Abstract
Recent results confirm that long-term expression of therapeutic transgenes can be achieved by using a transposon-based system in primary stem cells and in vivo. Transposable elements are natural DNA transfer vehicles that are capable of efficient genomic insertion. The latest generation, Sleeping Beauty transposon-based hyperactive vector (SB100X), is able to address the basic problem of non-viral approaches – that is, low efficiency of stable gene transfer. The combination of transposon-based non-viral gene transfer with the latest improvements of non-viral delivery techniques could provide a long-term therapeutic effect without compromising biosafety. The new challenges of pre-clinical research will focus on further refinement of the technology in large animal models and improving the safety profile of SB vectors by target-selected transgene integration into genomic “safe harbors.” The first clinical application of the SB system will help to validate the safety of this approach.
Keywords: integration, non-viral, SB100X transposon, therapy, trial
Introduction
The ability to efficiently deliver foreign genes into cells offers opportunities to use gene therapy to correct certain genetic diseases and to augment cellular process to achieve a therapeutic effect. The relatively short history of gene therapy started out with an initial phase of overwhelming optimism, resulting in the first successful correction of a genetically inherited disease phenotype [1, 2]. The promise of the field was originally built on adapting virus-derived vectors for clinical use based upon their inherent gene transfer capabilities. Indeed, the vast majority of gene delivery systems currently tested in clinical trials are based on viral vectors. Depending on the cell type, viral vectors can be capable of transducing most of the infected cells. Some viral vectors, including adenoviral or adeno-associated viral vectors, remain largely episomal, requiring re-administration. However, repeated delivery may compromise efficacy [3] and might induce a severe immune response (reviewed in ref. 4). In contrast, retroviruses stably integrate their genetic cargo into a target-cell genome, resulting in robust, and stable expression of the gene(s) of interest. However, a concern of using retroviral vectors is the potential for mutagenic effects associated with the site of genomic integration [5–7]. Such risk is especially pronounced with γ-retroviral vectors based on murine leukemia virus (MLV) that have a pronounced tendency to insert into transcription start sites [8]. Furthermore, lentiviral vectors (Lenti) based on HIV are also potential mutagens because they exhibit a bias toward insertions into transcription units [9, 10]. Indeed, insertional mutagenesis has been observed in clinical trials using an MLV-based vector for gene therapy of X-linked severe combined immunodeficiency (SCID-X1) [5, 7, 11, 12]. The second-generation recombinant retroviruses may be able to address some of the inadvertent side-effects such as insertional oncogenesis. Namely, the self-inactivating (SIN) vectors harbor deletions of the promoter/enhancer elements in the long terminal repeat (LTR) [13–15], enabling transgene expression via a cellular and perhaps more physiological promoter. Another concern is that expression of the integrated transgene can be reduced or lost due to transcriptional silencing over time (reviewed in ref. 16). The clinical use of retroviral vectors can be further curtailed due to the limited size of the payload, as multiple, or large transgenes compromise the efficiency of viral reverse transcription. Finally, regulatory issues and high costs associated with manufacture of clinical-grade retrovirus hamper their widespread translation into clinical practice. An ideal therapeutic vector would combine the favorable attributes of integrating viral vectors (i.e. stable chromosomal insertion) while significantly reducing the potential for adverse events. Transposable elements (transposons) could potentially offer such an alternative.
Transposons for gene therapy
Non-viral approaches to therapeutic gene transfer can be limited by low rates of delivery and fast declining transgene expression, due to the compromised physical stability of the transgene. Recent developments of non-viral delivery techniques, including nanotechnology, nucleofection, liposomes, and cell-penetrating peptides, can significantly enhance the efficacy of nucleic acid transfer into therapeutically relevant cell types, even in vivo. However, solving the introduction of the desired nucleic material into a cell does not guarantee long-term transgene expression.
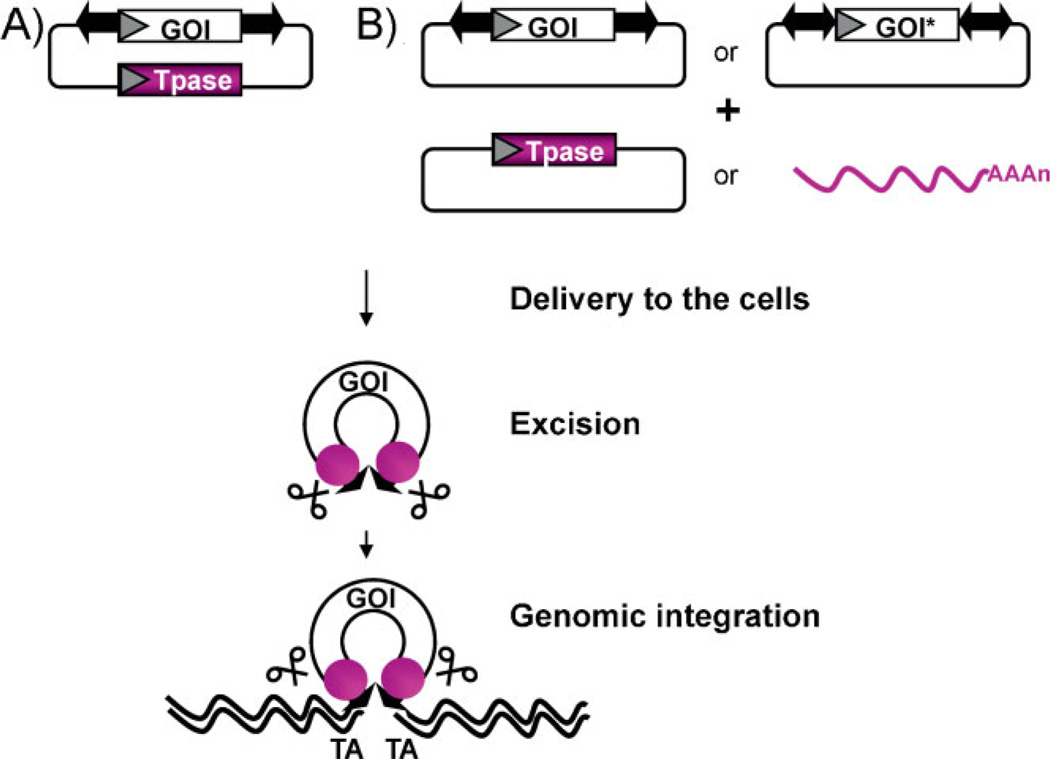
Transposable elements can be considered as natural gene delivery vehicles that, similar to retroviral vectors, are capable of efficient genomic insertion (Fig. 1). Based on transposons in fish that are presumed to have been active >10 million years ago, an ancient transposon was literally awakened after a long evolutionary “sleep” [17] in 1997, and named Sleeping Beauty (SB) after the Grimm brothers’ famous fairy tale. SB was the first transposon ever shown capable of gene transfer in mammalian cells, thereby opening up new possibilities for genetic manipulation in animal model species (reviewed in ref. 18) as well as for human gene therapy [19, 20]. As a non-viral alternative to viral vectors, the potential of the SB transposon [17] was thoroughly investigated during the last decade [20–23] (Table 1).
Figure 1.
The plasmid-based SB system. The SB plasmid-based transposon system combines the advantages of viral vectors with those of naked DNA molecules. The advantage of transposon-based gene delivery as compared to classical, plasmid-based, and non-viral delivery approaches is that, due to stable genomic insertion of expression cassettes, it can lead to both long-term and efficient transgene expression. However, in contrast to viral vectors, transposon vectors can be maintained and propagated as plasmid DNA. Making them simple and inexpensive to manufacture – an important issue regarding the implementation of future clinical trials. The SB transposon system has two functional components: a specific piece of DNA that frames a gene to be moved into the cell’s genome and a protein (the transposase) that mobilizes the transposon. Following DNA delivery to the cells, the transposase (purple) binds the terminal inverted repeats (arrows) flanking the gene of interest (GOI) and catalyzes the excision and subsequent genomic integration of the transposon. The transposase can be provided A: in cis (from the same plasmid molecule) or B: in trans in the form of a second plasmid. Notably, the transposase can be provided as mRNA as well, thereby reducing the risk of “re-hopping” of the transposon-based vector [109]. Integration occurs at TA dinucleotides, which are duplicated to flank the transposon at the site of insertion. Although the transposition efficiency decreases with increasing insert size [110], inserts of up to ~8 kb can be efficiently transferred by the SB-based vector. When the cargo is flanked by two transposons (two-ended arrows) in a “sandwich” configuration, transposition efficiencies of inserts over 8 kb (GOI*) can be significantly improved [24]. Since the transposition mechanism does not involve reverse transcription, DNA-based transposon vectors are not prone to incorporate mutations and can tolerate larger and more complex transgenes, including those containing repeated DNA motifs. Moreover, the use of SB-based gene delivery eliminates the risk of rearrangements of the expression cassette that, as part of a transposing unit of DNA, integrates into chromosomal DNA in an intact form [20].
Table 1.
Milestones in SB-mediated therapeutic approaches
| References | ||
|---|---|---|
| Vector development | ||
| V.1 | Establishment of the Sleeping Beauty (SB) transposon system | [17] |
| V.2 | The integration pattern of SB was found to be fairly random and declared to be inherently safer in comparison to other integrating vectors | [37, 40] |
| V.3 | First developments of improved vector components: hyperactive transposases and improvements to the inverted repeats | [24, 50–52, 112] |
| V.4 | The enhancer/promoter activity of the transposon vector was found negligible in comparison to viral LTR | [25–26] |
| V.5 | Proof of principle of targeted vector integration | [27, 85] |
| V.6 | Large-scale, in vitro evolution strategy yielded a hyperactive transposase mutant – comparable efficacy to viral vectors | [31] |
| Delivery technologies | ||
| D.1 | SB was proposed as a new generation non-viral therapeutic vector | [19] |
| D.2 | Sustainable gene expression in primary cells (Cd34+, primary T, and human embryonic stem cells) by nucleofection | [31, 43, 69–74] |
| D.3 | Complexing the transposon DNA with polyethylenimine (PEI) | [45, 47, 55, 59, 66] |
| D.4 | Hydrodynamic injection | [19, 46, 65, 67] |
| D.5 | Adaptation to the hydrodynamic delivery to a large animal (dog) | [103] |
| D.6 | Coated nanocapsule (intravenous injection) | [49] |
| D.7 | Proteo-liposome (intravenous injection) | [48] |
| D.8 | Hybrid transposon/viral vectors (adeno, lenti, and herpes virus) | [65, 76–78] |
| Phenotypic correction in disease models | ||
| P.1 | First demonstrations that a transposon could be used to correct patient’s cells (junctional epidermolysis bullosa) | [113] |
| P.2 | Hemophilia A | [47, 49, 58] |
| P.3 | Hemophilia B | [19, 31] |
| P.4 | Pulmonary hypertension | [45] |
| P.5 | Lung transplantation-associated complications | [47] |
| P.6 | Glioblastoma | [59] |
| P.7 | Inherited tyrosinemia | [65] |
| P.8 | Mucopolysaccharidosis | [46, 67] |
| P.9 | Crigler-Najjar syndrome type I | [48] |
| P.10 | B-lymphoid malignancies | [70, 71] |
| Clinical application | ||
| C.1 | First clinical trial approved; B-lymphoid malignancies | [80] |
The Sleeping Beauty-based non-viral, integrating system as an alternative to viral vectors
The advantages of the SB system include its reduced immunogenicity [19], relaxed limitation on the size of expression cassettes [24], and improved safety/toxicity profiles [23, 25–27]. In comparison to retroviral systems, the SB vectors have an inherently low enhancer/promoter activity [25, 26], and seem to trigger significantly milder epigenetic changes at the genomic insertion site [28]. Transgene silencing using SB-based expression vectors was found to be dependent on the cargo rather than on vector sequences [28–30], and SB is capable of supporting stable, long-term transgene expression both in vitro [30] and in vivo [19, 31]. Insertion of insulator sequences flanking the transcription units of the cargo to prevent accidental trans-activation of promoters of neighboring genes further increased the safety features of the SB system [25]. Chromosomal integration of SB transposons is precise, and no SB-associated adverse effects have been reported [20, 22]. Of note, negligible promoter/enhancer activity or precise integration mechanism do not appear to be general features of all recombinase/transposon systems. For example, the piggyBac transposon (PB) exhibits significant promoter/enhancer activity [32, 33], while the integration promoted by the bacteriophage-derived PhiC31 system was reported to generate chromosomal rearrangements [34–36].
Target-site selection of Sleeping Beauty
Similar to most other transposable elements, SB displays specificity in target site selection at the primary DNA sequence level [37]. Namely, SB exclusively integrates into TA dinucleotides. A palindromic AT-repeat consensus sequence associated with bendability and hydrogen bonding potential of the target DNA was found to affect SB’s target selection [37]. It was later shown that a unique deformation inherent in the sequence may be a recognition signal for target selection [38]. This deformation, and therefore the likelihood a particular TA will be targeted by SB within a piece of DNA, can be predicted by the V(step) algorithm [38]. Such predictions may allow us to assess theoretical risks associated with transposon insertions in particular genomic regions: an important parameter in the safety profile of SB-based gene vectors [39].
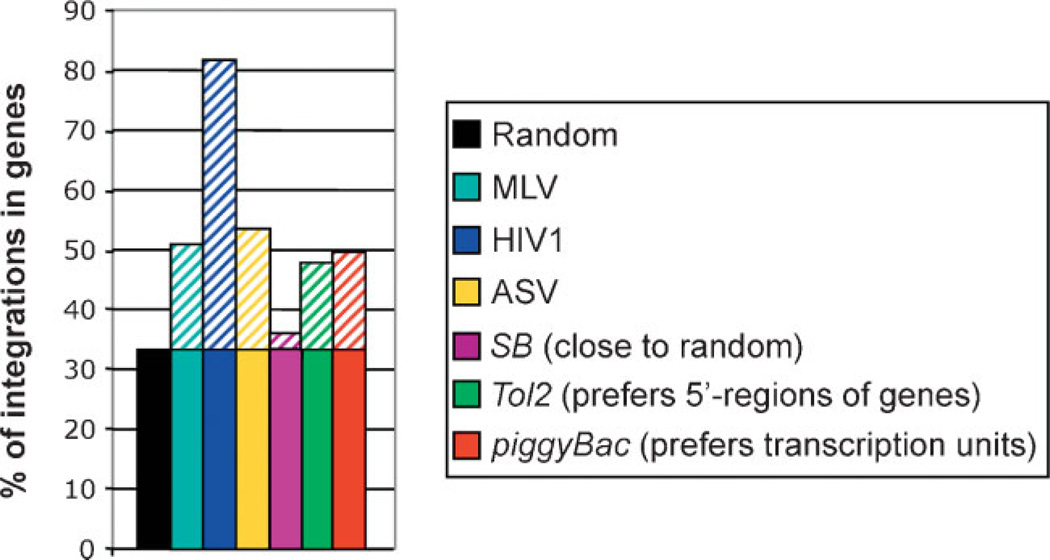
Even though SB transposition shows considerable specificity at the actual DNA sequence level, analysis of insertion sites recovered from primary and cultured mammalian cells (mouse and human) showed that SB integration can be considered fairly random on a genomic level [37, 40] (Fig. 2). However, because the SB transposon could theoretically insert into any of the ~108 TA sites in the human genome, it could potentially activate or inactivate cellular genes [6], as discussed above. Insertional mutagenesis by SB can be purposefully applied in genetic screens; however, it has to be stressed that the SB vectors used for such screens are deliberately modified to be as mutagenic as possible by incorporating strong transcriptional signals and splice sites. Such mutagenic SB vectors have been successfully utilized in cancer screens [41, 42]. As outlined above, the SB vectors utilized for gene therapy purposes are designed to avoid mutagenesis. As a possible strategy to further improve the safety profile of SB, targeted integration of the therapeutic gene to a “safe” site in the human genome would prevent possible hazards to the host cell and organism due to the problems mentioned above (see Challenges section: Target-selected transposition into genomic “safe harbors”).
Figure 2.
Genomic insertion preferences of integrating vector systems. Genomic integration of viral vector systems shows considerable preference for genes [8–10, 111]. In contrast, SB integration can be considered fairly random on a genomic level [37, 40]. Only about one-third of SB insertions occur in transcribed regions (the vast majority in introns), suggesting that SB might be a safer vector for therapeutic gene delivery than most viruses that are currently used. The genomic integration profile of piggyBac [33] or Tol2 resembles that of integrating viral vectors [30].
Despite the several advantageous features, the relatively low efficacy of plasmid-based, transposon-mediated gene transfer limited its applicability in most therapeutically relevant cell types. A recent breakthrough, however, has demonstrated that stable chromosomal insertion can be achieved from transposon-based, non-viral systems which results in robust, long-term expression of therapeutic transgenes in certain primary stem cells as well as in vivo [19, 23, 31, 43–49] (Table 1).
Hundred-fold increase in Sleeping Beauty’s activity
Unlike viruses, transposable elements have to co-exist with host cells in order to survive. Thus, transposable elements are not selected for maximal activity in nature in order to avoid insertional mutagenesis of essential genes. It was therefore likely that the reconstituted SB transposon [17] did not represent its most active form. Previously, it was known that replacing certain amino acids in the transposase could lead to a modest increase of its activity [24, 31, 50–52], and it was reasoned that perhaps a certain combination of these variants could yield a highly active enzyme. But guessing the right combinations of variants out of the millions that are possible is like finding the right combinations of numbers to win a lottery. A high throughput, PCR-based, DNA-shuffling strategy and screening in mammalian cells produced a variant of SB that was 100-fold more potent in chromosomal insertion of a transgene than the originally reconstructed protein [31]. The use of SB100X demonstrated that it is possible to establish a transposon-based, non-viral vector system, that is capable of stable gene transfer coupled with long-term gene expression at an efficiency comparable to that of viral strategies [31]. The SB100X hyperactive transposon system yields unprecedented stable gene transfer efficiencies following non-viral gene delivery into therapeutically relevant primary cell types, including stem or progenitor cells, or in vivo. Thus, the hyperactive SB100X transposase holds great promise for gene therapy by offering a likely broad utility in gene therapy, including clinical implementation of ex vivo and in vivo gene therapies (and was therefore awarded with the title “Molecule of the Year, 2009,” http://www.biotechniques.com/news/Sleeping-Beauty-named-Molecule-of-the-Year/biotechniques-187068.html?autnID=191663).
Gene transfer methods to deliver the non-viral, transposon-based vectors
Delivery in vivo
Since transposons are not capable of infection, it is necessary to provide efficient delivery of this plasmid-based system into cells. The major obstacles are the numerous barriers that the introduced DNA must traverse, including the endothelial lining of vessel walls, cellular plasma membranes, endosomal membranes, nuclear membranes, nuclease activity, and chromosomal integrity [53]. Robust delivery of naked therapeutic DNA in vivo can be particularly challenging (see Challenges section: Setting up large animal models for pre-clinical studies).
In order to exclude ectopic transgene expression, cell type-specific expression of the transgene is desirable. Using specific promoters inside the SB transposon, erythroid-specific expression of β-globin was reported for the sickle cell disease model [54]. Intravenous injection of DNA complexed with linear polyethylenimine (PEI) is one of the most effective non-viral gene delivery methods to deliver the therapeutic DNA to the lung. This approach has been used with the SB transposon to demonstrate long-term expression of luciferase in mouse lung [55]. Using the SB system, at least a 100-fold increase in expression was achieved primarily in type-2 alveolar pneumocytes. Liu et al. [56] further demonstrated that marker gene expression could be directed to endothelial cells of the lung by using the endothelian-1 promoter to regulate transgene expression from SB transposons delivered in a PEI complex. Low therapeutic levels of Hemophilia Factor VIII (FVIII) (10% of normal), but phenotypic correction of the bleeding disorder, were achieved in all animals that received the PEI-complexed FVIII-transposon [57]. In a therapeutic rat model of pulmonary hypertension, systemic administration of SB transposon harboring a constitutively active endothelial nitric oxide synthase (eNOS) gene resulted in physiological improvements, including the inhibition of induced pulmonary hypertension [45]. Furthermore, PEI-based systemic administration of SB vectors encoding human indoleamine-2,3-dioxygenase (hIDO) was evaluated in the context of lung transplantation-associated chronic complications [47]. SB gene delivery showed a remarkable therapeutic response, as evidenced by near normal pulmonary function histological appearance, and reduced collagen content in lung allografts [47]. Ohlfest et al. [58] reported that SB-mediated anti-angiogenic gene transfer significantly increased survival of mice bearing human glioblastoma xenografts. They further showed that SB-mediated gene transfer could improve the efficacy of immunotherapy by facilitating sustained cytokine expression in the glioma model [59], following direct intratumoral infusion of PEI/DNA complexes of the mIFN-γ/SB construct [59].
As an alternative to complexing with a chemical compound, the use of physical delivery systems, such as hydrodynamic injection, can overcome the low efficiency of cellular uptake [60, 61]. This procedure involves the injection of a large volume – equivalent to about 10% the weight of the mouse – of DNA/saline solution through the tail vein in less than 10 seconds, with 99% of transgene expression accumulating in the liver. The mechanism of DNA uptake is poorly understood, but appears to involve expanding liver endothelium (reviewed in ref. [62], see Challenges section: Setting up large animal models for pre-clinical studies). Thus, in comparison of using PEI-DNA conjugates, targeting of SB transposons to liver is more effective (~5-fold) by hydrodynamic delivery [63]. Indeed, hydrodynamic injection [64] in combination with SB gene transfer has been successfully applied in various animal models of monogenetic human diseases. The ability of SB to mediate stable, long-term expression in mouse liver was initially reported in the seminal work by Yant et al. [19], who reported extended expression of α1-anti-trypsin as a reporter in normal C57BL/6 mice and of human clotting factor IX as a therapeutic gene product in factor IX-deficient mice. Montini et al. [65] used fumarylacetoacetate hydrolase (FAH) mutant mice with inherited tyrosinemia to demonstrate selective outgrowth of hepatocytes that had been genetically corrected by hydrodynamics-based delivery of a FAH-encoding SB transposon. Additional, successful pre-clinical testing of the SB system has been established in disease models for hemophilia A [66] and mucopolysaccharidosis [46, 67].
Delivery systems may also take advantage of novel nanotechnology. The SB transposon system, expressing the B domain-deleted canine FVIII, has been encapsulated into hyaluronan- and asialoorosomucoid-coated nanoparticles that were successfully targeted to hepatocytes in vivo. This approach resulted in stably improved phenotype of hemophilia A mice using simple intravenous injection [49]. Similarly, successful in vivo delivery and sustainable therapeutic gene expression was demonstrated in the hepatocytes of the rat model of Crigler-Najjar syndrome type 1 following intravenous injection. In this approach, asialoglycoprotein receptor (ASGPR)-mediated endocytosis of proteo-liposomes incorporating a fusogenic glycoprotein targeted SB-based vectors to the liver [48]. Importantly, no significant host immune response was observed toward the gene delivery vehicle or the transgene product in either of the above methods [48, 49].
Delivery ex vivo
In this approach the therapeutic gene construct is introduced into a selected cell population removed from the patient, then the treated cells are infused back into the same patient. In principle, transposons can be combined with any delivery techniques developed for transferring nucleic acids into cells. The efficacy of DNA transfer is a rate-limiting factor in transposition, since transposition rate is initially dependent on the efficiency of uptake of the introduced plasmids into the cell nuclei. Although the delivery of nucleic acids into mammalian cells is assumed to be a routine procedure, there is no universal solution, and procedures must be refined for each cell type. In hard-to-transfect cells, the entry of transposon-based vectors could be significantly facilitated by nucleofection that electro-transfers nucleic acids directly to the nucleus. Nucleofection in conjunction with transposition was successfully applied for several types of stem cells, including cord blood-derived CD34+ hematopoietic progenitor cells [31, 43, 44, 68, 69], primary T cells [70–72], or human embryonic stem cells [73, 74]. Gene expression was stable and CD34+ cells could differentiate into erythroid, megakaryocytic granulocyte/ monocyte/macrophage [31] as well as to CD4+CD8+ T, CD19+ B, CD56+CD3− natural killer cells (NK), and CD33+ myeloid lineages [43]. Strikingly, the hyperactive, SB100X-transfected CD34+ cells achieved approximately 46% engraftment in NOD/SCIDγc(null) mice [31]. These results offer a new opportunity to establish transposon-based vectors for ex vivo gene therapy. Hybrid vectors that deliver the transposon components packaged into virions have also been explored [75–78]. In addition to addressing delivery of the transposon system into cells, these vectors might prove superior to either component of the hybrid. For example, the lentivirus/SB hybrid vector efficiently retargets vector insertions away from transcription units [77, 78], whereas the herpes simplex virus/SB hybrid successfully extends cargo capacity of SB [79]. In sum, the success of this non-viral, transposon-based procedure may significantly facilitate clinical realization of ex vivo gene therapy based on the genetic modification of stem cells for the treatment of hematopoietic disorders and cancer [80].
Over the past 20 years, the genetic modification of T cells using naked DNA has been inefficient, resulting in long in vitro culturing times to select for T cells bearing stable integrants. Ideally, culturing times should be short, otherwise the T cells might lose their proliferative character, compromising their therapeutic potential. Until recently, only retroviral vectors were capable of achieving stable gene transfer into T cells at efficiencies that were clinically relevant. It should be noted that T cells transduced with γ-retrovirus are typically propagated with OKT3 and interleukin (IL-2) for 2–3 weeks before harvesting for infusion. With the use of transposon-based vectors systems such as SB [71–73] or piggyBac [81, 82] stable gene transfer into T cells can now be achieved by non-viral means within approximately the same time frame. Significantly, it is possible to generate sufficiently high numbers of SB-modified T cells for clinical application within 3–4 weeks after nucleofection (see Challenges section: Translating Sleeping Beauty Transposition to cancer gene therapy by T cell engineering: Hurdles and potential solutions).
In combination with recent developments of non-viral vector delivery approaches, the SB transposon system shows considerable efficacy in reaching sustained therapeutic levels of gene expression as well as tissue-specific targeting both ex vivo and in vivo. In principle, the plasmid-based transposon system can be combined with any non-viral delivery method that is able to provide efficient, but non-toxic, plasmid DNA transfer. When combined with the transposon system, these approaches would be able to provide the desired long-term therapeutic effect without compromising biosafety, thereby setting a new standard in gene therapy.
Challenges
Target-selected transposition into genomic “safe harbors”
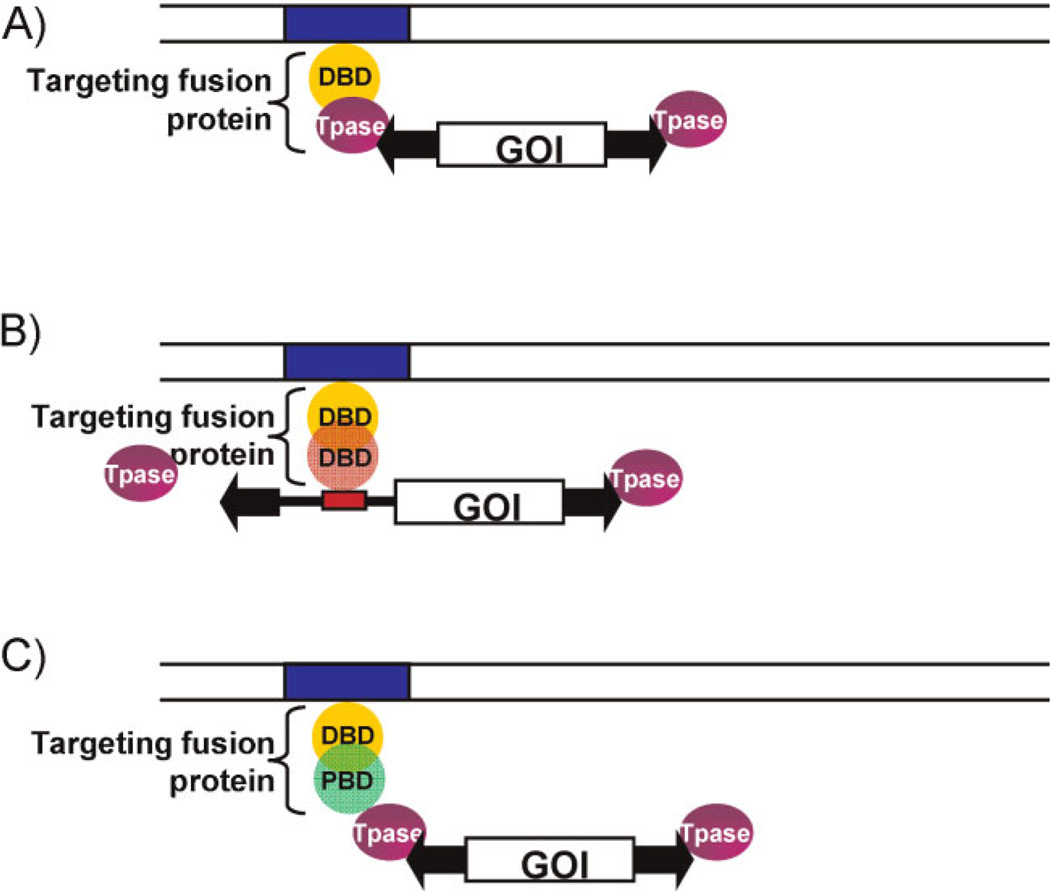
Transgene integration into selected sites in the genome could prevent hazardous effects to the organism due to insertional mutagenesis of cellular genes. We envision three distinct molecular strategies for targeted SB transposition (Fig. 3).
Figure 3.
Experimental strategies for targeting SB transposition. The common components of the targeting systems include a transposable element that contains the terminal inverted repeats (TIRs, black arrowheads) of the transposon and a GOI equipped with a suitable promoter. The transposase (purple oval) binds to the TIRs and catalyzes transposition. A DNA-binding protein domain (yellow sphere) recognizes a specific sequence (blue box) in the target DNA (parallel lines). A: Targeting with transposase fusion proteins. Targeting is achieved by fusing a specific DNA-binding protein domain to the transposase. B: Targeting with fusion proteins that bind the transposon DNA. Targeting is achieved by fusing a target-specific DBD to another DBD (red sphere) that binds to a specific DNA sequence within the transposable element (red box). In this strategy, the transposase is not modified. C: Targeting with fusion proteins that interact with the transposase. Targeting is achieved by fusing a target-specific DBD to a protein binding domain (PBD, green sphere) that interacts with the transposase. In this strategy, neither the transposase nor the transposon is modified.
The premise of the first approach is that, upon binding of the engineered transposase to a specific target site specified by the heterologous DNA-binding domain (DBD), transposon insertion may occur in adjacent regions (Fig. 3A). There is some evidence for the feasibility of using transposase fusions to target insertions to specific sites to a certain extent. Namely, fusions of the bacterial IS30 transposase with the λ repressor and with the DBD of the transcription factor Gli1 showed altered insertions profiles in Escherichia coli and in zebrafish embryos, respectively, using plasmid targets [83]. Furthermore, direct fusions of the Mos1 and piggyBac transposases with the GAL4 DBD have been shown to retain transpositional activity, and to result in site-selective transposon insertion in a plasmid-to-plasmid experimental setup in mosquito embryos [84]. The SB transposase only tolerates N-terminal fusions, and fusions of HSB5 (a third-generation improved SB transposase) with the GAL4 and E2C (a synthetic, zinc finger protein recognizing an 18-bp target site in the 5′-untranslated region of the human erbB-2 gene) DBDs resulted in a drop in transposition efficiency to ~20–26% of unfused HSB5 [85]. Nevertheless, these fusion transposases clustered around upstream activating sequence (UAS) sites and E2C binding sites, respectively, as compared with integration patterns mediated by unfused HSB5 in plasmid-based assays in cultured human cells. However, cell-based assays failed to detect targeting of the E2C binding site in the genomic context. Similarly, the artificial three-finger protein Jazz, binding to a 9-bp sequence in the promoter region of the human utrophin gene [86], failed to direct targeted insertion when fused to the SB transposase [27], suggesting lack of sufficient specificity at the level of binding to the target.
The second strategy is based on a fusion protein with dual DNA-binding activity that has the capacity to bind to two DNA molecules that contain binding sites of the respective fusion partners, thereby bringing them into close proximity (Fig. 3B). A similar mechanism of bridging of DNA molecules by proteins might act in targeting some P element transposon vectors in Drosophila. P element insertion is essentially random at the genome scale. However, P elements containing regulatory sequences from the engrailed gene show some insertional specificity by frequently inserting near the endogenous, parental gene [87, 88]. Potential targeting of SB by such a mechanism was assessed by engineering a LexA operator into a benign site within an SB transposon vector (Fig. 3A). Targeted transposition events into endogenous chromosomal MAR sequences, as well as a chromosomally integrated tetracycline response element, were recovered by employing targeting fusion proteins containing LexA and either the SAF-box, a protein domain that binds to chromosomal matrix attachment regions (MARs), or the tetracycline repressor (TetR) [27]. Thus, this strategy shows promise, because it does not measurably interfere with the transposition process.
The third strategy for targeted SB transposition is based on protein-protein interactions between a targeting protein and the SB transposase (Fig. 3C). Either naturally occurring or engineered transposase interactors may tether the transpositional machinery to specific DNA sites, potentially leading to integration into nearby regions. For example, based upon observations for a role of LEDGF/p75 in directing HIV integration into expressed transcription units, in vitro studies have shown increased integration near λ repressor binding sites by fusing either the full-length LEDGF/p75 or the LEDGF/p75 retroviral integrase (IN)-binding domain to the DBD of phage λ repressor protein [89]. In an analogous fashion, Sir4p (which mediates targeted insertion of the yeast Ty5 retrotransposon into heterochromatin) fused to the E. coli LexA DBD was shown to result in integration hot spots for Ty5 near LexA operators [90]. Such a strategy was successfully adapted for targeted SB transposition by co-expressing the SB transposase with a targeting fusion protein consisting of a specific DBD and a subdomain of the SB transposase (termed N57) that mediates protein-protein interactions between transposase subunits [27]. Targeted transposition into a chromosomally integrated tetracycline response element using a TetR-N57 fusion was monitored in human cells [27]. Using this strategy, >10% of cells receiving transposon insertions contained at least one transposition event within the targeted chromosomal region. A significant advantage of this technology as compared to direct transposase fusions is that the transposase polypeptide does not have to be modified; thus, potential negative effects on transposase activity are eliminated.
There are several factors affecting site-selectivity of integrating vector systems. These include primary sequence and physical structure of the DNA at the targeted region, accessibility of specific chromosomal sites determined by chromatin components, expression of endogenous proteins that may compete for binding, and the specificity as well as capacity of chimeric proteins in DNA-binding as well as in catalytic functions. Three major challenges remain to be dealt with during the development of this technology:
Future work will have to focus on the identification of applicable, endogenous chromosomal target sites that fulfill the criteria for a genomic “safe harbor,” and on the selection of DNA-binding proteins that can be exploited for efficiently targeting transposition into those sites in vivo. In this respect, artificial zinc fingers offer a potentially superior resource: their modular character in structure and function provides a key advantage in engineering of proteins that are able to recognize theoretically any sequence in the human genome [91].
Direct fusions of DBDs to transposase proteins appear to interfere with the biochemical activities of the transposase; thus, a systematic evaluation of protein spacer sequences linking the two fusion partners will be required in order to allow rational design of direct transposase fusions.
A major hurdle for targeting systems engineered from promiscuously integrating vectors (such as SB) are the considerable off-target insertions in the context of the human genome. Although these hurdles are yet to be overcome before technologies of targeted gene insertion can be considered for applications, recent evidence suggests that target-selected transgene insertion into desired regions in the human genome is a realistic goal.
Setting up large animal models for pre-clinical studies
The effectiveness of the hydrodynamic procedure for non-viral gene transfer in mice has led to its application in large animal models (Table 2). Using an ex vivo infusion approach in livers for transplantation, injection volumes of 40–70% the liver weight were found to be effective in gene delivery with expression that varied about 100-fold [92]. However, direct infusion without surgery is preferred. Owing to the large sizes of the animals, and the consequential huge volumes of fluid that would be required for rapid delivery, direct infusion of plasmid DNA into the liver has been the preferred route. For instance, assuming a dog’s liver is about 3% of its total body mass [63, 93], the scale-up from a 20 g mouse to a 6 kg dog with a liver weighing 200 g would only be about tenfold. Hydrodynamic delivery in the mouse requires injection of 10% the animal mass, which, if applied to the dog’s liver, would be an increase in volume from 2 to 20 mL. However, the mouse’s liver appears to take up a disproportionate amount of the injected solution, in which case the injected volume for the 200 g liver could be as high as 200 mL. The amount of DNA delivered must be also be scaled accordingly. Experimental results from hydrodynamic injections directly into the livers of larger animals including rats, rabbits [94, 95], dogs [96–99], and pigs [100] suggest that about 2 mg/kg total body weight of DNA in injection volumes nearly equivalent to the total liver volume are most effective (Table 2).
Table 2.
Hydrodynamic infusion to liver
| Model | Method | DNA mg (µg/g) | VOL (ml/sec) | RATE | References |
|---|---|---|---|---|---|
| Mouse (0.02–0.03 kg) | |||||
| Tail-vein | 0.025 (1)* | 2 ml | >0.2 ml/sec | [60, 61, 64] | |
| Rat (0.2–0.3 kg) | |||||
| IVC catheter | 0.8 (80) | 15 ml | 0.4 ml/sec | [97] | |
| IVC catheter | 0.15 (110) | 3 ml | 0.4 ml/sec | [112] | |
| IVC catheter | 0.8 (100) | 16 ml | ~2 ml/sec | [113] | |
| Rabbit (~3–4 kg) | |||||
| IVC catheter | 60 (2) | 60 ml | ~2 ml/sec | [94] | |
| Dog (~5–20 kg) | |||||
| IVC catheter | 10–30 (90) | 600 ml | ~2 ml/sec | [97, 98] | |
| IVC catheter | 11 (110) | 200 ml | 20 ml/sec | [103] | |
| Pig (~20–30 kg) | |||||
| IVC catheter | 3 (6) | 150 ml | 5 ml/sec | [114] | |
| IVC catheter | 4 (5) | 100 ml | 7.5 ml/sec | [100] | |
| SVC catheter | 20–50 (33–80) | 300 ml | 20 ml/sec | [115] | |
| IVC catheter | 18 (40) | 370 ml | 1.7 ml/sec | [116] | |
| SLL** catheter | 80 (320) | 800 ml | 40 ml/sec | [101] | |
| SLL** catheter | 60 (~300) | 600 ml | 40 ml/sec | [102] | |
| Human (n = 14) | |||||
| Hepatic vein catheter | 1–45* | 50–300 ml | >1–6 ml/sec | [115] |
Amounts of infused DNA are calculated per weight of the targeted tissue, except where denoted by an asterisk, in which case amounts are for the entire animal. Unless data are specifically reported, the values for DNA delivered per kg target organ weight were calculated assuming the liver was 3% the weight of rabbits, dogs and pigs.
Selected liver lobe.
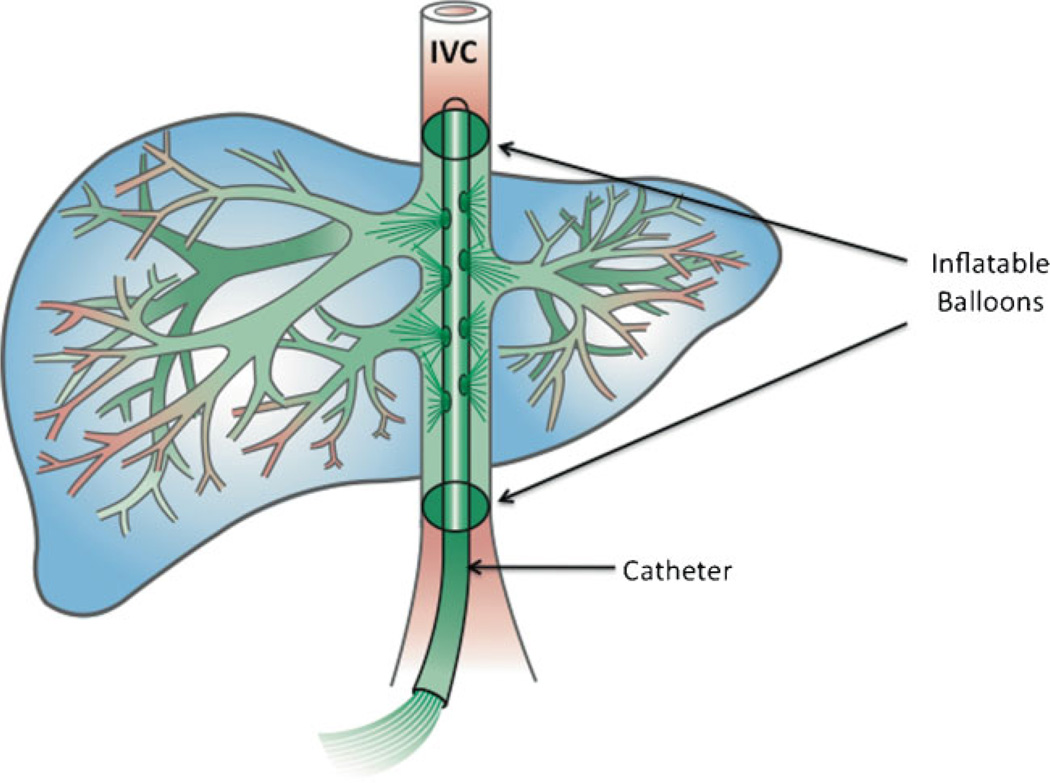
There are several potential access routes into the liver, including the hepatic veins (the route that is taken in hydro-dynamically infused mice via the tail vein), the portal vein (which is relatively large and can accommodate large volumes of fluid), and the hepatic arteries. Most studies have employed catheter-mediated delivery to the liver via the hepatic vein (Fig. 4). The problem has been a lack of sustained expression equivalent to that in mice. In general transgene expression in large animals has been transient and generally less than 1% that attained in the mouse following hydrodynamic tail-vein injection. Despite the number of variables that have been analyzed, it is not clear whether it is the pressure, impulse, volume, or some other parameter that is the critical variable required to attain high efficiency gene transfer to hepatocytes in larger animals. There is hope, however: relatively high levels of transient transgene expression in porcine liver have been achieved when hepatic-vein infusion into particular lobes was computer-controlled to maintain a constant pressure with occlusion of the inferior vena cava (IVC) and portal vein [101, 102]. Hydrodynamic infusion of SB transposons in dog liver via the hepatic veins has resulted in sustained transgene activity for several weeks [103].
Figure 4.
Balloon-catheter-based DNA delivery to liver in large animals. Catheter-mediated delivery to the liver is made via the hepatic vein with incisions into the femoral or jugular vein followed by snaking the catheter through the IVC. Inflatable balloons on the catheter(s) are used to occlude appropriate veins and arteries to restrict outflow from the liver. The most common approach (Table 2) has been to insert a one or more balloon catheters into the IVC from entry points in either a femoral or jugular vein. With a double-balloon catheter, the DNA solution can be infused into the entire liver following temporary occlusion of the IVC above and below the access sites of the hepatic veins. In addition to the catheter that is used to deliver the DNA solution, other balloon catheters may be introduced into vessels such as arteries and/or the portal vein to isolate or semi-isolate the liver by blocking outflow of the infused solution. The DNA is shown in green exiting from multiple ports in the catheter to the large hepatic veins and hence into the sinusoids. The infusion is retrograde to normal blood flow. Figure drawn by Lynn Fellman.
Extrapolating from methods used in in-bred lines of mice to out-bred large animals requires caution. Some lines of mice can produce unexpected findings such as the promoter-dependent gender effects on transgene expression [46]. We have found that high levels of canine erythropoietin (EPO) expressed from SB transposons in dog liver have unexpectedly transient expression compared to canine secreted alkaline phosphatase delivered under similar conditions [103]. As a result of the variations in promoters, reporter genes, and variations in methods, it is difficult to compare relative efficiencies of various strategies for gene delivery in larger animals. Nevertheless, the achievements of stable, therapeutic levels of cell type-specific transgene expression following systemic, intravenous injection using the combination of the non-viral SB vector with nanocapsule delivery or the proteo-liposome system are encoraging [48, 49]. The observation that, in addition to efficacy, these protocols did not provoke a massive immune response provides an important step toward successful human clinical trials of hepatic metabolic diseases.
Translating Sleeping Beauty transposition to cancer gene therapy by T cell engineering: Hurdles and potential solutions
The clinical application of gene therapy relies on the transfer of research outcomes from the laboratory bench to the patient’s bedside. Successful investigators practice translational science so that data collected in humans can be relayed back to the design of new pre-clinical experiments, which govern the planning of next-generation trials (Fig. 5). The bench → bedside → bench algorithm requires a gene therapy approach that is cost-effective, can be adapted to changing needs, minimizes risk of genotoxicity, and can be readily implemented. The SB system meets many, if not all of these requirements, and serves as a new approach to non-viral gene transfer for therapeutic human application.
Figure 5.
The circuit algorithm of clinical trials. The SB system lends itself to translation science, as gene therapy applications can be developed in the laboratory, assessed in humans and the results used to refine subsequent bench research for future clinical trials.
The first clinical application of the SB system will be tested using autologous T cells genetically modified to redirect specificity for B-lineage malignancies [80]. Lymphocytes are a suitable initial platform for testing new gene transfer systems as there have been hundreds of infusions of clinical-grade T cells genetically modified using viral and non-viral approaches without apparent toxicity stemming from the gene transfer event [104]. The SB transposon to be introduced in the first-in-human application carries a chimeric antigen receptor (CAR) to render the T cells specifically cytotoxic toward CD19+ B-lineage tumors [70, 71]. The second generation CD19-specific CAR (designated CD19RCD28) uses a murine monoclonal antibody (mAb) scFv region to bind to the antigen, which is fused in frame to a mAb Fc region to form the exodomain. This is coupled to a fusion of chimeric CD28 and CD3-ζ to provide T cells with an endodomain to achieve the CD19-specific activation events of cytolysis, cytokine production, and proliferation [105]. First, the two DNA plasmids coding for (i) the CD19-specific CAR and (ii) the SB transposase (early-generation version SB11 [50]), will be electro-transferred into T cells ex vivo. In a second step, CAR+ T cells will be selectively propagated on clinical-grade γ-irradiated CD19+ artificial antigen presenting cells (aAPC), derived from a tumor cell line, K562, in the presence of exogenous cytokines. The testing to release these genetically modified T cells for a phase I trial will include (i) measurements of sterility (bacteria, fungi, mycoplasma, and endotoxin), (ii) identity (presence of CD3+ T cells and absence of CD32+ K562-aAPC, and CD19+ contaminating tumor cells), (iii) transgene expression (presence of Fc+ cells), (iv) chain of custody (product matches donor using low resolution HLA class I typing), and (v) absence of autonomous cell growth (measured by a control of T cells cultured without aAPC and cytokines). These CAR+ T cells are to be infused after autologous hematopoietic stem-cell transplantation to try to prevent relapse in patients with advanced B-lymphoid disease.
Like all genetic therapy trials, this application of the SB system for therapeutic gene transfer has potential risks and benefits. To ready the SB system for clinical application in T cells, a PCR assay was established to demonstrate that the transposase was below minimal detection in the genomes of treated cells. Thus, the risk of “re-hopping” of the integrated CD19RCD28 transposon appears mitigated by the absence of continued SB transposase activity. Fluorescent in situ hybridization and quantitative PCR (Q-PCR) demonstrated that, following nucleofection and propagation, there is on average one stably integrated copy of the CD19RCD28 transposon detectable per T cell genome [81]. A full discussion of the potential for SB system to cause genotoxicity can be found in a recent review [106]. In summary, the SB system appears ready for human application infusing CAR+ T cells to express a CAR to target aggressive malignancies.
One upside of using the SB system for the genetic modification of clinical-grade T cells includes the reduced cost associated with manufacturing clinical-grade DNA plasmids compared with manufacturing cost for recombinant viral vectors. This reduced cost facilitates improvements to the CAR design and development of CARs targeting other tumor-associated antigens. Currently, aAPC can be used to select and numerically expand the genetically modified T cells from those that do not express CAR (Fig. 6). By inclusion of cytokines or expression of co-stimulatory molecules on the aAPC, T cells can be programmed for desired function, such as preservation of central memory or naive T cell phenotypes of improved in vivo persistence [107, 108]. Indeed, our approach to using the introduced CAR to drive T cell expansion and the enforced expression of T cell co-stimulatory molecules on the aAPC (CD86, CD137L, and membrane-bound IL-15), leads to outgrowth of T cells with central memory phenotype [81] with retained telomere length [81]. Furthermore, the protocol might be adapted for hyperactive SB100X [31] instead of the less active version of the SB transposase (SB11 [50]).
Figure 6.
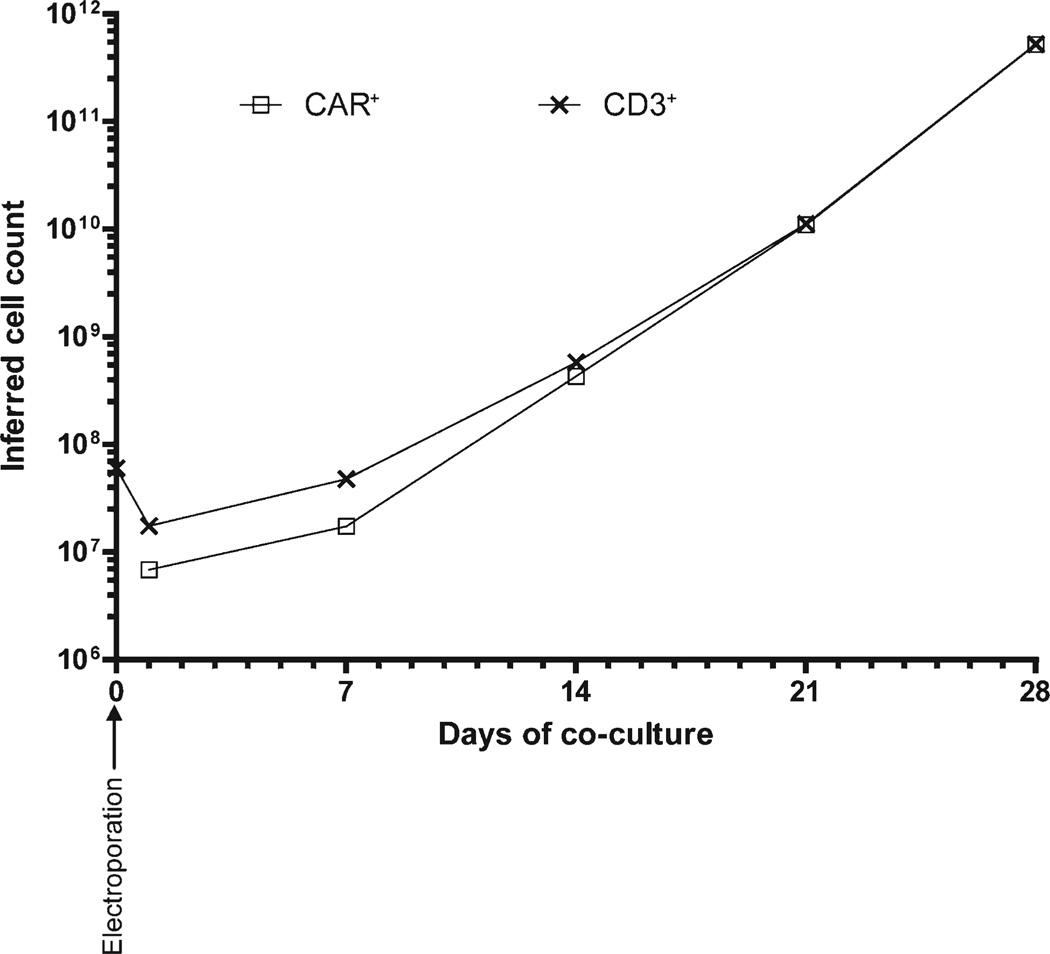
Kinetics of numeric expansion of genetically modified T cells. Primary human T cells from peripheral blood can be nucleofected ex vivo to express a CD19-specific CAR and the genetically modified T cells can be selectively propagated by recursive passaging (every 7 days) on γ-irradiated CD19+ aAPC in presence of soluble cytokines. The graph shows the kinetics (from 0 to 28 days) of numeric expansion of CD3+ and CAR+ T cells after electro-transfer of SB plasmids on day 0. This time of ex vivo propagation can lead to differentiation of T cells and potential for replicative senescence. However, this period of tissue culture also presents opportunities as the T cells can be monitored for undesired autonomous cell growth and the culturing environment can be manipulated. Within 28 days, almost all T cells express CAR at numbers suitable for adoptive transfer in clinical trials.
Outlook
Non-viral vectors equipped with an integrating feature are far more efficient for long-term therapeutic gene expression than the non-viral gene transfer approaches used previously. The desired transgenes expressed from DNA plasmids can be introduced using non-viral methods which should permit investigations of gene therapy to be undertaken by researchers that do not have ready access to production facilities to generate clinical-grade virus. Research in the past decade has developed enhanced vectors for SB-mediated gene transfer into primary cell types, both ex vivo and in vivo, that benefit from reduced immunogenicity, lack of a strict size limitation of the therapeutic expression cassette, improved safety/toxicity profiles, and can achieve long-term transgene expression. In particular, the SB100X hyperactive system has the potential to yield gene transfer, as well as expression at efficiencies previously unprecedented with any non-viral vector system. The next phase of pre-clinical research will focus on further refinement in large animal models to undertake SB-mediated transposition in vivo and improving the safety profile of SB vectors by target-selected transgene integration into genomic “safe harbors”. The first application of the SB system is currently undergoing regulatory review. No matter the outcome in terms of clinical efficacy, this trial will help validate the safety of this approach and allow investigators to revisit the design of DNA vectors in general to improve therapeutic effect in subsequent next-generation trials.
Acknowledgments
We acknowledge the financial support of EU FP6 (INTHER), EU FP7 (PERSIST), the Deutsche Forschungsgemeinschaft (SPP1230 “Mechanisms of gene vector entry and persistence”) to Z. Iz. and Z. Iv., NIH grants RO1CA124782 and R01CA120956 to L. J. C. and NIH grant 1R01DK082516 to P. B. H. P. B. H. has equity in Discovery Genomics, Inc., a small biotech company that receives funding from the NIH to explore the use of the Sleeping Beauty Transposon System for gene therapy.
Abbreviations
- aAPC
artificial antigen presenting cells
- ASGPR
asialoglycoprotein receptor
- CAR
chimeric antigen receptor
- DBDs
DNA-binding domains
- eNOS
endothelial nitric oxide synthase
- EPO
erythropoietin
- FAH
fumarylacetoacetate hydrolase
- FVIII
Hemophilia Factor VIII
- FIX
Hemophilia Factor IX
- hIDO
human indoleamine-2,3-dioxygenase
- IL
interleukin
- IN
retroviral integrase
- IVC
inferior vena cava
- Lenti
lentiviral vector
- LTR
long terminal repeat
- mAb
murine monoclonal antibody
- MAR
matrix attachment region
- MLV
murine leukemia virus
- NK
natural killer cells
- PB
piggyBac transposon
- PEI
polyethylenimine
- Q-PCR
quantitative PCR
- SB
Sleeping Beauty
- SB100X
hyperactive transposon system
- SCID-X1
X-linked severe combined immunodeficiency
- SIN
self-inactivating
- TetR
tetracycline repressor
- UAS
upstream activating sequence
References
- 1.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, Le Deist F, Carlier F, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 3.Riviere C, Danos O, Douar AM. Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Ther. 2006;13:1300–1308. doi: 10.1038/sj.gt.3302766. [DOI] [PubMed] [Google Scholar]
- 4.Hartman ZC, Appledorn DM, Amalfitano A. Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Res. 2008;132:1–14. doi: 10.1016/j.virusres.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 6.Baum C, von Kalle C, Staal FJ, et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Li Y, Crise B, et al. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 9.Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 10.Schroder AR, Shinn P, Chen H, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 11.Thrasher AJ, Gaspar HB, Baum C, et al. Gene therapy: X-SCID transgene leukaemogenicity. Nature. 2006;443:E5–E6. doi: 10.1038/nature05219. discussion E6–7. [DOI] [PubMed] [Google Scholar]
- 12.Kustikova O, Fehse B, Modlich U, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 13.Schambach A, Galla M, Maetzig T, et al. Improving transcriptional termination of self-inactivating gamma-retroviral and lentiviral vectors. Mol Ther. 2007;15:1167–1173. doi: 10.1038/sj.mt.6300152. [DOI] [PubMed] [Google Scholar]
- 14.Schambach A, Mueller D, Galla M, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- 15.Modlich U, Navarro S, Zychlinski D, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 17.Ivics Z, Hackett PB, Plasterk RH, et al. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 18.Ivics Z, Li MA, Mates L, et al. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yant SR, Meuse L, Chiu W, et al. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 20.Ivics Z, Izsvak Z. Transposons for gene therapy. Curr Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- 21.Izsvak Z, Ivics Z. Sleeping Beauty transposition: biology and applications for molecular therapy. Mol Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 22.Fernando S, Fletcher BS. Sleeping Beauty transposon-mediated nonviral gene therapy. BioDrugs. 2006;20:219–229. doi: 10.2165/00063030-200620040-00003. [DOI] [PubMed] [Google Scholar]
- 23.Vandendriessche T, Ivics Z, Izsvak Z, et al. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood. 2009;26:26. doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- 24.Zayed H, Izsvak Z, Walisko O, et al. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Walisko O, Schorn A, Rolfs F, et al. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther. 2008;16:359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- 26.Moldt B, Yant SR, Andersen PR, et al. Cis-acting gene regulatory activities in the terminal regions of Sleeping Beauty DNA transposon-based vectors. Hum Gene Ther. 2007;18:1193–1204. doi: 10.1089/hum.2007.099. [DOI] [PubMed] [Google Scholar]
- 27.Ivics Z, Katzer A, Stuwe EE, et al. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Park CW, Sjeklocha L, et al. High-level genomic integration, epigenetic changes, and expression of Sleeping Beauty transgene. Biochemistry. 2010;49:1507–1521. doi: 10.1021/bi9016846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrison BS, Yant SR, Mikkelsen JG, et al. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabundzija I, Irgang M, Mates L, et al. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mates L, Chuah MK, Belay E, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 32.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 34.Ehrhardt A, Engler JA, Xu H, et al. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Jeppesen I, Nielsen K, et al. PhiC31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 36.Chalberg TW, Genise HL, Vollrath D, et al. phiC31 integrase confers genomic integration and long-term transgene expression in rat retina. Invest Ophthalmol Vis Sci. 2005;46:2140–2146. doi: 10.1167/iovs.04-1252. [DOI] [PubMed] [Google Scholar]
- 37.Vigdal TJ, Kaufman CD, Izsvak Z, et al. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Geurts AM, Yae K, et al. Target-site preferences of Sleeping Beauty transposons. J Mol Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 39.Geurts AM, Hackett CS, Bell JB, et al. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 2006;34:2803–2811. doi: 10.1093/nar/gkl301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yant SR, Wu X, Huang Y, et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collier LS, Carlson CM, Ravimohan S, et al. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 42.Dupuy AJ, Akagi K, Largaespada DA, et al. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 43.Xue X, Huang X, Nodland SE, et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;4:4. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- 44.Izsvak Z, Chuah MK, Vandendriessche T, et al. Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods. 2009;49:287–297. doi: 10.1016/j.ymeth.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Liu H, Visner G, et al. Sleeping Beauty-mediated eNOS gene therapy attenuates monocrotaline-induced pulmonary hypertension in rats. FASEB J. 2006;20:2594–2596. doi: 10.1096/fj.06-6254fje. [DOI] [PubMed] [Google Scholar]
- 46.Aronovich EL, Bell JB, Khan SA, et al. Systemic correction of storage disease in MPS I NOD/SCID mice using the Sleeping Beauty transposon system. Mol Ther. 2009;17:1136–1144. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Liu L, Fletcher BS, et al. Sleeping Beauty-based gene therapy with indoleamine 2,3-dioxygenase inhibits lung allograft fibrosis. FASEB J. 2006;20:2384–2386. doi: 10.1096/fj.06-6228fje. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Sarkar DP, Mani P, et al. Long-term reduction of jaundice in Gunn rats by nonviral liver-targeted delivery of Sleeping Beauty transposon. Hepatology. 2009;50:815–824. doi: 10.1002/hep.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kren BT, Unger GM, Sjeklocha L, et al. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J Clin Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geurts AM, Yang Y, Clark KJ, et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 51.Yant SR, Park J, Huang Y, et al. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baus J, Liu L, Heggestad AD, et al. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 53.Wagner E, Culmsee C, Boeckle S. Targeting of polyplexes: toward synthetic virus vector systems. Adv Genet. 2005;53PA:333–354. doi: 10.1016/S0065-2660(05)53013-X. [DOI] [PubMed] [Google Scholar]
- 54.Zhu J, Kren BT, Park CW, et al. Erythroid-specific expression of beta-globin by the Sleeping Beauty transposon for Sickle cell disease. Biochemistry. 2007;46:6844–6858. doi: 10.1021/bi6024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belur LR, Frandsen JL, Dupuy AJ, et al. Gene insertion and long-term expression in lung mediated by the Sleeping Beauty transposon system. Mol Ther. 2003;8:501–507. doi: 10.1016/s1525-0016(03)00211-9. [DOI] [PubMed] [Google Scholar]
- 56.Liu L, Sanz S, Heggestad AD, et al. Endothelial targeting of the Sleeping Beauty transposon within lung. Mol Ther. 2004;10:97–105. doi: 10.1016/j.ymthe.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 57.Liu L, Mah C, Fletcher BS. Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice using an endothelial-targeted Sleeping Beauty transposon. Mol Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 58.Ohlfest JR, Demorest ZL, Motooka Y, et al. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the Sleeping Beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther. 2005;12:778–788. doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
- 59.Wu A, Oh S, Ericson K, et al. Transposon-based interferon gamma gene transfer overcomes limitations of episomal plasmid for immunogene therapy of glioblastoma. Cancer Gene Ther. 2007;14:550–560. doi: 10.1038/sj.cgt.7701045. [DOI] [PubMed] [Google Scholar]
- 60.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 61.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 62.Suda T, Kamimura K, Kubota T, et al. Progress toward liver-based gene therapy. Hepatol Res. 2009;39:325–340. doi: 10.1111/j.1872-034X.2008.00479.x. [DOI] [PubMed] [Google Scholar]
- 63.Podetz-Petersen KM, Bell JB, Steele TW, et al. Gene expression in lung and liver after intravenous infusion of polyethyleneimine complexes and hydrodynamic delivery of Sleeping Beauty transposons. Hum Gene Ther. 2010;21:210–220. doi: 10.1089/hum.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell JB, Podetz-Pedersen KM, Aronovich EL, et al. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montini E, Held PK, Noll M, et al. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol Ther. 2002;6:759–769. doi: 10.1006/mthe.2002.0812. [DOI] [PubMed] [Google Scholar]
- 66.Ohlfest JR, Frandsen JL, Fritz S, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 67.Aronovich EL, Bell JB, Belur LR, et al. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hollis RP, Nightingale SJ, Wang X, et al. Stable gene transfer to human CD34(+) hematopoietic cells using the Sleeping Beauty transposon. Exp Hematol. 2006;34:1333–1343. doi: 10.1016/j.exphem.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 69.Sumiyoshi T, Holt NG, Hollis RP, et al. Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells using the Sleeping Beauty transposon system. Hum Gene Ther. 2009;18:18. doi: 10.1089/hum.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang X, Guo H, Kang J, et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh H, Manuri PR, Olivares S, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang X, Wilber A, McIvor RS, et al. DNA transposons for modification of human primary T lymphocytes. Methods Mol Biol. 2009;506:115–126. doi: 10.1007/978-1-59745-409-4_9. [DOI] [PubMed] [Google Scholar]
- 73.Wilber A, Linehan JL, Tian X, et al. Efficient and stable transgene expression in human embryonic stem cells using transposon-mediated gene transfer. Stem Cells. 2007;25:2919–2927. doi: 10.1634/stemcells.2007-0026. [DOI] [PubMed] [Google Scholar]
- 74.Orban TI, Apati A, Nemeth A, et al. Applying a “double-feature” promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells. 2009;27:1077–1087. doi: 10.1002/stem.45. [DOI] [PubMed] [Google Scholar]
- 75.Yant SR, Ehrhardt A, Mikkelsen JG, et al. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- 76.Bowers WJ, Mastrangelo MA, Howard DF, et al. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 77.Staunstrup NH, Moldt B, Mates L, et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther. 2009;24:24. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vink CA, Gaspar HB, Gabriel R, et al. Sleeping Beauty transposition from nonintegrating lentivirus. Mol Ther. 2009;17:1197–1204. doi: 10.1038/mt.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Silva S, Mastrangelo MA, Lotta LT, Jr, et al. Extending the transposable payload limit of Sleeping Beauty (SB) using the herpes simplex virus (HSV)/SB amplicon-vector platform. Gene Ther. 2009;29:29. doi: 10.1038/gt.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williams DA. Sleeping Beauty vector system moves toward human trials in the United States. Mol Ther. 2008;16:1515–1516. doi: 10.1038/mt.2008.169. [DOI] [PubMed] [Google Scholar]
- 81.Raja Manuri PV, Wilson MH, Maiti SN, et al. piggyBac transposon/transposase system to generate CD19-specific T cells for treatment of B-lineage malignancies. Hum Gene Ther. 2009;21:427–437. doi: 10.1089/hum.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakazawa Y, Huye LE, Dotti G, et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–836. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szabo M, Muller F, Kiss J, et al. Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett. 2003;550:46–50. doi: 10.1016/s0014-5793(03)00814-7. [DOI] [PubMed] [Google Scholar]
- 84.Maragathavally KJ, Kaminski JM, Coates CJ. Chimeric Mos1 and piggyBac transposases result in site-directed integration. FASEB J. 2006;20:1880–1882. doi: 10.1096/fj.05-5485fje. [DOI] [PubMed] [Google Scholar]
- 85.Yant SR, Huang Y, Akache B, et al. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Corbi N, Libri V, Fanciulli M, et al. The artificial zinc finger coding gene ’Jazz’ binds the utrophin promoter and activates transcription. Gene Ther. 2000;7:1076–1083. doi: 10.1038/sj.gt.3301204. [DOI] [PubMed] [Google Scholar]
- 87.Hama C, Ali Z, Kornberg TB. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- 88.Kassis JA, Noll E, VanSickle EP, et al. Altering the insertional specificity of a Drosophila transposable element. Proc Natl Acad Sci USA. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ciuffi A, Diamond TL, Hwang Y, et al. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- 90.Zhu Y, Dai J, Fuerst PG, et al. Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mandell JG, Barbas CF. Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsoulfas G, Takahashi Y, Liu D, et al. Hydrodynamic plasmid DNA gene therapy model in liver transplantation. J Surg Res. 2006;135:242–249. doi: 10.1016/j.jss.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 93.Evans HE. Miller’s Anatomy of the Dog. 3rd edition. Philadelphia, PA: WB Saunders Co.; 1983. p. 451. [Google Scholar]
- 94.Eastman SJ, Baskin KM, Hodges BL, et al. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum Gene Ther. 2002;13:2065–2277. doi: 10.1089/10430340260395910. [DOI] [PubMed] [Google Scholar]
- 95.Dulak J, Schwarzacher SP, Zwick RH, et al. Effects of local gene transfer of VEGF on neointima formation after balloon injury in hyper-cholesterolemic rabbits. Vasc Med. 2005;10:285–291. doi: 10.1191/1358863x05vm630oa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chapman GD, Lim CS, Gammon RS, et al. Gene transfer into coronary arteries of intact animals with a percutaneous balloon catheter. Circ Res. 1992;71:27–33. doi: 10.1161/01.res.71.1.27. [DOI] [PubMed] [Google Scholar]
- 97.Zhang G, Vargo D, Budker V, et al. Expression of naked plasmid DNA injected into the afferent and efferent vessels of rodent and dog livers. Hum Gene Ther. 1997;8:1763–1772. doi: 10.1089/hum.1997.8.15-1763. [DOI] [PubMed] [Google Scholar]
- 98.Huang GM, Li GS, Zhu GY, et al. Safety and bioactivity of intracoronary delivery of naked plasmid DNA encoding human atrial natriuretic factor. Acta Pharmacol Sin. 2002;23:609–611. [PubMed] [Google Scholar]
- 99.Hagstrom JE, Hegge J, Zhang G, et al. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 100.Alino SF, Herrero MJ, Noguera I, et al. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007;14:334–343. doi: 10.1038/sj.gt.3302873. [DOI] [PubMed] [Google Scholar]
- 101.Suda T, Suda K, Liu D. Computer-assisted hydrodynamic gene delivery. Mol Ther. 2008;16:1098–1104. doi: 10.1038/mt.2008.66. [DOI] [PubMed] [Google Scholar]
- 102.Kamimura K, Suda T, Xu W, et al. Image-guided, lobe-specific hydrodynamic gene delivery to swine liver. Mol Ther. 2009;17:491–499. doi: 10.1038/mt.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hackett PB, Urness M, Bell JB, et al. Use of non-invasive baloon catheters placed under fluoroscopic guidance for delivery of the Sleeping Beauty transposon system under pressure to the liver of dogs. Mol Ther. 2008;16:S312. [Google Scholar]
- 104.Bonini C, Grez M, Traversari C, et al. Safety of retroviral gene marking with a truncated NGF receptor. Nat Med. 2003;9:367–369. doi: 10.1038/nm0403-367. [DOI] [PubMed] [Google Scholar]
- 105.Kowolik CM, Topp MS, Gonzalez S, et al. CD28 costimulation provided through a CD19-specific chimeric antigen receptor enhances in vivo persistence and antitumor efficacy of adoptively transferred T cells. Cancer Res. 2006;66:10995–11004. doi: 10.1158/0008-5472.CAN-06-0160. [DOI] [PubMed] [Google Scholar]
- 106.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berger C, Jensen MC, Lansdorp PM, et al. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J Clin Invest. 2008;118:294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hinrichs CS, Borman ZA, Cassard L, et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc Natl Acad Sci USA. 2009;106:17469–17474. doi: 10.1073/pnas.0907448106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilber A, Frandsen JL, Geurts JL, et al. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 110.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 111.Narezkina A, Taganov KD, Litwin S, et al. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cui Z, Geurts AM, Liu G, et al. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 113.Ortiz-Urda S, Lin Q, Yant SR, et al. Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 2003;10:1099–1104. doi: 10.1038/sj.gt.3301978. [DOI] [PubMed] [Google Scholar]