Abstract
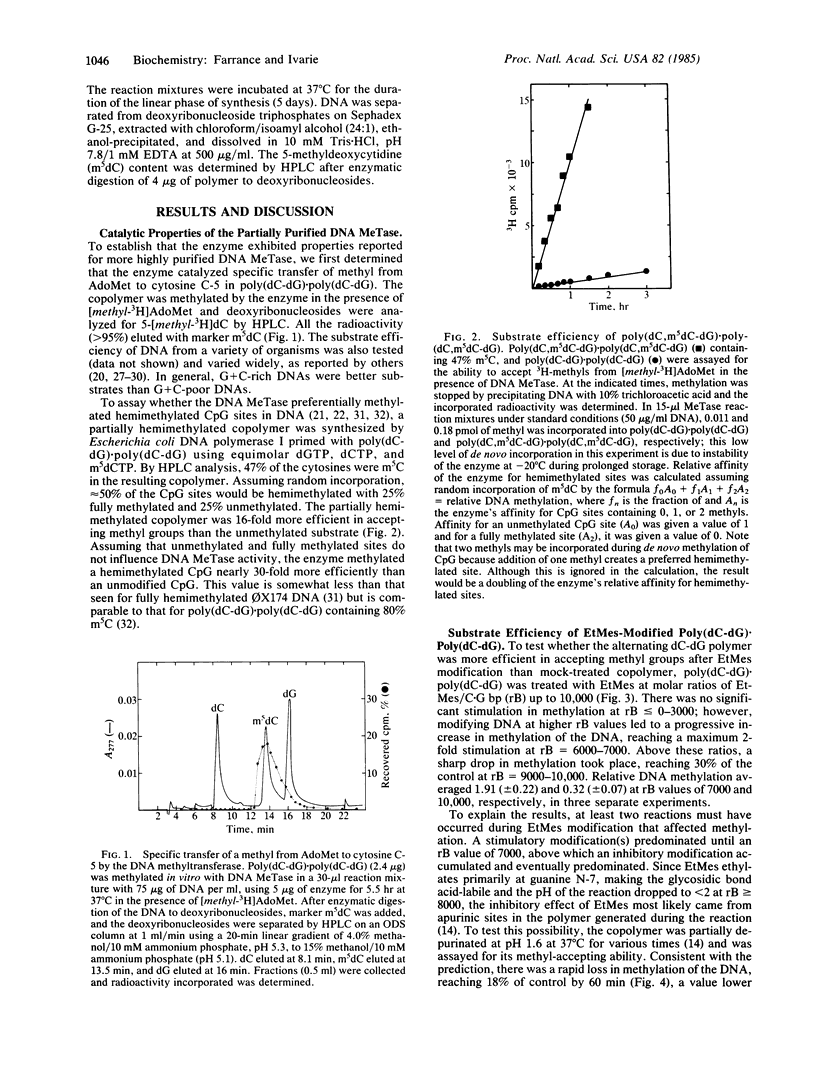
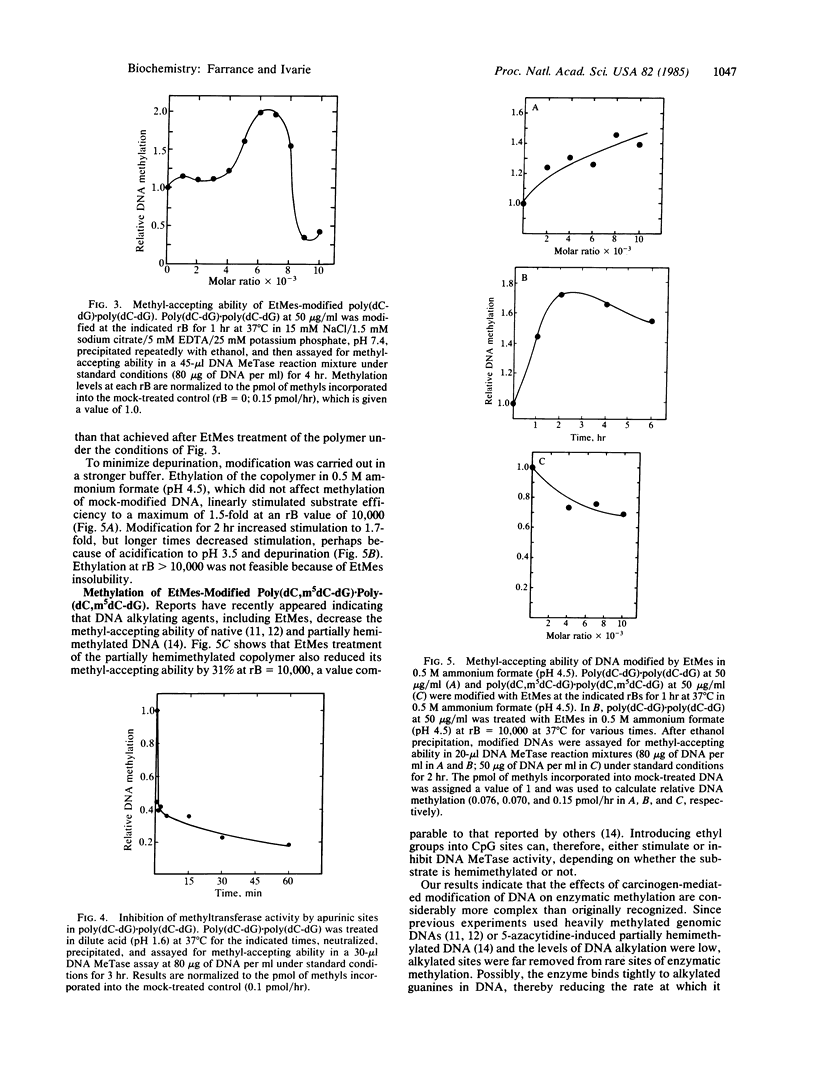
Ethylation of poly(dC-dG).poly(dC-dG) with ethyl methanesulfonate (EtMes), a known carcinogen, at increasing molar ratios of EtMes/C X G base pairs progressively stimulated the methyl-accepting ability of the DNA during in vitro methylation by partially purified rat DNA (cytosine-5)-methyltransferase (EC 2.1.1.37). Maximum stimulation was 2-fold over mock-treated DNA when 2.7% of the guanines were modified at the N-7 position, the major site of ethylation by EtMes in DNA. If a CpG site "hemiethylated" at guanine N-7 mimics a hemimethylated CpG site, we calculate that the enzyme has a relative affinity for hemiethylated CpG 18-fold above unmodified CpG. If ethylation of a dioxyphosphate oxygen of the phosphodiester bond is responsible for stimulation, the relative affinity could be much higher, up to 370-fold.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., McKay E. L., Craig L. M., Burdon R. H. Mouse DNA methylase: methylation of native DNA. Biochim Biophys Acta. 1979 Feb 27;561(2):345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T. L., Drahovsky D. Alteration of enzymatic DNA methylation by chemical carcinogens. Recent Results Cancer Res. 1983;84:212–225. doi: 10.1007/978-3-642-81947-6_16. [DOI] [PubMed] [Google Scholar]
- Boehm T. L., Drahovsky D. Hypomethylation of DNA in Raji cells after treatment with N-methyl-N-nitrosourea. Carcinogenesis. 1981;2(1):39–42. doi: 10.1093/carcin/2.1.39. [DOI] [PubMed] [Google Scholar]
- Boehm T. L., Drahovsky D. Inhibition of enzymatic DNA methylation by N-methyl-N-nitro-N-nitrosoguanidine in human Raji lymphoblast-like cells. Int J Biochem. 1981;13(12):1225–1232. doi: 10.1016/0020-711x(81)90068-9. [DOI] [PubMed] [Google Scholar]
- Craddock V. M., Henderson A. R. Effect of a single treatment with the alkylating carcinogens dimethylnitrosamine and methyl methanesulphonate on liver regenerating after partial hepatectomy. IV. Effect on methylase-mediated methylation of DNA. Chem Biol Interact. 1979 Sep;27(1):81–89. doi: 10.1016/0009-2797(79)90151-0. [DOI] [PubMed] [Google Scholar]
- Darlington G. J., Papaconstantinou J., Sammons D. W., Brown P. C., Wong E. Y., Esterman A. L., Kang J. Generation and chracterization of variants of mouse hepatoma cells with defects in hepato-specific gene expression. I. Albumin synthesis variants. Somatic Cell Genet. 1982 Jul;8(4):451–464. doi: 10.1007/BF01538707. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Farmer P. B., Foster A. B., Jarman M., Tisdale M. J. The alkylation of 2'-deoxyguanosine and of thymidine with diazoalkanes. Some observations on o-alkylation. Biochem J. 1973 Sep;135(1):203–213. doi: 10.1042/bj1350203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J. P., Coca-Prados M., Hsu M. T. Enzymatic analysis of 5-methylcytosine content in eukaryotic DNA. Study of intracellular Simian Virus 40 DNA. J Biol Chem. 1980 Aug 25;255(16):7544–7547. [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Holliday R. A new theory of carcinogenesis. Br J Cancer. 1979 Oct;40(4):513–522. doi: 10.1038/bjc.1979.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Ivarie R. D., Morris J. A. Induction of prolactin-deficient variants of GH3 rat pituitary tumor cells by ethyl methanesulfonate: reversion by 5-azacytidine, a DNA methylation inhibitor. Proc Natl Acad Sci U S A. 1982 May;79(9):2967–2970. doi: 10.1073/pnas.79.9.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarie R. D., Morris J. A., Martial J. A. Prolactin-deficient variants of GH3 rat pituitary tumor cells: linked expression of prolactin and another hormonally responsive protein in GH3 cells. Mol Cell Biol. 1982 Feb;2(2):179–189. doi: 10.1128/mcb.2.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Hemimethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981 Jun 25;9(12):2933–2947. doi: 10.1093/nar/9.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M. B., Gowans B. J., Lieberman M. W. Methylation of deoxycytidine incorporated by excision-repair synthesis of DNA. Cell. 1982 Sep;30(2):509–516. doi: 10.1016/0092-8674(82)90248-3. [DOI] [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Ivarie R. Asymmetrical distribution of CpG in an 'average' mammalian gene. Nucleic Acids Res. 1982 Dec 11;10(23):7865–7877. doi: 10.1093/nar/10.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A., Nordheim A., Nichols S. R., Rich A. 7-Methylguanine in poly(dG-dC).poly(dG-dC) facilitates z-DNA formation. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4777–4781. doi: 10.1073/pnas.78.8.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Grünwald S., Boehm T. L., Drahovsky D. Isolation and characterization of DNA cytosine 5-methyltransferase from human placenta. Biochim Biophys Acta. 1983 Aug 2;740(3):323–330. doi: 10.1016/0167-4781(83)90141-0. [DOI] [PubMed] [Google Scholar]
- Pfohl-Leszkowicz A., Fuchs R. P., Keith G., Dirheimer G. Enzymatic methylation of chicken erythrocyte DNA modified by two carcinogens, 2-(N-acetoxyacetylamino) fluorene and methylnitrosourea. Recent Results Cancer Res. 1983;84:193–201. doi: 10.1007/978-3-642-81947-6_14. [DOI] [PubMed] [Google Scholar]
- Pfohl-Leszkowicz A., Salas C., Fuchs R. P., Dirheimer G. Mechanism of inhibition of enzymatic deoxyribonucleic acid methylation by 2-(acetylamino)fluorene bound to deoxyribonucleic acid. Biochemistry. 1981 May 26;20(11):3020–3024. doi: 10.1021/bi00514a005. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Sharp J. D., Capecchi N. E., Capecchi M. R. Altered enzymes in drug-resistant variants of mammalian tissue culture cells. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3145–3149. doi: 10.1073/pnas.70.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. Reaction of guanosine with ethylating agents. Biochemistry. 1972 Oct 10;11(21):3939–3947. doi: 10.1021/bi00771a017. [DOI] [PubMed] [Google Scholar]
- Sun L., Singer B. The specificity of different classes of ethylating agents toward various sites of HeLa cell DNA in vitro and in vivo. Biochemistry. 1975 Apr 22;14(8):1795–1802. doi: 10.1021/bi00679a036. [DOI] [PubMed] [Google Scholar]
- Turnbull J. F., Adams R. L. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976 Mar;3(3):677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wigler M. H. The inheritance of methylation patterns in vertebrates. Cell. 1981 May;24(2):285–286. doi: 10.1016/0092-8674(81)90317-2. [DOI] [PubMed] [Google Scholar]
- Wilson V. L., Jones P. A. Inhibition of DNA methylation by chemical carcinogens in vitro. Cell. 1983 Jan;32(1):239–246. doi: 10.1016/0092-8674(83)90514-7. [DOI] [PubMed] [Google Scholar]