Abstract
miRNAs regulate gene expression through translational repression and/or mRNA deadenylation/decay. As translation, deadenylation and decay are closely linked processes, it is important to establish their ordering and thus to define the molecular mechanism of silencing. We have investigated the kinetics of these events in miRNA-mediated gene silencing using a Drosophila S2 cell-based controllable expression system and show that mRNAs with both natural and engineered 3′UTRs with miRNA target sites are first subject to translational inhibition, followed by effects on deadenylation and decay. We next use a natural translational elongation stall to show that miRNA-mediated silencing inhibits translation at an early step, potentially translation initiation.
Keywords: miRNA, Argonaute, mechanism
MicroRNAs (miRNAs) are short endogenous RNAs that regulate protein expression from targeted genes by pairing to sites in the 3′ untranslated region (3′UTR)(1). While some studies showed a strong correlation between the diminution of protein and mRNA levels of miRNA-targeted genes (2-6), other studies showed that miRNAs principally affect protein expression of miRNA-targeted genes without obvious effects on mRNA abundance (7-10). By simultaneously measuring translational efficiencies (thus indirectly levels of protein synthesis) and mRNA abundance, global analyses have shown evidence of significant mRNA destabilization and translational repression (11, 12). Since only slightly more translational repression is observed than mRNA destabilization, it is possible that most of the loss in protein synthesis could directly result from effects on mRNA stability. Most of these studies have not, however, evaluated the kinetics of the miRNA-related cellular processes (5, 10, 13, 14). Exceptions include several analyses in in vitro systems which concluded that the effects of miRNAs on translational repression precede effects on mRNA target deadenylation or decay (15-17) but concerns remain that the in vitro reactions may not fully recapitulate the in vivo situation.
Here we used an in vivo luciferase-based reporter system in Drosophila melanogaster S2 cells under the control of an inducible metallothionein promoter (Mtn) (18). The reporter constructs consist of one of the luciferase reporter genes (Firefly (F-Luc) or Renilla (R-Luc)) fused at its 5′ end to the Mtn promoter and at its 3′ end to synthetic or natural 3′UTRs that contain miRNA binding sites responsive to either endogenously expressed (bantam and miR-279) or ectopically introduced miRNAs (miR-9b) (Supplementary Figure 1); control constructs not subject to miRNA-mediated gene regulation are detailed in Supplementary Figure 1. We first asked whether miRNA-mediated responsiveness is limited by endogenous components of the miRISC complex (Argonaute protein or miRNAs). Using a previously characterized bantam-responsive synthetic construct and ectopically expressed additional miRISC components (bantam and Ago1), we followed repression levels of the target mRNAs 24 hours after induction (Figure 1A, (19)). The results indicate that Ago1 is not limiting for repression in Drosophila S2 cells, while bantam is limiting since greater repression was observed when it was over-expressed (Figure 1A). In the remaining experiments we express additional amounts of endogenously present miRNAs (bantam and miR-279) from plasmids under the constitutive actin promoter.
Figure 1.
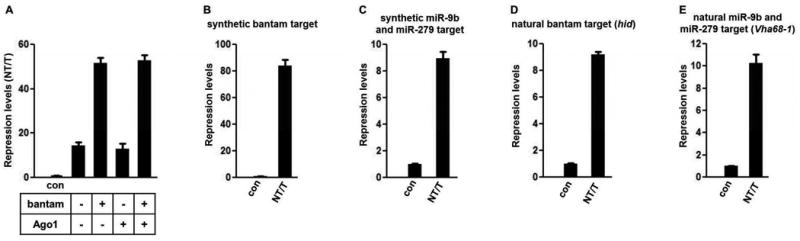
Steady state evaluation of miRNA-mediated gene silencing using copper inducible in vivo reporter system. (A) Measured protein amounts (luminescence) from transfected non-targeted (NT), targeted (T) and control constructs 24 hours after induction. Additional expression of bantam miRNA, not Argonaute 1, results in increased repression for synthetic bantam targeted constructs (Supplementary Fig. 1). (B-E) Ratios of steady state protein amounts for synthetic and natural miRNA-targeted constructs 48 hours after induction. In each case, mean values ± standard deviation from three independent triplicate experiments are shown as a normalized ratio of protein amounts (NT/T).
We next used a set of synthetic and natural 3′ UTRs with miRNA binding sites responding to either bantam or miR-9b and miR-279, and their corresponding controls (Supplementary Figure 1, Supplementary table 1 and 2). We chose natural (endogenous) 3′ UTRs from hid and Vh68-1 and a pair of synthetic 3′ UTR targets that contain 6 natural tandem sites for either bantam or miR-9b and miR-279 miRNAs (19, 20). Following transfection and constitutive induction, luciferase expression levels were measured after 48 hours and normalized to assess the endpoint effects of miRNA-mediated repression (Figure 1B-E). The synthetic bantam 3′ UTR exhibited repression levels up to 80-fold when compared to the control (Figure 1B); the reporter containing the 3′UTR of the hid gene with as many as 8 miRNA binding sites exhibited ∼9-fold repression (Figure 1D) (19). Additionally, both synthetic and natural (20) 3′ UTRs responding to ectopically expressed miR-9b and miR-279 exhibited strong repression 48 hours after induction (Figure 1C, E). The addition of antagomirs to the cell cultures induced similar levels of derepression for the reporter constructs as did comparison to the construct with altered miRNA binding sites (Supplementary Figure 2).
For regulated induction, transcriptional shut-off of the Mtn promoter was accomplished using a specific copper chelator, bathocuproine disulphonate (BCS). While BCS chelates any residual copper in the S2 cell medium, it does not penetrate the cell membrane and thus does not affect normal cellular homeostasis (21). A 90 minute pulse induction by copper(II)–sulfate induced expression of the various reporter constructs to levels that could be monitored over the subsequent 48 hours. Using this optimized pulse induction protocol (Supplementary Figure 3) we determined how our reporter pairs with synthetic and natural 3′ UTRs respond to miRNAs during an extended period of time. Normalized levels of luciferase luminescence for both miRNA-targeted (T) and non-targeted (NT) constructs were used to assess miRNA-mediated gene silencing at the protein level. mRNA levels for reporter and control genes were determined using quantitative reverse transcriptase PCR (qRT-PCR) from RNA samples isolated using oligo(dT)25 resin as well as from total RNA. Both values were normalized with respect to the parallel transfected control genes and were used to calculate repression ratios between miRNA-targeted and non-targeted constructs (Figure 2A-D). We emphasize that there is substantial mRNA degradation (for all reporters) during the experiment (Supplemental Figure 4), but here we are interested in the relative amount of decay of the targeted and non-targeted mRNAs. The results of the pulse-induction experiments were consistent and show that the repressive effects of miRNAs on synthesis of all four proteins precede any effects on mRNA deadenylation or decay (Figure 2A-D).
Figure 2.
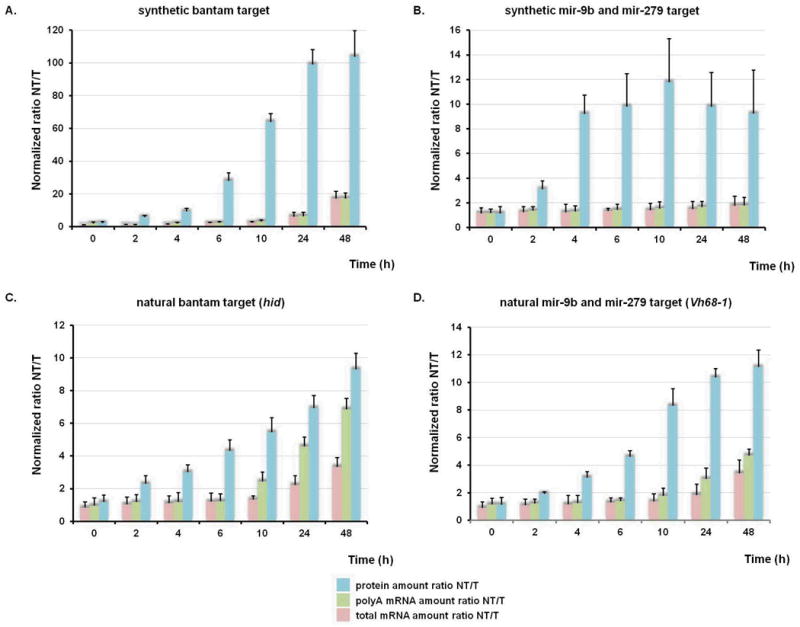
Time resolved progression of miRNA-mediated gene silencing establishes that repression of protein synthesis precedes mRNA deadenylation and decay. (A-D) Normalized levels of protein amounts for both miRNA-targeted (T) and non-targeted (NT) constructs; normalized mRNA levels for reporter genes from oligo(dT)25 resin pull down or from total RNA presented as ratios of poly(A) and total mRNA, respectively. Each data point represents the mean value ± standard deviation calculated from three independent experiments,
These four examples include effects on protein and mRNA levels that are both well correlated and poorly correlated (5, 10-12, 19, 20). For reporters with natural 3′ UTRs, the repression effects on protein and mRNA levels are correlated at later (but not early) time points (Fig 2C and 2D), consistent with global studies on miRNA-mediated silencing (11, 12). For reporters with synthetic 3′ UTRs, the repression effects on protein and mRNA levels are uncorrelated at both early and late time points (Fig 2A and 2B) (5, 10, 19, 20). The differences in overall stability of the mRNAs likely reflect the complexity of RNA degradation as specified by various 3′UTR located sequence motifs. In all experiments here, repression of protein synthesis is consistently seen just 2 hours after induction while mRNA destabilization comes later. Additionally, in experiments where less bantam miRNA was present (by not supplying an exogenous source), the observed timing of protein repression and mRNA deadenylation/decay were unaffected (Supplementary Figure 5A and B).
We performed a similar set of experiments using actinomycin D to more rapidly shut off transcription (Supplementary Figure 6A-D). At a single time point after induction, all constructs showed a substantial reduction in protein production but no reduction in mRNA abundance (Figure 2A-D). Consistent with BCS shut-off data, translation rates for targeted constructs are substantially reduced with respect to non-targeted ones (Supplementary Figure 6A-D).
mRNA polyA tails are involved in translation initiation and thus in determining translational efficiency (22, 23). Potential subtle changes in poly(A) tail length not detectable using an oligo(dT)25 isolation procedure were analyzed for the complete set of RNA samples using a poly(A) tail-length assay that allows for specific amplification of poly(A) containing mRNAs. Subtle changes in poly(A) tail length could be documented using this assay when cells were treated with puromycin (Supplementary Figure 7)(24). However, we did not observe any shortening of poly(A) tails in reporter constructs responding to bantam or mir-9b and mir-279 (Figure 3A-D). As no intermediates are observed, these data suggest that deadenylation in vivo is processive and closely coupled to mRNA decay. These data argue that translational inhibition is not triggered by deadenylation, either partial or complete, in the system that we have established (in contrast with earlier studies (e.g.(6, 12, 25)).
Figure 3.
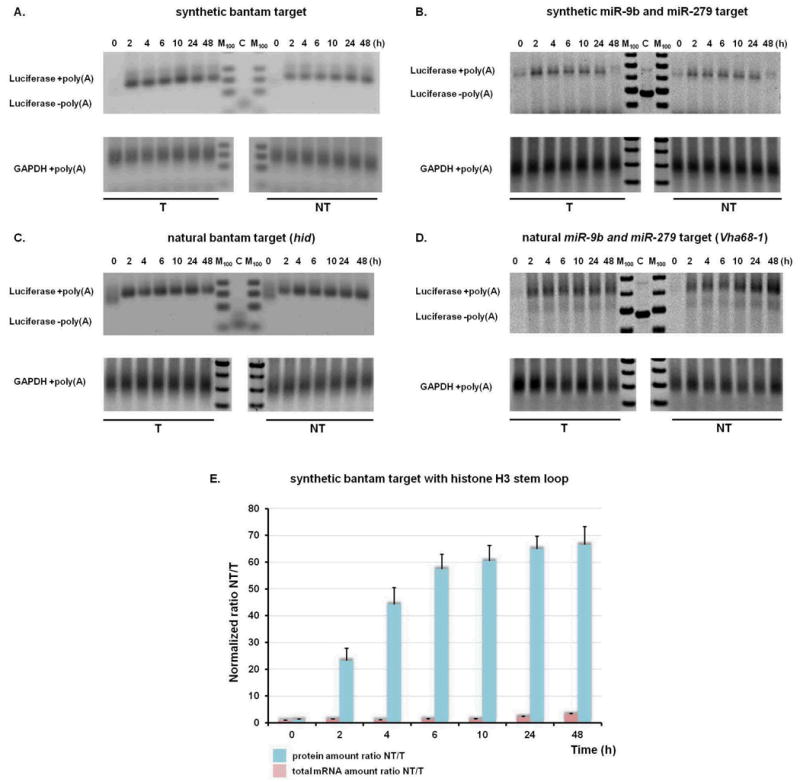
mRNA deadenylation is not required for miRNA-mediated translational repression. (A-D) Length of poly(A) tail was determined using G/I tailing PCR based amplification (Materials and Methods). Positions of over-amplified products with or without (C) poly(A) tail are indicated. GAPDH mRNA poly(A) tail length is shown as a control. M lane represents 100 bp markers. (E) Time resolved progression of miRNA-mediated gene silencing for histone H3 constructs. Normalized levels of protein and mRNA amounts for miRNA-targeted (T) and non-targeted (NT) constructs are shown. Each data point represents an average value from three independent experiments.
We also evaluated the timing and extent of silencing of reporter gene pairs carrying histone H3 terminal stem-loops (and no poly(A) tail) (26)(Supplementary Figure 8). This reporter pair is translationally repressed rapidly (Figure 3E) and at levels comparable to those observed with equivalent constructs carrying a poly(A) tail (Figure 2A vs. 3E). These data provide further support for the idea that deadenylation is not required for translational repression by miRNAs (5). However, deadenylases may play a direct role in translational repression independent of their deadenylation activity (27-29) and deadenylation may consolidate the observed translational repression.
To define where in the translational cycle the miRNA-mediated stalling occurs, we developed an approach that uses as a read-out the specific cleavage of mRNAs when ribosomes stall during translation elongation (30) (Supplementary Figure 9). We inserted 12 lysine codons at position 6 in both the natural and synthetic reporters; as a control, we inserted 12 arginines and 12 more neutral glutamine residues (Supplementary Figure 10). Using qRT primers that encompassed the predicted cleavage site as well as the stall sequence, we determined the extent of cleavage of both the targeted and non-targeted reporter mRNAs over time (Figure 4A-D and Supplementary Figure 11A-C). Ratios of the targeted and non-targeted proteins and mRNAs were normalized to the amount of a parallel control reporter. In all cases, mRNAs of non-targeted reporters were rapidly cleaved and degraded, while mRNAs of the targeted reporters were relatively stabilized. Similar results were obtained in a stalling experiment with histone H3 constructs, confirming that the poly(A) tail is not essential for miRNA-mediated translational repression (Figure 4E). These data establish that miRNA-mediated translational silencing happens in the Drosophila system during the initiation or early elongation phase of protein synthesis.
Figure 4.
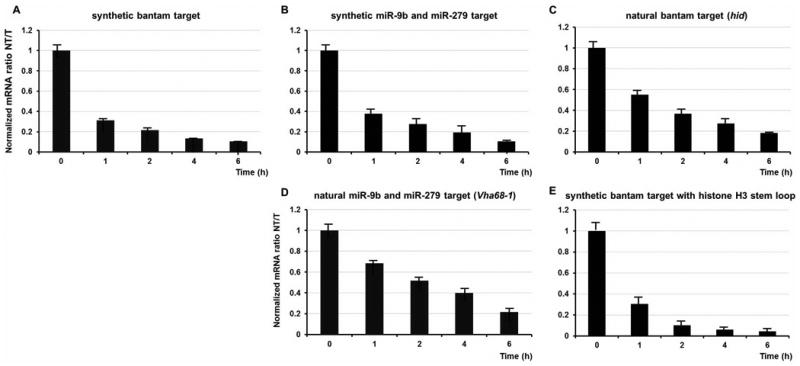
Translational elongation stalling assay indicates that miRNA-mediated gene silencing targets early steps in translation. (A-E) Time resolved progression of miRNA-mediated effects on the stability of various mRNA constructs containing the lysine-induced elongation stall. Normalized mRNA amounts for the miRNA-targeted (T) and non-targeted (NT) constructs are shown as the ratio of NT/T; note that values are less than one and decreasing. Each data point represents an average from three independent experiments.
We find that miRNA-mediated gene silencing in Drosophila S2 cells is first manifested through effects on translation, and in particular the early events thereof, and is subsequently consolidated by mRNA deadenylation and decay. While it is possible that the order of events is different in other systems or in a fashion that is mRNA-specific, the consistency of our data in Drosophila is striking. Moreover, these observations are consistent with earlier studies on miRNA-mediated silencing in vitro (15, 17) and with previous studies of translation as affected by iron levels (24). With these insights into the relative timing of the events involved in miRNA-mediated gene silencing, we can now focus subsequent molecular mechanistic analysis on these earliest triggering steps.
Supplementary Material
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 3.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 4.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 5.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beilharz TH, et al. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PloS one. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Developmental biology. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 8.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 9.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Molecular cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. The EMBO journal. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS biology. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & development. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 16.Zdanowicz A, et al. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Molecular cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Molecular cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansen H, et al. Regulated expression at high copy number allows production of a growth-inhibitory oncogene product in Drosophila Schneider cells. Genes & development. 1989;3:882–889. doi: 10.1101/gad.3.6.882. [DOI] [PubMed] [Google Scholar]
- 19.Nahvi A, Shoemaker CJ, Green R. An expanded seed sequence definition accounts for full regulation of the hid 3′ UTR by bantam miRNA. RNA. 2009;15:814–822. doi: 10.1261/rna.1565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & development. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selvaraj A, et al. Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes & development. 2005;19:891–896. doi: 10.1101/gad.1301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & development. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 23.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muckenthaler M, Gunkel N, Stripecke R, Hentze MW. Regulated poly(A) tail shortening in somatic cells mediated by cap-proximal translational repressor proteins and ribosome association. RNA. 1997;3:983–995. [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Molecular cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nature reviews Genetics. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun Joerg E, Huntzinger E, Fauser M, Izaurralde E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Molecular cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nature structural & molecular biology. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nature structural & molecular biology. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 30.Kuroha K, et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO reports. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.We thank K. Wehner, J. Doudna, M. Jinek, N. Guydosh and J. Coller for helpful comments. Funding from HHMI.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


