctuations may be significant, e.g. tumour growth, drug-resistant cell populations. Finally, it is important to note that by measuring not just the average population, but also the population fluctuation, important information can be obtained about the underlying growth and control mechanism. We have seen that the population fluctuation can be quite different, even when the average population growth rate is the same. Indeed, the full theory must consider all moments of the population distribution. Therefore, statistical fluctuations in population dynamics contain vital information about the biological mechanisms of growth and evolution.
Acknowledgements
This work was supported by NIH 1U54CA143868-01, 1R01GM075305 and NSF PHY-1205795.
Glossary
| t (arb. units) | time |
| a (arb. units) | age of a cell |
| ω(a) (arb. units)–1 | PDF (probability density function) of distribution of generation times |
| α (dimensionless value) | shape parameter for ω(a), equation (2.17) |
| β (arb. units) | scale parameter for ω(a), equation (2.17) |
| a0 (arb. units) | shift parameter for ω(a), equation (2.17) |
| k(a) (arb. units)–1 | probability of mitosis per unit time |
| kb (arb. units)–1 | constant cell division rate |
| τ (arb. units) | mean generation time |
| cv | coefficient of variation of ω(a) |
| kd (arb. units)–1 | cell death rate |
| n(a, t) (arb. units)–1 | cell population density |
| nh(a, t) (arb. units)–1 | cell population density from a particular stochastic simulation |
| N0 | initial number of cells |
| N(t) | average number of cells at time t |
| Nlost/Nalive | ratio of numbers of lost to live cells |
| τ* (arb. units) | average doubling time of cell population |
| v (arb. units)–1 | average growth rate of cell population |
| g(a) (arb. units)–1 | PDF of age distribution at steady state |
| σ(t) | standard deviation of the mean N(t) |
| (σ/N)lim | asymptotic value of σ(t)/N(t) at long t |
| γ0 (arb. units)–1 | cell death rate for competitive population |
| Nss | steady-state size of competitive population |
| σss | standard deviation of N(t) at steady state |
| p(n;t) (dimensionless value) | PDF of population size distribution at time t |
| p(tn) (arb. units)–1 | PDF of distribution of times tn to reach n cells |
References
- 1.Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, Jun S. 2010. Robust growth of Escherichia coli. Curr. Biol. 20, 1099–1103 10.1016/j.cub.2010.04.045 (doi:10.1016/j.cub.2010.04.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. 2009. Cell growth and size homeostasis in proliferating animal cells. Science 325, 167–171 10.1126/science.1174294 (doi:10.1126/science.1174294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halter M, Elliott JT, Hubbard JB, Tona A, Plant AL. 2009. Cell colume distributions reveal cell growth and division times. J. Theor. Biol. 257, 124–130 10.1016/j.jtbi.2008.10.031 (doi:10.1016/j.jtbi.2008.10.031) [DOI] [PubMed] [Google Scholar]
- 4.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. 2010. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 10.1126/science.1192588 (doi:10.1126/science.1192588) [DOI] [PubMed] [Google Scholar]
- 5.Sun Z, Komarova NL. 2012. Stochastic modeling of stem-cell dynamics with control. Math. Biosci. 240, 231–240 10.1016/j.mbs.2012.08.004 (doi:10.1016/j.mbs.2012.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowak MA. 2006. Evolutionary dynamics: exploring the equations of life. Cambridge, MA: Belknap Press of Harvard University Press [Google Scholar]
- 7.Horowitz J, Normand MD, Corradini MG, Peleg M. 2010. Probabilistic model of microbial cell growth, division, and mortality. Appl. Environ. Microbiol. 76, 230–242 10.1128/AEM.01527-09 (doi:10.1128/AEM.01527-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pin C, Baranyi J. 2006. Kinetics of single cells: observation and modeling of a stochastic process. Appl. Environ. Microbiol. 72, 2163–2169 10.1128/AEM.72.3.2163-2169.2006 (doi:10.1128/AEM.72.3.2163-2169.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutalik Z, Razaz M, Baranyi J. 2005. Connection between stochastic and deterministic modeling of microbial growth. J. Theor. Biol. 232, 285–299 10.1016/j.jtbi.2004.08.013 (doi:10.1016/j.jtbi.2004.08.013) [DOI] [PubMed] [Google Scholar]
- 10.Lu T, Volfson D, Tsimring L, Hasty J. 2004. Cellular growth and division in the Gillespie algorithm. Syst. Biol. 1, 121–128 10.1049/sb:20045016 (doi:10.1049/sb:20045016) [DOI] [PubMed] [Google Scholar]
- 11.Smith JA, Martin L. 1973. Do cells cycle? Proc. Natl Acad. Sci. USA 70, 1263–1267 10.1073/pnas.70.4.1263 (doi:10.1073/pnas.70.4.1263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell EO. 1956. Growth rate and generation time of bacteria, with special reference to continuous culture. J. Gen. Microbiol. 15, 492–511 10.1099/00221287-15-3-492 (doi:10.1099/00221287-15-3-492) [DOI] [PubMed] [Google Scholar]
- 13.Rubinow SI. 1968. A maturity-time representation for cell populations. Biophys. J. 8, 1055–1073 10.1016/S0006-3495(68)86539-7 (doi:10.1016/S0006-3495(68)86539-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bremer H. 1982. Variation of generation times in Escherichia coli populations: its cause and implications. J. Gen. Microbiol. 128, 2865–2876 10.1099/00221287-128-12-2865 (doi:10.1099/00221287-128-12-2865) [DOI] [PubMed] [Google Scholar]
- 15.Bremer H, Chuang L. 1981. The cell cycle in Escherichia coli B/r. J. Theor. Biol. 88, 47–81 10.1016/0022-5193(81)90328-3 (doi:10.1016/0022-5193(81)90328-3) [DOI] [PubMed] [Google Scholar]
- 16.Engelberg J. 1964. The decay of synchronization of cell division. Exp. Cell Res. 36, 647–662 10.1016/0014-4827(64)90320-9 (doi:10.1016/0014-4827(64)90320-9) [DOI] [PubMed] [Google Scholar]
- 17.von Foerster H. 1959. Some remarks on changing populations. In The kinetics of cellular proliferation (ed. Stohlman F., Jr), pp. 382–407 New York, NY: Grune and Stratton [Google Scholar]
- 18.Trucco E. 1965. Mathematical models for cellular systems. The von Foerster equation. Part I. Bull. Math. Biohys. 27, 285–304 10.1007/BF02478406 (doi:10.1007/BF02478406) [DOI] [PubMed] [Google Scholar]
- 19.Trucco E. 1965. Mathematical models for cellular systems. The von Foerster equation. Part II. Bull. Math. Biohys. 27, 449–471 10.1007/BF02476849 (doi:10.1007/BF02476849) [DOI] [PubMed] [Google Scholar]
- 20.Kutalik Z, Razaz M, Elfwing A, Ballagi A, Baranyi J. 2005. Stochastic modeling of individual cell growth using flow chamber microscopy images. Int. J. Food Microbiol. 105, 177–190 10.1016/j.ijfoodmicro.2005.04.026 (doi:10.1016/j.ijfoodmicro.2005.04.026) [DOI] [PubMed] [Google Scholar]
- 21.Metris A, George SM, Peck MW, Baranyi J. 2003. Distribution of turbidity detection times produced by single cell-generated bacterial populations. J. Microbiol. Meth. 55, 821–827 10.1016/j.mimet.2003.08.006 (doi:10.1016/j.mimet.2003.08.006) [DOI] [PubMed] [Google Scholar]
- 22.Baranyi J, Pin C. 2001. Parallel study on bacterial growth and inactivation. J. Theor. Biol. 210, 327–336 10.1006/jtbi.2001.2312 (doi:10.1006/jtbi.2001.2312) [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Zhuang X. 2007. Kinetic analysis of sequential multistep reactions. J. Phys. Chem. B 111, 13 600–13 610 10.1021/jp073708+ (doi:10.1021/jp073708+) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floyd DL, Harrison SC, van Oijen AM. 2010. Analysis of kinetic intermediates in single-particle dwell-times. Biophys. J. 99, 360–366 10.1016/j.bpj.2010.04.049 (doi:10.1016/j.bpj.2010.04.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto H, Zeuthen E, Rasmussen L. 1973. Clonal growth of mouse cells (strain L). J. Cell Sci. 13, 879–888 [DOI] [PubMed] [Google Scholar]
- 26.Prescott DM. 1959. Variations in the individual generation times of Tetrahymena geleii HS. Exp. Cell Res. 16, 279–284 10.1016/0014-4827(59)90255-1 (doi:10.1016/0014-4827(59)90255-1) [DOI] [PubMed] [Google Scholar]
- 27.Shehata TE, Marr AG. 1970. Synchronous growth of enteric bacteria. J. Bacteriol. 103, 789–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kampen NG. 1981. Stochastic processes in physics and chemistry. Amsterdam, The Netherlands: Elsevier Science Ltd [Google Scholar]