Abstract
Contrary to other studies, here we describe cysteine (Cys) pseudoproline-containing peptides with short deprotection times in TFA. The deprotection times fell in the same range as other protecting groups commonly used in SPPS (e.g., 1–3 h). Moreover, when using Cys pseudoprolines as peptide macrocyclization-enhancing moieties a considerable reduction in reaction time was observed compared to a peptide containing trityl protected Cys.
Since the introduction of solid-phase peptide synthesis (SPPS) and its subsequent maturation through the continuous refinement of reagents, linkers, resins, and protocols, many complex peptides are readily accessible.1,2 SPPS now allows the preparation of complex peptides on a large scale, thus facilitating the global commercialization of complex peptide drugs, such as prialt and ziconotide.3,4 The prevalence of complex peptides is increasing, exemplified by the fact that several multiple disulfide-containing peptides are undergoing clinical trials.5 However, there are still many obstacles to overcome in the synthesis of complex peptides, such as addressing difficult sequences and orthogonal protecting groups, and solving low coupling efficiencies and solubility issues. In this regard, the development of novel tools, protocols, and techniques in peptide synthesis are required to tackle these difficulties. Developed by Mutter et al., pseudoproline dipeptide building blocks are a prime example of innovation in complex peptide synthesis.6
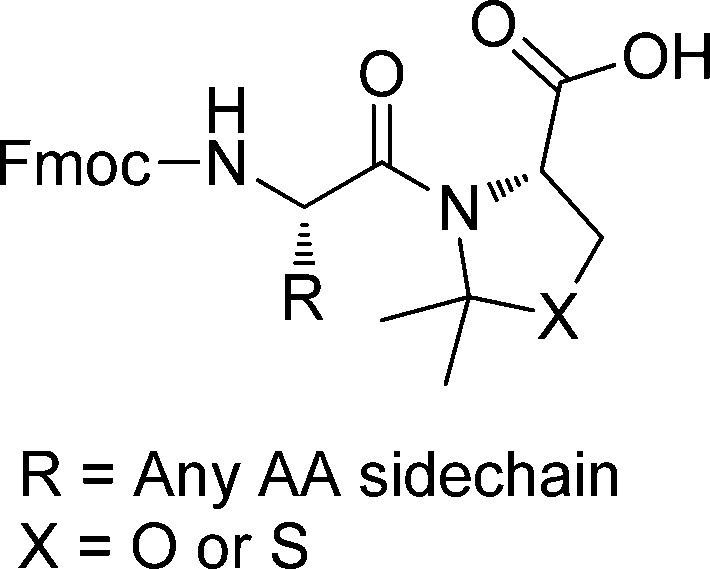
Pseudoproline dipeptides have become powerful tools for the synthesis of peptides containing difficult sequences.7 Mechanistically, pseudoprolines act by disrupting the secondary structure and increase the solubilization of protected peptides. The disruption of the secondary structure, such as β-sheet formation, is caused by the cisoid amide conformation of the oxazolidine- or thiazolidine-based pseudoproline rings, which introduce a “kink” in the backbone and disrupt backbone hydrogen bonding analogous to Pro (Figure 1).8 The cisoid pseudoproline conformation has also been utilized for reducing aspartimide formation during Lansbury aspartylation.9−11
Figure 1.
Structure of a pseudoproline (ψMe,Mepro) dipeptide.
Oxazolidine-based dipeptides derived from Ser or Thr are extensively used and commercially available as Fmoc-protected dipeptides. Typically, the oxazolidine ring can be deprotected to the parent amino acid within several hours using TFA based cleavage cocktails, as illustrated in the synthesis of human amylin.12
Conversely, thiazolidine based dipeptides have not gained widespread use because of their high stability to TFA. In the seminal paper by Mutter et al., deprotection times of 32 h were reported for the removal of Cys pseudoprolines in linear peptides to the corresponding deprotected Cys.6 This observation was confirmed in a recent publication, where treatment with TFA/TIS/H2O (95:2.5:2.5) for 36 h was required to remove Cys pseudoproline in a linear peptide.13 In addition, this publication claimed Cys pseudoproline removal in head-to-tail cyclic peptides takes 13 days in TFA/TIS/H2O (95:2.5:2.5) and requires the use of harsh acids, such as neat trifluoromethanesulfonic acid, to achieve quantitative deprotection in a reasonable time frame. Furthermore, our group has prepared head-to-tail cyclic peptides containing up to four Cys pseudoprolines.14 In this case, we observed high stability to TFA, thereby confirming the long deprotection times for thiazolidine-based pseudoprolines in head-to-tail cyclic peptides.
Pseudoprolines have been described as turn inducers.15 This has been substantially developed by Jollifffe et al., which focused mainly on Ser and Thr pseudoprolines as turn inducers.16−18 In one study Cys pseudoprolines were used as turn inducers.13 These studies noted a yield increase upon macrocyclization; however, none of the previous studies described the effect of pseudoprolines on macrocyclization kinetics.
To widen the applicability of Cys pseudoprolines, we initiated a study as part of our ongoing research into Cys protection. Herein, we report on the remarkable Cys pseudoproline lability in the same time scale as oxazolidine-based pseudoprolines in a series of peptides. Moreover, we identify a considerable increase in kinetics when using Cys pseudoprolines as macrocyclization-enhancing moieties, which was not observed in previous studies. In this regard, these moieties significantly reduced the coupling time required for complete peptide macrocyclization.
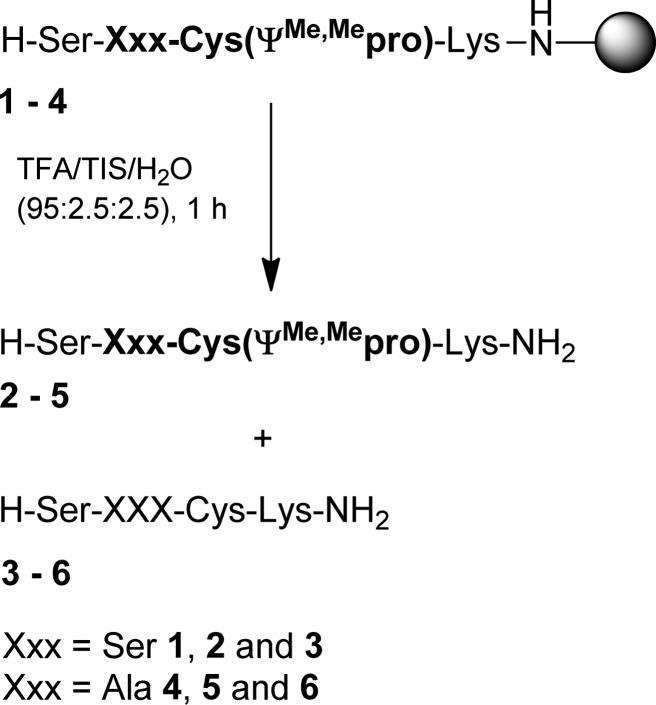
We first synthesized two random tetrapeptides using the commercial Cys pseudoproline building blocks Fmoc-Ser(tBu)-Cys(ψMe,Mepro)-OH and Fmoc-Ala-Cys(ψMe,Mepro)-OH (Scheme 1). With the exception of Cys pseudoproline dipeptides, standard protected Fmoc-amino acids were used for all residues in peptide synthesis. Model peptides 2 and 5 were prepared on a Rink Amide resin using standard SPPS protocols and were then cleaved from the resin using a 1 h treatment with TFA/TIS/H2O (95:2.5:2.5). Chromatographic and spectroscopic analysis of peptide 2 revealed an unusual lability of the Cys pseudoproline to acidolysis. Only 65% of the expected Cys pseudoproline was observed, and 35% of the peptide was fully deprotected to the parent Cys (3). Analysis of peptide 5 revealed a similar lability to acidolysis with 85% of the Cys pseudoproline remaining after 1 h of TFA treatment. Complete Cys pseudoproline removal in peptide 2 required treatment with TFA/TIS/H2O (95:2.5:2.5) for 4 h while peptide 5 required 6 h. These deprotection times are significantly shorter than the previously reported deprotection times of 32–36 h for Cys pseudoproline-containing linear peptides.
Scheme 1. Acidolysis of Peptides 1 and 2 (1 h in TFA/TIS/H2O (95:2.5:2.5)).
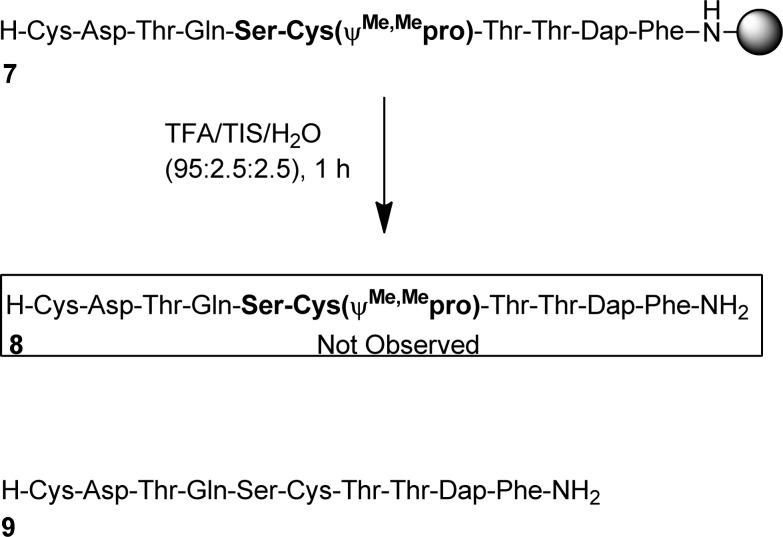
Peptide 7 which contained the Ser-Cys pseudoproline sequence was prepared using standard SPPS and subsequently cleaved from the resin (Scheme 2). Analysis of the cleaved peptide revealed complete Cys pseudoproline removal (9) after 1 h of TFA treatment.
Scheme 2. Acidolysis of Peptide 3 (1 h in TFA/TIS/H2O (95:2.5:2.5)).
Based on these results we prepared a small peptide library with different sequences (Figure 2). For design considerations in the library we used a series of nonapeptides, one octapeptide, and one decapeptide in which we focused on diversifying the N-terminal residues of the pseudoproline dipeptide building bock, i.e., Xxx(PG)-Cys(ΨMe,Mepro).
Figure 2.
Cys pseudoproline containing peptides.
The peptides were cleaved from the resin, and 2 h or less were required for complete removal of the Cys pseudoprolines at room temperature, except for peptides 23 and 26 which required 45 °C for complete removal (Table 1). Peptide 23 showed near-quantitative deprotecton whereas peptide 26 was deprotected 57% in 3 h at rt and both peptides required 2–3 h in TFA at 45 °C for complete removal. These results contrast strongly with Cys pseudoproline deprotection times of 32–36 h, using the same cleavage cocktail, on different model peptides.6,13
Table 1. Deprotection Times of Cys Pseudoproline Peptides.
| peptide | Ψ pro | temp (°C) | deprotection time (h) | deprotection % |
|---|---|---|---|---|
| 11 | Ser-Cys | 25 | 1 | 98 |
| 14 | Ala-Cys | 25 | 2 | 94 |
| 17 | Asp-Cys | 25 | 2 | 95 |
| 20 | Asp-Cys | 25 | 2 | 91 |
| 23 | Lys-Cys | 45 | 2 | 93 |
| 26 | Lys-Cys | 45 | 3 | 91 |
Our results strongly indicate that Cys pseudoproline removal is sequence-dependent. The factors governing the Cys pseudoproline deprotection times are not well understood. This will be the focus of our ongoing research efforts, which will be reported in due course. In addition, we noted that the deprotection times fell in the same range as those for oxazolidine pseudoprolines, thereby making Cys pseudoprolines more applicable as they do not require harsh acids for deprotection.
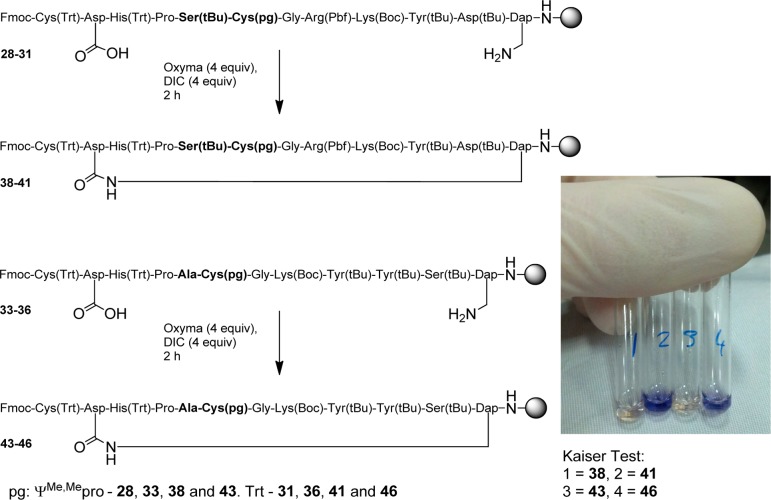
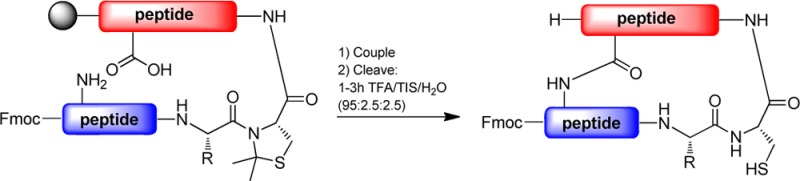
After observing that Cys pseudoprolines were removed using short TFA treatments, we addressed the capacity of thiazolidine rings to enhance peptide macrocyclization. This experiment was based on the cisoid amide conformation of the thiazolidine ring, through which the “kink” in the backbone would bring the two reacting groups closer and consequently speed up the macrocyclization. We chose two α-conotoxins (CnIB and A1.4) from the ConoServer database that contained the Ser-Cys or Ala-Cys sequences, which were used as suitable model systems for Cys pseudoproline macrocyclization studies.19 In these peptides two of the four Cys residues were replaced with allyl protected Asp and a diaminopropionic acid (Dap) residue. The experiment required four peptides, two conotoxin derivatives with either a Ser-Cys or an Ala-Cys pseudoproline, and two peptides with standard protected amino acids. The allyl protecting groups were removed using palladium catalysis, and the on-resin macrocyclization was performed using diisopropylcarbodiimide (DIC) and Oxyma Pure as the coupling system.20 After 2 h the coupling was stopped. A negative Kaiser test showed the disappearance of amine in the pseudoproline containing peptides, which indicated completion of macrocyclization (Scheme 3, vials 1 and 3).21 This observation contrasts with the incomplete macrocyclization for the peptides that did not contain Cys pseudoprolines as indicated by the presence of an amine in a positive Kaiser test (Scheme 3, vials 2 and 4). Analysis of the peptides after cleavage from the resin determined complete macrocyclization of those containing Cys pseudoprolines (38 and 43), while peptides 41 and 46, which did not contain Cys pseudoprolines, showed 52% and 70% macrocyclization, respectively. These results show that the thiazolidine ring in Cys pseudoprolines enhance macrocyclization by significantly decreasing the coupling time.
Scheme 3. Macrocyclization Experiments of Peptides with and without Pseudoprolines.
Thiazolidine based Cys pseudoproline dipeptides have previously been described to be highly stable to TFA and require harsh acids for removal. As a result, these protecting groups are not widely used. We observed markedly different behavior to TFA, with deprotection times of 1 h for peptides 7 and 11 and several hours for the other examples. Our findings show that Cys pseudoproline removal can be equally efficient as that of the widely used oxazolidine pseudoprolines. Cys pseudoprolines were shown to enhance the on-resin macrocyclization efficiency of two conotoxin derivatives by significantly decreasing the coupling time required for completion of the reaction. Thus, we envisage that the combined use of Cys pseudoprolines and diphenylmethyl (Dmp) protection will render a pair of Cys protecting groups compatible with trimethoxyphenylthio (S-Tmp), trityl (Trt), or phenylacetamido (Phacm) protecting groups, thus amplifying the repertoire of Cys protection for the synthesis of complex peptides. In conclusion, the short deprotection times and enhanced macrocyclization efficiency of Cys pseudoprolines significantly increase their applicability as Cys protecting groups.
Acknowledgments
We thank Dr. Thomas Bruckdorfer and Mrs. Annick Vancolen from Iris Biotech GmbH for providing us the Cys pseudoproline and for useful discussions. This work was partially supported by the Marie Curie ITN MEMTIDE (FP7-PEOPLE-ITN08), CICYT (CTQ2012-30930), and the Generalitat de Catalunya (2009SGR 1024).
Supporting Information Available
Detailed experimental procedures, characterization, spectroscopic and chromatographic data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Merrifield R. B. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar]
- Amblard M.; Fehrentz J.-A.; Martinez J.; Subra G. Mol. Biotechnol. 2006, 33, 239–254. [DOI] [PubMed] [Google Scholar]
- Prommer E. Drugs Today 2006, 42, 369–378. [DOI] [PubMed] [Google Scholar]
- Dolgin E. Nat. Med. 2012, 18, 1308–1309. [DOI] [PubMed] [Google Scholar]
- Essack M.; Bajic V. B.; Archer J. A. C. Mar. Drugs 2012, 10, 1244–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhr T.; Wahl F.; Nefzi A.; Rohwedder B.; Sato T.; Sun X.; Mutter M. J. Am. Chem. Soc. 1996, 118, 9218–9227. [Google Scholar]
- White C. J.; Yudin A. K. Nat. Chem. 2011, 3, 509–524. [DOI] [PubMed] [Google Scholar]
- Dumy P.; Keller M.; Ryan D. E.; Rohwedder B.; Wöhr T.; Mutter M. J. Am. Chem. Soc. 1997, 119, 918–925. [Google Scholar]
- Anisfeld S. T.; Lansbury P. T. J. Org. Chem. 1990, 55, 5560–5562. [Google Scholar]
- Ullmann V.; Rädisch M.; Boos I.; Freund J.; Pöhner C.; Schwarzinger S.; Unverzagt C. Angew. Chem., Int. Ed. 2012, 51, 11566–11570. [DOI] [PubMed] [Google Scholar]
- Wang P.; Aussedat B.; Vohra Y.; Danishefsky S. J. Angew. Chem., Int. Ed. 2012, 51, 11571–11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. R.; Kowalczyk R.; Hay D.; Brimble M. Int. J. Pept. Res. Ther. 2013, 19, 147–155. [Google Scholar]
- Wong M. S. Y.; Jolliffe K. A. Aust. J. Chem. 2010, 63, 797–801. [Google Scholar]
- Pelay M.; Melli A.; Tulla-Puche J.; Albericio F. J. Med. Chem. 2013, 56, 9780–9788. [DOI] [PubMed] [Google Scholar]
- Sager C.; Mutter M.; Dumy P. Tetrahedron Lett. 1999, 40, 7987–7991. [Google Scholar]
- Skropeta D.; Jolliffe K. A.; Turner P. J. Org. Chem. 2004, 69, 8804–8809. [DOI] [PubMed] [Google Scholar]
- Fairweather K. A.; Sayyadi N.; Luck I. J.; Clegg J. K.; Jolliffe K. A. Org. Lett. 2010, 12, 3136–3139. [DOI] [PubMed] [Google Scholar]
- Davis M. R.; Singh E. K.; Wahyudi H.; Alexander L. D.; Kunicki J. B.; Nazarova L. A.; Fairweather K. A.; Giltrap A. M.; Jolliffe K. A.; McAlpine S. R. Tetrahedron 2012, 68, 1029–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas Q.; Yu R.; Jin A.-H.; Dutertre S.; Craik D. J. Nucleic Acids Res. 2012, 40, D325–D330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subirós-Funosas R.; Prohens R.; Barbas R.; El-Faham A.; Albericio F. Chem.—Eur. J. 2009, 15, 9394–9403. [DOI] [PubMed] [Google Scholar]
- Kaiser E.; Colescott R. L.; Bossinger C. D.; Cook P. I. Anal. Biochem. 1970, 34, 595–598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.