Abstract
Purpose: Effective dose (ED) is a widely used metric for comparing ionizing radiation burden between different imaging modalities, scanners, and scan protocols. In computed tomography (CT), ED can be estimated by performing scans on an anthropomorphic phantom in which metal-oxide-semiconductor field-effect transistor (MOSFET) solid-state dosimeters have been placed to enable organ dose measurements. Here a statistical framework is established to determine the sample size (number of scans) needed for estimating ED to a desired precision and confidence, for a particular scanner and scan protocol, subject to practical limitations.
Methods: The statistical scheme involves solving equations which minimize the sample size required for estimating ED to desired precision and confidence. It is subject to a constrained variation of the estimated ED and solved using the Lagrange multiplier method. The scheme incorporates measurement variation introduced both by MOSFET calibration, and by variation in MOSFET readings between repeated CT scans. Sample size requirements are illustrated on cardiac, chest, and abdomen–pelvis CT scans performed on a 320-row scanner and chest CT performed on a 16-row scanner.
Results: Sample sizes for estimating ED vary considerably between scanners and protocols. Sample size increases as the required precision or confidence is higher and also as the anticipated ED is lower. For example, for a helical chest protocol, for 95% confidence and 5% precision for the ED, 30 measurements are required on the 320-row scanner and 11 on the 16-row scanner when the anticipated ED is 4 mSv; these sample sizes are 5 and 2, respectively, when the anticipated ED is 10 mSv.
Conclusions: Applying the suggested scheme, it was found that even at modest sample sizes, it is feasible to estimate ED with high precision and a high degree of confidence. As CT technology develops enabling ED to be lowered, more MOSFET measurements are needed to estimate ED with the same precision and confidence.
Keywords: effective dose, MOSFET, computer tomography, dosimetry, sample size
INTRODUCTION
Computed tomography (CT) has advanced substantially in recent years, introducing scanners with more detector-rows, novel scan modes, new image reconstruction methods, and numerous other technological advances.1 This has resulted in a CT market characterized by dramatic variations in technology and consequently substantive differences in the physics of radiation exposure, underscoring a need for individualized scanner and protocol dosimetry to better quantify radiation burden. Effective dose (ED) is an important radiation protection quantity2 which reflects relative biological risk. ED is presently defined in accordance with a formulation deriving from International Commission on Radiological Protection (ICRP) Publication 103,2 as the sum over all specified organs of doubly weighted organ absorbed doses, where weights reflect both the relative radiosensitivity of each organ, and the type of radiation. Its special unit, shared with equivalent dose and weighted equivalent dose, is the Sievert, equal to 1 J/kg. While ED has been criticized as a dosimetry parameter for having a high degree of uncertainty and generality3, 4 its great virtue is that it enables comparison of radiation burden not only between different scanners and protocols, but even between different imaging modalities. This has led to its great popularity in the clinical literature and clinical practice. Given ED's widespread use, it is desirable to minimize error in its assessment.
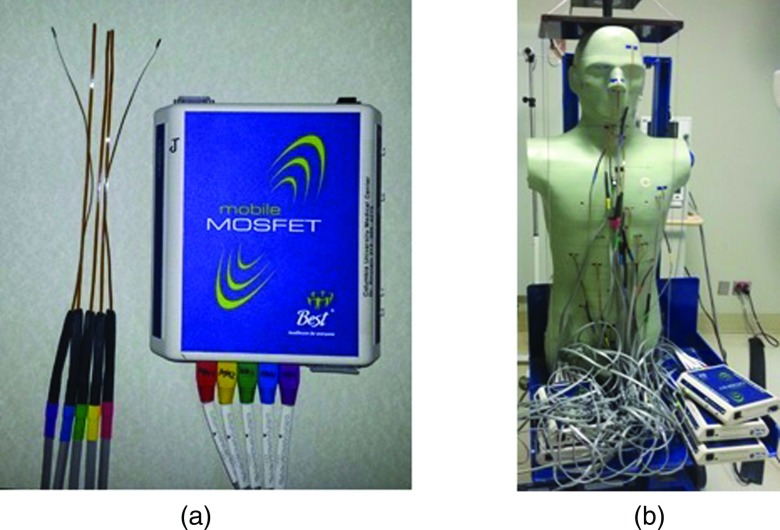
There exists a wide variety of CT scanner models, on each given scanner a number of protocols can be performed, and for each protocol many parameters can be adjusted. Estimation of ED for a given scanner, protocol, and set of parameters is based on organ dosimetry. One popular approach to estimating organ doses is the use of solid-state metal-oxide-semiconductor field-effect transistor (MOSFET) dosimeters placed in an anthropomorphic phantom5, 6, 7, 8, 9, 10, 11, 12 [Figs. 1a, 1b]. MOSFETs are eminently practical for dosimetry, being small in size, simple to use, and providing immediate dose measurements that can be repeated without removal or annealing. The use of MOSFETs has been reported to be a valid method for organ dose assessment in CT scanning.10, 12 Anthropomorphic phantoms simulate human male and female reference individuals, both physically and radiographically. Such phantoms are sectional with drilled holes, located at spots that are optimized for dosimetry of internal organs.
Figure 1.
(a) Five MOSFETs attached to a reader and (b) anthropomorphic phantom and set up of MOSFETs.
MOSFET detectors are placed within these holes. The estimation of ED using an anthropomorphic phantom and MOSFETs involves measurements of organ doses in each of the spots in which MOSFETs are placed. The estimated ED is affected by uncertainties inherent to these measurements. This includes the variation of the voltage readings of the various MOSFETs representing each organ. Examples of sources for the variation are the CT scanner x-ray tube start angle,13 electronic noise, and between-scan MOSFET variability. Another uncertainty is due to variation of the calibration factor (CF) which translates voltage (in units of mV) into dose (in units of mGy), affecting the dose reading and consequently the ED. Although the sole current manufacturer of medical high-sensitivity MOSFETs supplies a general CF, it is still recommended to calibrate the MOSFETs, hence to integrate variation that it introduces. Various calibration methods have been described in the literature such as those of Yoshizumi et al.12 and Brady et al.14 In this work, we address the impact of the CF variation on the estimation of ED, regardless of the calibration method. In particular, we show that the calibration sample size (calibration procedure repetitions) influences the sample size for ED.
Statistically, the more measurements performed for a given scanner and protocol, the more precise the organ doses and ED that are obtained, i.e., the better the estimate of the “true” value of the ED. This is the case as well for calibration scans performed to determine the CF. Practical considerations, however, often limit the number of measurements that can be performed for a given scan protocol. These limitations can include the cost of CT scanner time, the limited availability of the scanner for phantom scans due to clinical needs, and the tendency of many scanners to overheat when operated continuously for repetitive scans. Often, it is desired to estimate and compare dosimetry between numerous scan protocols using the same scanner,5 thus limiting the number of measurements that can be performed for a single protocol. Moreover, a MOSFET dosimeter has limited life span. Specifically, a large accumulated voltage reduces the linearity of the MOSFET detector's response, and hence the credibility of its measurements, whereas replacement of MOSFETs requires more scanner time and costs, as well as disassembly of the anthropomorphic phantom and repositioning, which can decrease the reproducibility of measurements. Accordingly, optimal determination of sample size, i.e., the number of repeated scans, for MOSFET scanning experiments is both critical to ensure reliable dosimetry and an important practical consideration.
Thus, this work is motivated by the need to design experiments for estimating ED using anthropomorphic phantoms and MOSFET dosimeters, to predefined precision, referring to factors that can be monitored and that affect the estimation of ED, yet minimizing the measurements’ number and scan time. To the best of our knowledge, this paper represents the first work aiming to provide a rigorous statistical framework for the design of such dosimetry experiments. Our objective here is to devise and illustrate a scheme to statistically determine the sample size required for estimating ED to desired degrees of precision and confidence, subject to practical limitations.
METHODOLOGY
Anthropomorphic phantom for estimating the effective dose
ED is estimated by measuring organ doses in an anthropomorphic phantom which physically and radiographically simulates a male or female human, using MOSFETs as shown in Fig. 1b. The phantom is made of tissue-equivalent polymers and resins that simulate soft tissue, lung, brain, bone, spinal cord, and spinal disks. It is sectional, consisting of a series of 25-mm-thick contiguous sections. In each section, holes are located at positions (spots) optimized for dosimetry. These holes are 5 mm in diameter, and are drilled in the craniocaudal direction; in total, the holes are located at positions optimized for dosimetry in internal organs. When MOSFET detectors are fixed in these holes, they are placed in tissue-equivalent holders. Our phantom has been modified from a commercially available anthropomorphic phantom (ATOM 701; CIRS, Norfolk, VA), with additional holes drilled to measure absorbed dose for each organ with a tissue weighting factor in the definition of ED in ICRP Publication 103, excluding skin. Modular breast phantoms are attached for the female phantom. These breast phantoms are medium-sized, constructed based on CT data from an actual patient imaged in the supine position.
MOSFET dosimeters and placements
We use a mobile MOSFET dose verification system (TN-RD-70W; Best Medical, Ottawa, Canada), with multiple readers, coupled with high-sensitivity MOSFETs (TN-1002RD-H; Best Medical, Ottawa, Canada). Each reader can be used to read up to five MOSFETs, as displayed in Fig. 1a. Two Bluetooth wireless devices are used to communicate voltage data from the MOSFET readers to two PC laptops with MOSFET software installed.
MOSFETs are positioned in spots corresponding to organs contributing to the computation of ED, as specified in ICRP Publication 103.2 In this ICRP report, tissue weighting factors are assigned to 28 different organs, including the group of “remainder” organs which have tissue weighting factors of less than 0.01. For larger organs and those with high tissue weighting factors, such as the lungs and female breast, it is desirable to place MOSFETs in multiple spots and use an average so as to better estimate the organ absorbed dose. Our group's approach in the estimation of ED is to employ MOSFETs in a total of 43 and 46 spots in the male and female phantoms, respectively, to cover these 28 organs. Different approaches can be used to obtain dose estimates for spots.
A common hardware configuration available to MOSFET users is four readers, each capable of reading 5 MOSFETS, for a total of 20 spots read for a single scan. For this configuration, three distinct MOSFET placements are required to cover 43 or 46 spots. These placements can most simply be performed by dividing the anthropomorphic phantom into three anatomic regions, viz. cranial (region 1), thoracic (region 2), and caudal (region 3), each of which receives up to 20 MOSFET placements when scanned. In this approach, MOSFETs are placed in one region, scans are performed and measurements obtained, and then the phantom is disassembled and MOSFETs repositioned prior to performing the identical scan protocol; this is repeated so that MOSFETs are placed in each region and scans performed in a consecutive manner. An advantage of this approach is that it permits different numbers of scans to the three different regions, which receive markedly different radiation exposure related to their being primarily in-field or out-of-field in terms of a scan's radiation. This offers the potential to optimize accumulated MOSFET voltage, and thus maximize MOSFET lifespans.
Nevertheless, it may be advantageous, when many MOSFET readers and MOSFETs are available, to simultaneously place MOSFETs in all spots used in the estimation of ED. We typically use this latter approach in our laboratory, with up to ten MOSFET readers simultaneously. This single region approach has the great advantage of obviating the need for phantom disassembly and MOSFET repositioning in the midst of a scan protocol, which can introduce greater variability in measurements at a given spot and inaccuracies in organ dose estimation. In this paper, we consider sample size determination for ED under both of these scenarios, i.e., for both three-region MOSFET placement and for single-region placement.
Statistical problem setting
ED is defined in ICRP Publication 103 (Ref. 2) as a weighted sum of equivalent doses HT:
| (1) |
where wT is the tissue weighting factor for tissue or organ T (), DT,R is an average absorbed dose, and wR is the radiation weighting factor for radiation R. For x-ray radiation, only a single radiation weighting factor is used which is equal to unity, so this simplifies to
This sum is performed over all organs and tissues of the human body that are considered as sensitive to the induction of stochastic effects. ED is determined as a single averaged value for both sexes, computed from the absorbed doses assessed for organ or tissue T of the Reference Male and Reference Female, respectively,
| (2) |
Using a physical phantom, the absorbed dose is calculated by translating the MOSFET voltage readings, in mV, into absorbed dose, in mGy, using a calibration factor (CF), β, in units of mGy/mV. We let μT and νT be the mean voltage readings for each organ T in male and in female, respectively, thus the ED can be written as
| (3) |
We particularize the organ-based definition of ED into a problem setting that refers to the specific MOSFET locations (spots), as spread in the phantom organs, as well as the phantom three-region partition as described above. Let i denote a region number in the anthropomorphic phantom (i = 1, 2, or 3 for three-region placement), j denote the spot number of a given MOSFET in a region, and wi, j be the corresponding portion of a tissue weighting factor assigned to that spot. The transition from tissue weighting factor wT, to tissue weighting factor per spot, wi, j, is according to the number of MOSFETs that are used to measure each organ within a specific region and is displayed in Table 1.
Table 1.
List of organs as defined in ICRP 103 (Ref. 2) with tissue weighting factors translated into tissue weighting factors for MOSFET spots associated with the organ. The three region locations i = 1, 2, and 3 of the spots within the phantom correspond, respectively, to MOSFET placements cranial-to-thorax, thorax, and caudal-to-thorax.
| Organ weight wT | Region of | Number of MOSFET | Tissue weighting | |
|---|---|---|---|---|
| Organ1 | [ICRP 103 (Ref. 2)] | organ i | spots per organ | factor per spot wi, j |
| Brain | 0.01 | 1 | 6 | 0.0016 |
| Salivary glands | 0.01 | 1 | 1 | 0.01 |
| Bone marrow | 0.12 | 1,2,3 | 7 | According to Eckerman et al. (Ref. 15), see text |
| Bone surface | 0.01 | 1,2,3 | 7 | |
| Thyroid | 0.04 | 1 | 1 | 0.04 |
| Lung2 | 0.12 | 2 | 5 | 0.0045, 0.0343, 0.0310, 0.0310, 0.0191 |
| Esophagus | 0.04 | 2 | 3 | 0.013 |
| Stomach | 0.12 | 3 | 1 | 0.12 |
| Liver | 0.04 | 3 | 2 | 0.02 |
| Breast | 0.12 | 2 | Female: 4 | 0.03 |
| Male: 1 | 0.12 | |||
| Colon | 0.12 | 3 | 3 | 0.04 |
| Bladder | 0.04 | 3 | 1 | 0.04 |
| Gonads | 0.08 | 3 | 1 | 0.08 |
| Remainder3 | 0.12 | 1,2,3 | 11 | 0.0109 |
Organ list according to ICRP 103, excluding skin.
Lung weights are according to the relative volumes surrounding each spot.
Remainder organs include all organs as listed in ICRP 103 (Ref. 2), except for lymph nodes and muscles.
If an organ is represented by a single spot, and vice versa, then wi, j for that spot would be identical to the organ's tissue weighting factor in ICRP Publication 103,2 e.g., for thyroid, in Table 1. If an organ is represented by a few spots, the weighting factor is set according to the computation defined for that organ. For example, if an organ is represented by three spots for which readings are averaged equally, the weight for each spot would be a third of the ICRP-defined tissue weighting factor for that organ, as in esophagus, in Table 1.
For the lungs, we use a weighted average which was calculated using weights determined by the percentage of the organ's volume nearest to each MOSFET. For bone and bone marrow, the percentage in different locations was estimated using mass weighting factors for 13 skeletal regions (e.g. ribs, ossa coxae, etc.) as specified by Eckerman et al.15 MOSFETs were placed in the phantom in bone corresponding to each of the 13 regions, except for mandible, where a MOSFET is placed in the adjacent oral mucosa, and cranium, where it is placed in adjacent brain tissue; together these constitute only a small proportion of both bone and marrow. The MOSFET readings from these placements were then weighted according to these weighting factors for bone surface as well as for bone marrow, and summed to obtain composite bone and bone marrow doses, which were used in calculating the ED.
Let si denote the total number of spots in a region, excluding spots corresponding to the breast, and let denote the number of spots in the female breast in the region containing the breast. Thus, the total number of spots in which MOSFETs are read is s1+s2+s3+1 for the male phantom, with one breast spot, and s1+s2+s3+ for the female phantom. For the male phantom in the region containing the breast, the single spot j = s2 + 1 corresponds to the breast, whereas for the female phantom in the region containing the breast, spots j = s2 + 1, …, j = s2 + correspond to the breast. Let μi,j and νi,j be the mean voltage over all measurements taken at the jth spot in the ith region of a male anthropomorphic phantom and a female phantom, respectively. The ED for a CT scan protocol is then determined by
| (4) |
where represents the tissue weighting factor for the breasts. Note, that while β, the CF, is used to translate voltage readings (in units of mV) into dose readings (in units of mGy or mSv), we can analogously define e as an uncalibrated ED with units of mV.
Suppose we are going to take ni repeated MOSFET readings at each spot in the ith region, i = 1, 2, 3 for a three-region placement, of which half will be taken from a male phantom and the other half from a female phantom. Let and be the average MOSFET readings, in mV, at the jth spot in the ith region, for the male and female phantoms, respectively. We assume that has mean μi, j and variance σi, j2/(ni/2), and has mean νi, j and variance τi, j2/(ni/2). For the breast, we assume that for male has mean and variance , and for female, … have mean …, and variance …, . Then, the ED is estimated by
| (5) |
where is estimated based on a separate calibration data set that consists of m independent calibration processes, and is assumed to have mean β and variance . The estimate is unbiased; since and are independent:
Note that in this paper, the term “sample size” alone is used to refer to the sample size for MOSFET experiments, i.e., n1, n2, and n3, or n, whereas when we refer to the sample size for calibration experiments, we specifically use the term “calibration sample size.”
The variance of the ED is computed as
| (6) |
where
| (7) |
The values of can be estimated from pilot data as described in Sec. 2F.
Now suppose we wish to ensure that our estimate of the ED, determined using MOSFET measurements, is within a given proportion q of its true value E, with a desired degree of confidence 1 − α, by convention termed a confidence level. For example, say we wish for our estimate to be within q = 20% of the “true” ED with 1 − α = 95% confidence, i.e., α = 0.05. Assuming normality for this implies that were we to repeat our dosimetry measurements 100 times, we would expect the true E to be no more than q = 20%, less than or 20% greater than the estimated value , in at least 95 instances. Let z1 − α/2 denote the (1 − α/2)th percentile of the standard normal distribution, and let ρ = q/z1 − α/2. Then our goal is to minimize the total sample size N = n1 + n2 + n3 subject to
| (8) |
where ρ is an upper bound of the coefficient of variation (CV) for . For example, for a choice of precision of q = 20% and a 95% confidence level, ρ = q/z1 − α/2 = 0.2/z1 − 0.05/2 = 0.2/1.96 ≈ 0.1. Inequality 8 is equivalent to
| (9) |
We set
| (10) |
where r is the CV for . This yields the sample size for estimating the CF:
| (11) |
Plugging Eq. 10 into Eq. 9, we get
| (12) |
Note that in order for Eq. 12 to have a solution, the right hand side of Eq. 12 must be positive, or equivalently, r must be chosen such that r < ρ.
As an illustration of Eq. 11, choosing β = 0.33, σβ = 0.05β = 0.017 (based on historical or manufacturer's data), and r = 0.02 (r < ρ = 0.025), then we have m = 6.3. This means that we need at least 6.3 measurements for the calibration process, which we should round up to the next highest integer 7. Deriving as an average of m determinations of the CF, in turn, guarantees a desired confidence level of ED in view of Eq. 8.
Sample size calculation
We now derive the smallest sample sizes for MOSFET measurements that satisfy Eq. 12. Let x = 1/n1, y = 1/n2, z = 1/n3. We minimize the total sample size N(x, y, z) = n1 + n2 + n3 = 1/x + 1/y + 1/z subject to constraint ax + by + cz = t for a fixed t, where [from Eq. 12]
| (13) |
Let . By the Lagrange multiplier method, the minimizer of N(x, y, z) satisfies the following system of equations:
| (14) |
which yields
| (15) |
The minimal sample size is achieved when
| (16) |
noting t ⩽ t0.
When we use a single-region approach, with all MOSFETs placed simultaneously, we assume that n1 = n2 = n3 = n, and the above inequality 12 becomes
| (17) |
which yields the minimal sample size as
| (18) |
In summary, the determination of sample size requirements for estimating ED is dependent upon three important user-selectable parameters, namely, the degree of confidence desired (1 − α), the precision of the estimated ED (q), and the CV for the calibration factor (r). In addition, the in the expression of a, b, and c are true (thus unknown) parameters hence are not available and must be replaced by based on some historical data.
Determining sample size for protocols
Sample size determinations were performed, in accordance with the scheme described, for protocols of two different CT scanners: a 320 detector-row scanner (Aquilion ONE, Toshiba Medical Systems) and a 16 detector-row scanner (Precedence 16P, Philips Healthcare). For the 320-row scanner, the sample size was determined for cardiac, abdomen–pelvis, and chest protocols, whereas for the 16-row scanner, a chest protocol was examined. The cardiac protocol employed volume scanning in target mode with target phase of 75% and exposure time 400 ms, using a collimation of 280 × 0.5 mm, tube potential of 100 kVp, tube current of 500 mA, craniocaudal coverage of 140 mm, and a simulated heart rate of 60 beats/min. The abdomen–pelvis protocol used helical scanning with collimation of 80 × 0.5 mm, tube potential of 120 kVp, tube current of 300 mA, pitch of 0.65, and 455 mm of craniocaudal coverage in the abdomen–pelvis region. For the chest protocol on the 320-row scanner, we used a helical scanning with collimation of 80 × 0.5 mm, tube potential of 120 kVp, tube current of 300 mA, and pitch of 1.1, with 335 mm coverage of the chest, whereas for the 16-row scanner we used a helical scanning with collimation of 16 × 0.5 mm, 120 kVp, 250 mA, pitch of 0.75 and 336 mm of coverage.
Pilot data for estimating variance of spot doses and expected effective dose
To use Eq. 16 or 18, we need to know and e. These parameters are typically unknown in reality but can be estimated by conducting a pilot study where are estimated by the sample variance and e by Eq. 5 or prior experience. We estimated these values using the MOSFETs placed in the anthropomorphic phantoms, for each scanner and protocol described in Sec. 2E. The MOSFETs were placed in similar configuration for measuring ED, in the different organs. Scanning was performed repeatedly 10 times for the male and for the female phantoms. The variance was calculated per spot for each of the phantoms, and the quantities a, b, and c could then be estimated, according to Eq. 7.
RESULTS
Pilot data results
Pilot data were obtained from measurements performed using each protocol. Results for the helical chest protocol, on the 320-row scanner, are illustrated in Table 2, along with organ dose per each gender and their average. The results include the values of , , and , which are estimated weighted composites of the spot variances for regions 1, 2, and 3, respectively. Regions 1, 2 and 3 represent areas of different radiation exposure, where region 2 is “in-field,” receiving the most radiation for the chest protocol. , which is associated with region 2, is greater than and , likewise, the weighted voltage is highest for region 2, which is also reflected by the high weighted equivalent doses to organs in that region.
Table 2.
Pilot data for three regions for helical chest protocol on 320-row scanner, along with weighted equivalent dose per gender.
| Region of | Female weighted | Male weighted | Average weighted | |
|---|---|---|---|---|
| organ i | equivalent dose (mSv) | equivalent dose (mSv) | equivalent dose (mSv) | |
| Brain | 1 | 0.01 | 0.01 | 0.01 |
| Salivary glands | 1 | 0.13 | 0.13 | 0.13 |
| Thyroid |
1 |
0.84 |
0.89 |
0.87 |
| Region 1 | ||||
| , weighted voltage1 mV | ||||
| Cranial-to-thorax | ||||
| Lung | 2 | 1.77 | 1.78 | 1.78 |
| Esophagus | 2 | 0.56 | 0.58 | 0.57 |
| Breast |
2 |
1.72 |
2.25 |
1.98 |
| Region 2 | ||||
| , weighted voltage1 mV | ||||
| Thorax | ||||
| Stomach | 3 | 2.06 | 2.06 | 2.06 |
| Liver | 3 | 0.52 | 0.54 | 0.53 |
| Colon | 3 | 0.13 | 0.13 | 0.13 |
| Bladder | 3 | 0.00 | 0.00 | 0.00 |
| Gonads |
3 |
0.02 |
0.01 |
0.01 |
| Region 3 | ||||
| , weighted voltage1 mV | ||||
| Caudal-to-thorax | ||||
| Bone marrow | 1,2,3 | 0.44 | 0.47 | 0.45 |
| Bone surface | 1,2,3 | 0.04 | 0.04 | 0.04 |
| Remainder | 1,2,3 | 1.05 | 1.10 | 1.08 |
Weighting based on tissue weighting factors, per spots in the same region.
The result of the estimated uncalibrated ED is which, assuming β of 3, suggests 9.2 mSv. Analogous findings regarding in-field regions were noted for all other protocols. Note that for the abdominopelvic protocol, two of the three regions, viz. regions 2 and 3, are in-field.
Effects of precision and confidence parameters on sample size
Having at hand the pilot data for a given protocol, we need to choose three parameters which influence the required ED measurement sample size. These are (i) the required confidence of the ED, 1 − α, (ii) the precision of the ED, q, and (iii) the CV of the CF, r. Table 3 demonstrates different choices for these parameters and the effect on the ED sample size for both the three-region approach according to Eq. 16, and for the single-region approach according to Eq. 18, for the helical chest protocol on the 320-row scanner. Table 3 also demonstrates the effect of the choice of r on the calibration sample size, m. Given historical or manufacturer's data for initial values of β and σβ, according to Eq. 11, the calibration sample size is decided according to the value of r (subject to the constraint r < ρ). In turn, r has an impact on the final sample sizes n1, n2, and n3, or n.
Table 3.
The effect of the choice of the parameters 1 − α, q(ρ) and r on m, the number of repetitions required for calibration, and on the ED sample size estimates n1, n2, n3, or n for a helical chest protocol on the 320-row scanner with parameter set a = 0.035, b = 1.180, c = 0.002, e = 27.7 mV, β = 1/3, and σβ = 0.05% β.
| Three-region approach |
Single-region approach |
||||||
|---|---|---|---|---|---|---|---|
| Precision | Calibration | Region 1 | Region 2 | Region 3 | Sample | ||
| Confidence | of ED | CV of | sample size | sample size | sample size | sample size | size |
| 1− α (%) | q (%) | r < ρ | m | n1 | n2 | n3 | n |
| 95 | 5 | 0.01 < 0.026 | 25 | 0.6 | 3.3 | 0.1 | 2.8 |
| 95 | 5 | 0.024 < 0.026 | 4.3 | 3.3 | 18.7 | 0.8 | 15.9 |
| 95 | 10 | 0.024 < 0.051 | 4.3 | 0.2 | 0.9 | 0.04 | 0.7 |
| 95 | 10 | 0.05 < 0.051 | 1 | 3 | 17 | 0.8 | 14.4 |
| 90 | 5 | 0.02 < 0.031 | 6.3 | 0.5 | 3.1 | 0.1 | 3 |
| 90 | 5 | 0.03 < 0.031 | 2.8 | 4.9 | 28.1 | 1.3 | 23.9 |
| 90 | 10 | 0.024 < 0.061 | 4.3 | 0.1 | 0.6 | 0.02 | 0.5 |
| 90 | 10 | 0.06 < 0.061 | 0.7 | 2.5 | 14.2 | 0.6 | 12.1 |
A number of interesting findings related to the relationships between these various quantities are illustrated in Table 3. When either the desired confidence parameter is higher or desired precision of ED is greater (i.e., lower q), for the same r, the ED sample sizes n1, n2, n3, or n are larger, as would be expected. For lower r, implying higher precision of the CF, the calibration sample size m, is larger. The lower the calibration sample size m, the higher the sample sizes n1, n2, n3, or n. This type of table, thus, can be constructed as a decision tool, to illustrate the tradeoff between sample size(s), calibration sample size, and the desired precision and confidence of the ED estimate.
Also of note, for a three-region approach, we consistently observe sample size n2 to be greater than n1 and n3. Sample size n2 refers to the thoracic region, which receives the highest dose when a chest protocol is used. Moreover, using a single-region approach, the sample size n is smaller than the values of n2 for similar conditions. Similar behavior is observed for other protocols.
Sample size estimates for effective dose
Sample sizes n1, n2, and n3, for the three-region approach, and n, for the one-region approach, were determined according to Eqs. 16, 18, respectively, and based on the calculated CF, , and the pilot data of , , , for each of the four protocols. Tables 4, 5, 6, 7 display the highest sample size required for either the three-region approach or the one-region approach, for a given set of parameters, rounded up to the nearest integer; in general for the four protocols considered, these correspond to n2. Sample sizes are displayed for both 95% and 90% confidence levels for different expected ED in the range of 1–15 mGy (mSv) and for different precision levels with q ranging from 2% to 50% of the ED. For all protocols, these tables assume r = 0.017 and m = 9 for all precisions, except for q of 2%, in which case r was chosen to be 0.009, m = 31, in order to ensure that the criterion r < ρ is still met.
Table 4.
Highest sample size required for three- or one-region approach for cardiac volume protocol performed on 320-row scanner, as a function of anticipated effective dose, precision, and desired confidence level. Sample sizes displayed are for confidence level of 95%, with sample sizes for 90% confidence level in parentheses.
| Precision | Anticipated effective dose (mSv) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (q [%]) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| 2 | 8803(7273) | 2201(1819) | 979(809) | 551(455) | 353(291) | 245(203) | 180(149) | 138(114) | 109(90) | 89(73) | 73(61) | 62(51) | 53(44) | 45(38) | 40(33) |
| 5 | 498(274) | 125(69) | 56(31) | 32(18) | 20(11) | 14(8) | 11(6) | 8(5) | 7(4) | 5(3) | 5(3) | 4(2) | 3(2) | 3(2) | 3(2) |
| 10 | 76(51) | 19(13) | 9(6) | 5(4) | 4(3) | 3(2) | 2(2) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 15 | 32(22) | 8(6) | 4(3) | 2(2) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 20 | 18(12) | 5(3) | 2(2) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 50 | 3(2) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
Table 5.
Highest sample size required for three- or one-region approach for helical chest protocol performed on 320-row scanner, as a function of anticipated effective dose, precision, and desired confidence level. Sample sizes displayed are for confidence level of 95%, with sample sizes for 90% confidence level in parentheses.
| Precision | Anticipated effective dose (mSv) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (q [%]) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| 2 | 8402(6941) | 2101(1736) | 934(772) | 526(434) | 337(278) | 234(193) | 172(142) | 132(109) | 104(86) | 85(70) | 70(58) | 59(49) | 50(42) | 43(36) | 38(31) |
| 5 | 476(262) | 119(66) | 53(30) | 30(17) | 20(11) | 14(8) | 10(6) | 8(5) | 6(4) | 5(3) | 4(3) | 4(2) | 3(2) | 3(2) | 3(2) |
| 10 | 73(49) | 19(13) | 9(6) | 5(4) | 3(2) | 3(2) | 2(1) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 15 | 30(21) | 8(6) | 4(3) | 2(2) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 20 | 17(12) | 5(3) | 2(2) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 50 | 3(2) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
Table 6.
Highest sample size required for three- or one-region approach for helical chest protocol performed on 16-row scanner, as a function of anticipated effective dose, precision, and desired confidence level. Sample sizes displayed are for confidence level of 95%, with sample sizes for 90% confidence level in parentheses.
| Precision | Anticipated effective dose (mSv) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (q [%]) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| 2 | 2896(2392) | 724(598) | 322(266) | 181(150) | 116(96) | 81(67) | 60(49) | 46(38) | 36(30) | 29(24) | 24(20) | 21(17) | 18(15) | 15(13) | 13(11) |
| 5 | 164(91) | 41(23) | 19(11) | 11(6) | 7(4) | 5(3) | 4(2) | 3(2) | 3(2) | 2(1) | 2(1) | 2(1) | 1(1) | 1(1) | 1(1) |
| 10 | 25(17) | 7(5) | 3(2) | 2(2) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 15 | 11(8) | 3(2) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 20 | 6(4) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 50 | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
Table 7.
Highest sample size required for three- or one-region approach for abdomen–pelvis protocol performed on 320-row scanner, as a function of anticipated effective dose, precision, and desired confidence level. Sample sizes displayed are for confidence level of 95%, with sample sizes for 90% confidence level in parentheses.
| Precision | Anticipated effective dose (mSv) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (q [%]) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| 2 | 6727(5558) | 1682(1390) | 748(618) | 421(348) | 270(223) | 187(155) | 138(114) | 106(87) | 84(69) | 68(56) | 56(46) | 47(39) | 40(33) | 35(29) | 30(25) |
| 5 | 381(210) | 96(53) | 43(24) | 24(14) | 16(9) | 11(6) | 8(5) | 6(4) | 5(3) | 4(3) | 4(2) | 3(2) | 3(2) | 2(2) | 2(1) |
| 10 | 58(39) | 15(10) | 7(5) | 4(3) | 3(2) | 2(2) | 2(1) | 2(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 15 | 24(17) | 6(5) | 3(2) | 2(2) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 20 | 14(10) | 4(3) | 2(2) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| 50 | 2(2) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
In these tables, it can be observed that with higher precision, the sample sizes are greater and that for low expected ED the sample size is also greater. For example, in Tables 5, 6, for a helical chest protocol, for 95% confidence that the estimate is within ±5% of the true value of the ED, we require up to 30 measurements on the 320-row scanner but only 11 on the 16-row scanner when the anticipated ED is 4 mSv. These sample sizes are 5 and 2, respectively, when the anticipated ED is 10 mSv. For a lower, 90% confidence level, and less precision of ±10%, sample sizes are 4 and 2 for an anticipated ED of 4 mSv, respectively, and 1 for anticipated ED of 10 mSv, for both scanners. For very low doses, the sample size needed for high precision with high confidence level is prohibitively large.
DISCUSSION
To ensure a fair comparison of radiation burden between diagnostic imaging modalities and protocols, it is critically important to understand how trustworthy the estimated dosimetry quantities are. This is especially the case for ED, which is currently the only single metric enabling such comparison across modalities. In this work we have developed a statistical tool to assess the sample size required for estimating ED using a CT scan protocol, to a desired precision and confidence, while minimizing the number of measurements due to practical constraints. ED is calculated by a weighted average of organ absorbed doses,2 which can be determined from MOSFETs positioned in the organs of an anthropomorphic phantom (male and female). Scanner time in daily routine is a limited resource and MOSFET dosimetry is a costly and a nontrivial task. Despite these hurdles, the goal still remains to maintain a reliable ED.
Applying this statistical scheme, we have shown that, in general, a total of ten scans, divided between male and female phantoms, are sufficient to ensure high precision (±5%) with high confidence (90%–95% confidence level) for a range of different protocols when the ED is at least 6 mSv. Still, for some scan protocols, considerably less than ten scans are needed to ensure this precision and confidence even for lower EDs than 6 mSv. When the expected ED is higher than 6 mSv, only a single set of measurements is needed, which effectively translates into two sets of measurements as both male and female anthropomorphic phantoms should be used to estimate ED. These observations point at the feasibility of determining ED with high precision and confidence using MOSFET-based dosimetry.
In practice, the desirable levels of precision and confidence for MOSFET CT experiments will depend on the experimental or clinical scenario. For an approximate understanding of the ED of a clinical protocol, so as to gauge its radiation burden, precision of 10% and confidence of 90% will generally be ample. For lower-dose protocols, where sample size requirements to attain the same precision and confidence are larger, a lower precision will often be acceptable. For example, for a 2 mSv scan, 25% precision still translates to an estimate within half a mSv of its true value, a low level of radiation which, on a patient level, is not very meaningful biologically. In other MOSFET CT experiments, however, such as those to obtain reliable conversion factors to estimate ED from dose–length product, higher degrees of precision and confidence may be desirable.
Our work demonstrates the interrelationship and tradeoff between MOSFET reading sample sizes, calibration sample size, and the desired precision and confidence of the ED estimate. Most importantly, it enables one to gauge variability and then determine suitable sample sizes to ensure desired precision and confidence, subject to practical considerations. For example, if the calibration procedure is expensive, one might prefer to reduce calibration sample size m by choosing the CV for calibration (r) to be close to the CV for ED (ρ), thus requiring greater MOSFET reading sample size(s), for a desired confidence level and precision of the ED. Alternatively, in many circumstances it is preferred to minimize the MOSFET reading sample size(s), due to, for example, costs of scans or desire to maximize MOSFET lifespan; in such an instance choosing r ≪ ρ is a preferred approach.
On helical chest CT scans performed on the two different scanners, we noted considerable differences in sample size requirements for the same ED, precision, and degree of confidence. This reflects differences between the scanners in the interscan variability of individual MOSFET measurements, and underscores the importance of using scanner-specific pilot data as a part of sample size determination.
Our scheme is not dependent upon a particular calibration approach, but rather provides sufficient generality to ensure that the chosen calibration procedure permits the desired precision and confidence of the estimated ED. Our framework, thus, is applicable to any MOSFET calibration approach, examples of which have been suggested by Yoshizumi et al.12 and Brady et al.14
When partitioning the anthropomorphic phantom into three spatial regions for limited numbers of MOSFETs, we found that the sample size specified for the region primarily irradiated (e.g., for a chest protocol this would be n2, the sample size for the thoracic region) was the highest. When the whole phantom is taken as a single region, the sample size (n) is reduced in comparison.
As it is impossible in one scheme to incorporate all sources of variations in dosimetry estimates, our approach is necessarily subject to limitations. For example, for many organs we only use a single MOSFET to determine organ dose, whereas in actuality there can be lack of uniformity in point doses within an organ or tissue. Nevertheless, we have tried to address this by placing multiple MOSFETs in organs which are large and/or have high tissue weighting factor. Another limitation is that we did not incorporate all possible sources of variations, for example, we assume that the ionization chamber used to calibrate MOSFETs is itself well calibrated and provides reproducible measurements. However, ion chamber readings can vary depending upon, for example, room temperature and pressure.16 Even so, pilot data suggest that variability within and between ionization chambers is quite small and considerably less than the variability we considered in our model. While our approach is general in that it allows for a variety of different calibration methods, it still assumes a common, averaged CF for all MOSFETs, whereas some investigators may choose individual CFs for each MOSFET. In addition, our sample size calculation was not directly based on a hypothesis test but rather based on estimation precision. Once the sample size is determined using our formula, data can be collected, ED can be estimated, and its (1 − α) × 100% confidence interval will be constructed to the desired precision. The (1 − α) × 100% confidence interval can be used to generate useful hypotheses for future studies. Finally, it should be noted that ED is calculated for a reference phantom, which is based on an idealized male or female person.2 It is not tailored per patient, nor does it include adjustments for gender or anatomy variation.17 Its use is not meant for an individual person, and yet its value lies in evaluation of relative biological risk and in its usage for comparing different modalities.
In conclusion, this paper has established a statistical scheme for the determination of ED with specified confidence and precision for use in design of experiments using MOSFET dosimeters on adult phantoms. Based upon pilot data obtained for a specific protocol, and three user-selectable parameters, sample size determination should be a straightforward yet rigorous process.
ACKNOWLEDGMENTS
This work was supported by NIH-NHLBI R01 Grant Nos. HL109711, HL098237, and HL098305, by a Herbert Irving Associate Professorship, and as a Victoria and Esther Aboodi Cardiology Researcher (Dr. Einstein). Dr. Trattner and Mr. Pieniazek have received funding for other research from Philips Healthcare. Dr. Einstein has received funding for other research from GE Healthcare and Philips Healthcare.
References
- Halliburton S., Arbab-Zadeh A., Dey D., Einstein A. J., Gentry R., George R. T., Gerber T., Mahesh M., and Weigold W. G., “State-of-the-art in CT hardware and scan modes for cardiovascular CT,” J. Cardiovasc. Comput. Tomogr. 6(3), 154–163 (2012). 10.1016/j.jcct.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 2007 recommendations of the International Commission on Radiological Protection. ICRP publication 103, Ann. ICRP 37(2–4), 1–332 (2007). [DOI] [PubMed] [Google Scholar]
- Martin C. J., “Effective dose: How should it be applied to medical exposures?,” Br. J. Radiol. 80(956), 639–647 (2007). 10.1259/bjr/25922439 [DOI] [PubMed] [Google Scholar]
- McCollough C. H., Christner J. A., and Kofler J. M., “How effective is effective dose as a predictor of radiation risk?,” Am. J. Roentgenol. 194(4), 890–896 (2010). 10.2214/AJR.09.4179 [DOI] [PubMed] [Google Scholar]
- Einstein A. J., Elliston C. D., Arai A. E., Chen M. Y., Mather R., Pearson G. D., Delapaz R. L., Nickoloff E., Dutta A., and Brenner D. J., “Radiation dose from single-heartbeat coronary CT angiography performed with a 320-detector row volume scanner,” Radiology 254(3), 698–706 (2010). 10.1148/radiol.09090779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein A. J., Elliston C. D., Groves D. W., Cheng B., Wolff S. D., Pearson G. D., Robert Peters M., Johnson L. L., Bokhari S., Johnson G. W., Bhatia K., Pozniakoff T., and Brenner D. J., “Effect of bismuth breast shielding on radiation dose and image quality in coronary CT angiography,” J. Nucl. Cardiol. 19(1), 100–108 (2012). 10.1007/s12350-011-9473-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frush D. P. and Yoshizumi T., “Conventional and CT angiography in children: Dosimetry and dose comparisons,” Pediatr. Radiol. 36(Suppl. 2), 154–158 (2006). 10.1007/s00247-006-0190-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Mukonoweshuro W., and Loader R., “A phantom-based evaluation of three commercially available patient organ shields for computed tomography X-ray examinations in diagnostic radiology,” Radiat. Protect. Dosim. 155(2), 161–168 (2013). 10.1093/rpd/ncs327 [DOI] [PubMed] [Google Scholar]
- Hurwitz L. M., Yoshizumi T. T., Goodman P. C., and Frush D. P., “Effective dose determination using an anthropomorphic phantom and metal oxide semiconductor field effect transistor technology for clinical adult body multidetector array computed tomography protocols,” J. Comput. Assist. Tomogr. 31(4), 544–549 (2007). 10.1097/RCT.0b013e31802d3dd2 [DOI] [PubMed] [Google Scholar]
- Jones A. K., Pazik F. D., Hintenlang D. E., and Bolch W. E., “MOSFET dosimeter depth-dose measurements in heterogeneous tissue-equivalent phantoms at diagnostic x-ray energies,” Med. Phys. 32(10), 3209–3213 (2005). 10.1118/1.2047827 [DOI] [PubMed] [Google Scholar]
- Sessions J. B., Roshau J. N., Tressler M. A., Hintenlang D. E., Arreola M. M., Williams J. L., Bouchet L. G., and Bolch W. E., “Comparisons of point and average organ dose within an anthropomorphic physical phantom and a computational model of the newborn patient,” Med. Phys. 29(6), 1080–1089 (2002). 10.1118/1.1481516 [DOI] [PubMed] [Google Scholar]
- Yoshizumi T. T., Goodman P. C., Frush D. P., Nguyen G., Toncheva G., Sarder M., and Barnes L., “Validation of metal oxide semiconductor field effect transistor technology for organ dose assessment during CT: Comparison with thermoluminescent dosimetry,” Am. J. Roentgenol. 188(5), 1332–1336 (2007). 10.2214/AJR.06.0742 [DOI] [PubMed] [Google Scholar]
- Zhang D., Savandi A. S., DeMarco J. J., Cagnon C. H., Angel E. A., Turner A. C., Cody D. D., Stevens D. M., Primak A. N., McCollough C. H., and McNitt-Gray M. F., “Variability of surface and center position in MDCT: Monte Carlo simulations using CTDI and anthropomorphic phantoms,” Med. Phys. 36(3), 1025–1038 (2009). 10.1118/1.3078053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S. L. and Kaufman R. A., “Establishing a standard calibration methodology for MOSFET detectors in computed tomography dosimetry,” Med. Phys. 39(6), 3031–3040 (2012). 10.1118/1.4712221 [DOI] [PubMed] [Google Scholar]
- Eckerman K. F., Bolch W. E., Zankl M., and Petoussi-Henss N., “Response functions for computing absorbed dose to skeletal tissues from photon irradiation,” Radiat. Protect. Dosim. 127(1–4), 187–191 (2007). 10.1093/rpd/ncm468 [DOI] [PubMed] [Google Scholar]
- IAEA, “Status of computed tomography dosimetry for wide cone beam CT scanners,” IAEA Human Health Reports, 2011.
- AAPM, “Size-specific dose estimates (SSDE) in pediatric and adult body CT examinations,” Report of AAPM TG 204, 2011. [DOI] [PubMed]