Abstract
Aim
This study compared the changes on the subgingival microbiota of subjects with “refractory” periodontitis (RP) or treatable periodontitis (GR) before and after periodontal therapy by using the Human Oral Microbe Identification Microarray (HOMIM).
Methods
Individuals with chronic periodontitis were classified as RP (n=17) based on mean attachment loss (AL) and/or >3 sites with AL ≥2.5 mm after scaling and root planing, surgery and systemically administered amoxicillin and metronidazole or as GR (n=30) based on mean attachment gain and no sites with AL ≥2.5 mm after treatment. Subgingival plaque samples were taken at baseline and 15 months after treatment and analyzed for the presence of 300 species by HOMIM analysis. Significant differences in taxa before and after therapy were sought using the Wilcoxon test.
Results
The majority of species evaluated decreased in prevalence in both groups after treatment; however, only a small subset of organisms was significantly affected. Species that increased or persisted in high frequency in RP but were significantly reduced in GR included Bacteroidetes sp., Porphyromonas endodontalis, Porphyromonas gingivalis, Prevotella spp., Tannerella forsythia, Dialister spp., Selenomonas spp., Catonella morbi, Eubacterium spp., Filifactor alocis, Parvimonas micra, Peptostreptococcus sp. OT113, Fusobacterium sp. OT203, Pseudoramibacter alactolyticus, Streptococcus intermedius or Streptococcus constellatus and Shuttlesworthia satelles. In contrast, Capnocytophaga sputigena, Cardiobacterium hominis, Gemella haemolysans, Haemophilus parainfluenzae, Kingella oralis, Lautropia mirabilis, Neisseria elongata, Rothia dentocariosa, Streptococcus australis and Veillonella spp. were more associated with therapeutic success.
Conclusion
Persistence of putative and novel periodontal pathogens, as well as low prevalence of beneficial species was associated with chronic “refractory” periodontitis.
Keywords: Refractory Periodontal Disease, Non-surgical periodontal therapy, antimicrobials, DNA microarrays, Microbiology
Introduction
The major goal of periodontal therapy is to promote a shift from a predominantly pathogenic microbiota to a host-compatible one in order to achieve clinical and microbiological stability.1,2 In most cases, therapeutic success is obtained for long periods of time.3 However, a small proportion of treated subjects, referred to as “refractory” periodontitis (RP) patients does not respond well to properly performed conventional therapy and continues to show loss of periodontal attachment.4,5 RP subjects have been shown to comprise a very heterogeneous group in terms of clinical, microbiological and immunological characteristics.6–14 Among several factors that may contribute to “refractory” disease is the existence of specific pathogenic microbiotas.12–17 In fact, studies have reported different microbial profiles associated with RP. 6,11,14,16 In general, putative periodontal pathogens are detected in high frequency and levels in these subjects,10,14,17 although “unusual” species have also been observed in relatively high prevalence.6,10,16 In a previous paper,16 we analyzed the baseline subgingival microbiota of subjects with periodontal health and severe chronic periodontitis who were successfully treated (Good Responders; GR) or who responded poorly (RP) to combined mechanical and antimicrobial treatment as measured by using the Human Oral Microbe Identification Microarray (HOMIM) technique. Our data showed that chronic periodontitis subjects who were “refractory” to treatment harbored significantly higher proportions of putative periodontal pathogens including members of the orange and red complexes, as well as microorganisms not usually associated with periodontitis in comparison to GR or periodontally healthy individuals. Ideally, lowering classical periodontal pathogens should lead to clinical improvement and long-lasting periodontal stability in most periodontitis patients.1,2,8,15,18 In some RP individuals, however, reduced levels of pathogens may not be sufficient to achieve periodontal stability.8,15 This may be due to some immunodeficiency in the host and/or to the predominance of an unusual pathogenic microbiota comprising of newly recognized or “uncultivable” species. Combined mechanical and antimicrobial periodontal therapies may be very effective in reducing classical periodontal pathogens, but the impact of these therapies on unusual microbial profiles of periodontitis is unknown. Thus, the current investigation evaluated the post-therapy changes on the subgingival microbiota of periodontitis patients who were RP or GR as measured by using the HOMIM technique reported in a previous paper.16
Material and methods
Subject Population
As described in Colombo et al.,16 the study subject population was comprised of 47 chronic periodontitis patients in good general health, recruited between April 2003 and February 2007, from the Clinical Center for Periodontal Research at The Forsyth Institute, and the Clinical Research Center at the Boston University Goldman School of Dental Medicine. Informed consent was obtained from all enrolled individuals. The study protocol was reviewed and approved by the Institutional Review Boards at The Forsyth Institute and Boston University Medical Center. All subjects had at least 20 natural teeth and were over 20 years of age. Chronic periodontitis was defined as ≥5 sites with pocket depth (PD) and attachment loss (AL) ≥6 mm at baseline. Exclusion criteria included pregnancy, lactation, systemic conditions that could affect the progression or treatment of periodontal diseases, and any known allergy to amoxicillin and/or metronidazole. In addition, subjects who had received systemic antibiotics or periodontal therapy in the previous 6 months were excluded.
Clinical Monitoring
Periodontal clinical measurements were performed at 6 sites per tooth at all teeth excluding third molars.19 They included PD and clinical attachment level (CAL) measured using a North Carolina periodontal probe#, presence or absence of gingival redness, suppuration, bleeding on probing, and supragingival plaque accumulation. PD and CAL measurements were repeated within a subject at each visit. Duplicate measurements were compared to ensure a standard deviation of <1.0 mm, and the means of pairs of CAL measurements at different visits used to determine disease progression. Examiners at Boston University (A. Serrenho and M. Martins) and at The Forsyth Institute (C. Roberts, L. Murray, and G. Torresyap) were trained and calibrated at every 6 months on periodontal measurements. The same examiner performed all measurements for each subject at every visit to further minimize the inter-examiner variability. Clinical measurements were taken at baseline and at 3, 6, 9, 12 and 15 months post-therapy.
Treatment Protocol
After completion of baseline monitoring, subjects with evidence of destructive periodontal disease received full mouth scaling and root planing (SRP) under local anesthetic,18 and instruction in proper home care procedures within a 4-week period. Approximately 2 months after the completion of SRP, the subjects were re-examined and received modified Widman flap surgery in quadrants with residual pockets >4 mm. At the first surgical visit, subjects were asked to take 500 mg amoxicillin and 250 mg metronidazole 3 times daily for 14 days. The surgical/antibiotic phase was completed within 4 weeks. Subjects then entered a maintenance and clinical monitoring phase with visits at 3, 6, 9, 12 and 15 months where clinical measurements, maintenance scaling and reinforcement of home care procedures were performed. GR had full mouth mean CAL gain and no sites with AL ≥2.5 mm during the one year maintenance period. Any subject showing mean AL, and/or >3 sites with AL ≥2.5 mm from the baseline visit to any monitoring visit within 1 year post-therapy was defined as having RP. 10
Subgingival Sample Taking and Isolation of Bacterial DNA
When a subject was diagnosed as “refractory”, subgingival samples were taken from the same sites evaluated at baseline and from sites that lost attachment post-therapy. The subject was then withdrawn from the study protocol to receive other periodontal treatments. For all other subjects, samples were taken for microbiological analysis at baseline and at the 15 month visit. After removal of supragingival biofilm with sterile gauze, individual subgingival biofilm samples were taken from the mesio-buccal aspect of up to 14 teeth in different quadrants (1 and 3 or 2 and 4) per subject using sterile periodontal curettes#. Each sample was placed in separate 1.5 ml tubes containing 50 μl of TE (50 mM Tris-HCl, 1 mM EDTA, pH 7.6). For bacterial DNA extraction, 44 μl of each sample was taken, and 0.5% Tween 20 ** and 1 μl of Proteinase K (10 mg/ml) †† were added. The samples were heated at 55°C for 2 h, and at 95°C for 5 min for inactivation of Proteinase K. All samples were stored in a freezer at −80°C prior to PCR amplification.
Human Oral Microbe Identification Microarray (HOMIM)
The HOMIM methodology here employed was described in detail in our previous study.16 Briefly, a total of 400 16S rRNA-based, reverse-capture oligonucleotide probes‡‡ targeting over 300 bacterial taxa 20,21 (Table S1 in the supplemental material) were printed on aldehyde-coated glass slides§§. 16S rRNA genes were PCR amplified from DNA extracts|||| using 16S rRNA universal forward and reverse primers and labeled via incorporation of Cy3-dCTP¶¶ in a second nested PCR. The labeled 16S amplicons were hybridized overnight to probes on the slides. After washing, the microarray slides were scanned## and crude data was extracted using a software for microarray image analysis***. Data were normalized by comparing individual signal intensities to the average of signals from universal probes.
Statistical Analysis
Statistical analyses were performed using a statistical package software†††. Full-mouth clinical measurements were computed for each subject and then averaged across subjects within the 2 clinical groups at each visit. Differences in demographic and clinical parameters between groups were sought using Mann-Whitney and Chi-square tests at baseline, and Wilcoxon signed rank test between pre- and post-therapy. The microbial data were generated from image files of scanned arrays using a HOMIM online analysis tool.22 Detection of a particular species in a sample was determined by the presence of a fluorescent spot for that unique probe. Qualified and adjusted spots of the same probe were then summed and a mean intensity calculated to represent the signal intensity for each specific probe. Signals were normalized by comparing individual signal intensities to the average of signals for the universal probes. Any original signal <2 times the background value was reset to 1 and was assigned to the signal level 0 (no signal for the corresponding probe). All the values >1 were categorized into scores 1 to 5, corresponding to different signal levels. The prevalence of each species (frequency of scores 0 to 5) was computed within groups using sites and patients as units of analysis. Significant differences between groups were sought using the Mann-Whitney and Chi-square tests, and between pre- and post-therapy using the Wilcoxon Signed Rank test. The significance level was set at 5%.
Results
The demographic features of the subject groups have been described in an earlier paper (Colombo et al. 2009).16 No significant differences regarding age, gender or race were observed between GR (48 ± 10 years of age; 60% males; 40% White, 43% African-Americans, 17% others, respectively) and RP (51 ± 11 years of age; 82% males; 53% White, 24% African-Americans, 23% others, respectively). In contrast, a significantly higher frequency of smokers was observed in the RP group (59%) compared to GR (23%; p < 0.01, Chi-square test). Table 1 shows the full-mouth clinical characteristics of the 2 groups at baseline and post-therapy. All clinical parameters of periodontal tissue destruction and inflammation were similar between groups at baseline, except for mean CAL and visible plaque which were significantly greater in the RP than GR group (p < 0.05; Mann-Whitney test). The GR showed significant improvement in all clinical parameters, except suppuration (p<0.05, Wilcoxon test). In the RP group, significant mean reductions were detected only for PD and bleeding on probing. In contrast, a slight but significant increase in mean CAL was observed in this group (p<0.05, Wilcoxon test). A total of 488 subgingival plaque samples were obtained at baseline (n=254 samples) and post-therapy (n=234) from both clinical groups for HOMIM analysis. The clinical features of these selected sites are presented in Table 2. Sites from the GR group showed a better clinical outcome compared to sites from RP patients. Significant reductions were observed for all periodontal parameters (p<0.05, Wilcoxon and Chi-square tests), except gingival redness. Conversely, sites from subjects in the RF group presented an increase in CAL (p<0.05, Wilcoxon test) and visible plaque (p<0.05, Chi-square test) at post-therapy. Out of the 91 samples from the RP group that were evaluated after treatment, 66 were sites that lost ≥2.5 mm of attachment; i.e. they were considered as “refractory” sites. These sites presented significantly higher mean PD (4.8±1.9) and CAL (6.5±2.6) than the other sites from the GR or RP groups (p<0.05, Mann-Whitney test; data not shown).
Table 1.
Full-mouth clinical parameters (mean ± SD) of good responders (GR) and “refractory” periodontitis (RP) subjects of the study population, at baseline and post-therapy.
| Clinical Parameters | Baseline | Post-therapy | ||
|---|---|---|---|---|
|
| ||||
| GR (n = 30) | RP (n = 17) | GR (n = 30) | RP (n = 17) | |
| Probing depth (mm) †‡§ | 3.7 ± 1.3 | 3.9 ± 0.9 | 2.7 ± 0.7 | 3.5 ± 1.0 |
| Clinical attachment (mm) *†‡§ | 3.2 ± 1.5 | 4.2 ± 1.2 | 2.6 ± 1.4 | 4.5 ± 1.2 |
| Mean % of sites with: | ||||
| Bleeding on probing ‡§ | 56 ± 25 | 53 ± 28 | 25 ± 15 | 37 ± 23 |
| Visible plaque *†§ | 46 ± 30 | 65 ± 29 | 28 ± 24 | 61 ± 29 |
| Gingival redness § | 85 ± 20 | 88 ± 18 | 75 ± 30 | 82 ± 29 |
| Suppuration | 0.2 ± 0.7 | 0.3 ± 0.6 | 0 | 0.4 ± 0.9 |
p < 0.05 refers to significant differences between GR and RP groups at baseline, and
p < 0.05 at post-therapy (Mann-Whitney test);
p < 0.05, refers to significant differences within the RP group pre- and post-therapy, and
p < 0.05, refers to significant differences within the GR group pre- and post-therapy (Wilcoxon Signed Rank test).
Table 2.
Clinical parameters of the sites sampled from the two clinical groups, good responders (GR) and “refractory” periodontitis (RP) subjects, at baseline and post-therapy.
| Clinical Parameters | Baseline | Post-therapy | ||
|---|---|---|---|---|
|
| ||||
| GR (n = 143) | RP (n = 111) | GR (n = 143) | RP (n = 91) | |
| Probing depth (mm, mean±SD) †‡ | 4.3 ± 2.2 | 4.9 ± 2.1 | 3.0 ± 1.3 | 4.4 ± 1.8 |
| Clinical attachment (mm, mean±SD) *†‡§ | 3.7 ± 2.4 | 4.7 ± 2.6 | 2.6 ± 1.5 | 5.8 ± 2.7 |
| % of sites with: | ||||
| Bleeding on probing †‡ | 63.6 | 69.4 | 31.2 | 59.7 |
| Visible plaque †‡§ | 47.6 | 59.5 | 33.5 | 81.0 |
| Gingival redness | 83.0 | 85.6 | 78.8 | 88.1 |
| Suppuration ‡ | 2.8 | 0 | 0 | 0 |
p < 0.05 refers to significant differences between GR and RP groups at baseline; and
p < 0.05 at post-therapy (Mann-Whitney or Chi-square test);
p < 0.05 refers to significant differences within the GP group pre- and post-therapy (Wilcoxon Signed Rank or Chi-square test);
p < 0.05 refers to significant differences within the RP group pre- and post-therapy (Wilcoxon Signed Rank or Chi-square test).
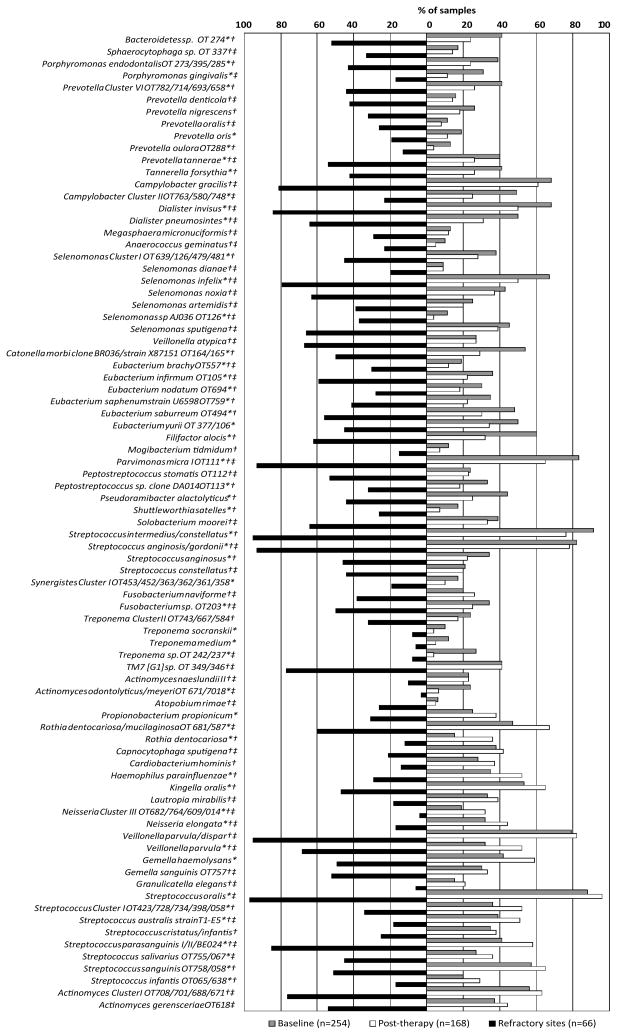
Overall changes in the frequency of bacterial species/clusters between baseline and 15 months post-therapy, as well as the frequency of these organisms in “refractory” sites are presented in figure 1. Most species/cluster decreased significantly in prevalence after treatment (p<0.05, Chi-square test), particularly Bacteroidetes sp. OT 274, Porphyromonas gingivalis, Prevotella spp., Tannerella forsythia, Filifactor alocis, Dialister spp., Selenomonas spp., Eubacterium spp., Treponema spp., Parvimonas micra, Peptostreptococcus spp., Pseudoramibacter alactolyticus, Shuttleworthia satelles, Fusobacterium sp. OT 203, Streptococcus anginosus/gordonii and Synergistes Cluster I OT453/452/363/362/361/358. Conversely, significant increases in frequency of detection post-therapy were observed for Propionibacterium propionicum, Rothia dentocariosa or mucilagenosa, Haemophilus parainfluenzae, Kingella oralis, Neisseria Cluster III OT682/764/609/014, Neisseria elongata, Veillonella parvula, Gemella haemolysans, and Streptococcus spp. (p<0.05, Chi-square test). After therapy, when “refractory” sites were compared to “non-refractory” sites, many species including various periodontal pathogens were detected significantly more often in “refractory” sites, except for R. dentocariosa, Capnocytophaga sputigena, Cardiobacterium hominis, H. parainfluenzae, K. oralis, Lautropia mirabilis, Neisseria Cluster III, N. elongata, Granulicatella elegans, Streptococcus Cluster I OT423/728/734/398/058, Streptococcus australis, Streptococcus cristatus/infantis, Streptococcus sanguinis and Streptococcus infantis which were found in higher frequency in “non-refractory” sites (p<0.05, Chi-square test).
Figure 1.
Bar chart of the frequency of bacterial species/cluster in subgingival samples from all subjects at baseline and 15 months post-therapy, and from sites that lost ≥2.5 mm of attachment (“refractory” sites) post-therapy in “refractory” periodontitis patients. OT means oral taxon designation. *Refers to significant differences between all samples at baseline and post-therapy. †Refers to significant differences between samples from “refractory” sites and sites at 15 months post-therapy. ‡Refers to significant differences between samples from “refractory” sites and sites at baseline (p<0.05, Chi-square test).
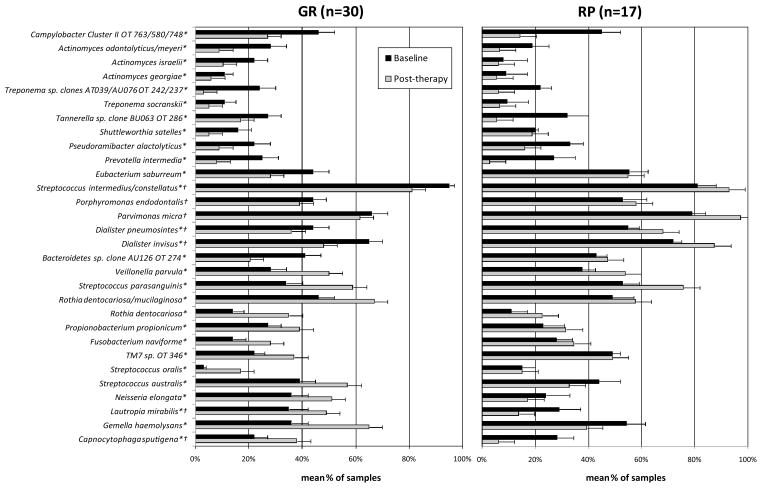
Figure 2 shows the significant changes in mean frequency of samples colonized by bacterial species at baseline and post-therapy in the RP and GR groups. In this analysis, the post-therapy samples from RP subjects included samples from “refractory” and “non-refractory” sites. On average, only a small subset of species was significantly affected after treatment in both groups. Interestingly, some species that increased significantly after therapy in GR such as C. sputigena, G. haemolysans, L. mirabilis, N. elongata, Streptococcus australis (p<0.05, Wilcoxon test) diminished in the RP group. On the other hand, Bacteroidetes sp. clone AU126 OT 274, Dialister spp., P. micra, S. satelles, Eubacterium saburreum, Porphyromonas endodontalis, and S. intermedius/constellatus reduced in frequency in GR and increased or did not changed in RP subjects.
Figure 2.
Bar chart of changes in mean frequency (±SD) of bacterial species from baseline to post-therapy in samples from “good responders” (GR) and “refractory” periodontitis (RP) subjects. OT means oral taxon designation. *Refers to significant differences between baseline and post-therapy in the GR group. †Refers to significant differences between baseline and post-therapy in the RP group (p<0.05; Wilcoxon test).
Discussion
In our earlier paper,16 we used the HOMIM technique to discriminate subjects with “refractory” periodontitis from individuals who were successfully treated or who were periodontally healthy based upon their baseline subgingival microbial profiles. We showed that periodontitis patients presented a greater diversity in the subgingival microbiota compared to healthy individuals at baseline, and that major putative periodontal pathogens were found more commonly in RP than in GR. Moreover, RP individuals harbored higher frequencies of several species not commonly associated with periodontitis. The authors concluded that RP patients present a distinct microbial profile compared to patients in the GR and periodontally healthy groups. This observation led us to the question of how periodontal treatment would impact these different microbial profiles, resulting in “refractory” or successfully treated periodontitis. Thus, in the current investigation, we evaluated the changes in the subgingival microbiota of the 47 severe chronic periodontitis patients between baseline and up to 15 months after combined mechanical (SRP and Widman flap surgery)/antimicrobial (500 mg amoxicillin plus 250 mg metronidazole 3 times daily for 14 days) periodontal therapy. Of these patients, 30 subjects were considered GR and 17 were RP subjects. “Refractory” disease was defined as presence of mean attachment loss, and/or > 3 sites with attachment loss ≥ 2.5 mm from the baseline visit to any monitoring visit within 1 year post-therapy.10 As reported in other studies,1,2,18,23–25 species highly associated with periodontitis decreased significantly after treatment in the subgingival microbiota of both periodontitis groups, whereas host-compatible microorganisms increased (Fig. 1). When post-therapy data comparing “refractory” and “non-refractory” sites were analyzed, we found that several known pathogens such as P. gingivalis, T. forsythia, Prevotella spp., E. nodatum, E. saburreum, P. micra, the “milleri” streptococci group (S. anginosus, S. constellatus and S. intermedius) persisted in high frequency in “refractory” sites after treatment (Fig. 1). Likewise, a significant post-therapy increase in the mean frequency of P. micra and S. constellatus/S. intermedius was observed in RP individuals, whereas these species decreased in GR patients (Fig. 2). These findings are in accord with other investigations that reported an association between the persistence of these pathogenic species and treatment failure or periodontal attachment loss.1,6,8,9,10,14,16,17,23,27,28 Based on our data and data reported in the literature, one could speculate that significant reductions in major pathogens took place shortly after treatment (commonly at 3 months post-therapy) in both periodontitis groups.1,2,15,18,24,25 GR were able to maintain low frequency and proportions of these organisms, whereas RP patients were more rapidly colonized by these species.1,14,15 Conceivably, RP may be incapable of maintaining low levels of putative periodontal pathogens due to host impairment29,30 and/or colonization by a more virulent periodontal microbiota, including unknown pathogenic taxa.10,16 On the other hand, the high frequency of pathogenic species in RP subjects may be due to a poor plaque control and persistence of deeper pockets after treatment in this group. Although all patients received mechanical and antimicrobial therapy, and were maintained in a supportive periodontal therapy, RP individuals presented high scores of supragingival plaque accumulation at the final monitoring visit. It is well documented that supragingival plaque removal may directly affect the counts and composition of the subgingival microbiota, leading to reductions in major periodontal pathogens, even in deep pockets.31,32 Other authors, however, have reported that instrumentation of supragingival plaque alone is not enough to prevent disease recurrence and to lead to a microbiota compatible with periodontal health.33 Thus, in RP subjects therapeutic approaches including a more intense regimen of professional mechanical and/or chemical plaque control may be warranted since they have been shown to be more effective in reducing pathogens and controlling long-term progression of disease.8,15
The subgingival habitat is colonized by a great diversity of microorganisms, including novel species, species that cannot yet be grown or are difficult to grow in culture.34–37 Although studies have correlated species not commonly considered disease-associated,10,16,36–40 the role of these organisms in periodontitis is still unknown. Some species that we previously reported to be more prevalent at baseline in RP than GR subjects16 decreased significantly after treatment in responsive sites/patients but were kept in high frequencies in “refractory” disease. Among those are Bacteroidetes sp. OT 274, Prevotella tannerae, Dialister invisus, Dialister pneumosintes, F. alocis, Selenomonas spp., Eubacterium spp., Fusobacterium sp. OT203, S. satelles, Peptostreptococcus sp. OT113, and Catonella morbi OT 164/165. Some of these spcies have been previously recognized,41 whereas others are novel species or phylotypes.16,35–37,42 Investigators have strongly indicated F. alocis and D. pneumosintes as candidate periodontal pathogens.36,40,43 In a recent paper, Heller and co-workers40 evaluated the 6-month impact of combined mechanical and antimicrobial therapy on many bacterial species of the subgingival microbiota in generalized aggressive periodontitis subjects. The authors found D. pneumosintes and P. tannerae in significantly higher mean levels in sites that lost attachment than successfully treated sites after therapy, corroborating our results. Moreover, the authors reported that most of the non-periodontal species tended to increase in mean counts after treatment. Based on these data, it is reasonable to state that individuals “refractory” to therapy are even more heterogeneous in terms of their periodontal microbiota than previously reported.14 Different microbial profiles, including not only combinations of known species but also novel species may exist, and consequently, significant differences in treatment response.
Species associated with periodontal health and/or clinical improvement 16,34,38,41,44 were also differently affected in subjects from the GR and RP groups. While some species increased significantly in responsive sites but were detected in lower frequency in RP after treatment, others, particularly Veillonella parvula or V. dispar, Gemella sanguinis, Streptococcus parasanguinis and Actinomyces Cluster I were more prevalent in “refractory” sites. In other studies, Actinomyces species, S. sanguinis and C. sputigena have been detected in lower prevalence in RP compared to healthy, well-maintained elderly and periodontitis subjects.6,10,14–16 In contrast, some “refractory” patients harbor high proportions of Actinomyces spp. and V. parvula.14,15 Among the species we found to be associated with periodontal health,16 limited information is available regarding the role of L. mirabilis, C. hominis, K. oralis, H. parainfluenzae, N. elongata, G. haemolysans and G. elegans in oral/periodontal homeostasis. Many of these species are common colonizers of the normal human oropharyngeal microbiota and are considered opportunistic pathogens associated with bacteremia/septicemia and endocarditis in immunocompromised individuals.35,36,44–50
In summary, combined mechanical and antimicrobial therapy was effective in reducing the prevalence of the majority of subgingival species, particularly putative periodontal pathogens in GR subjects. In contrast, periodontal pathogens and other potential pathogenic novel species or phylotypes persisted in high prevalence in sites and/or subjects with RP. New therapeutic approaches consisting of intensive mechanical therapy and different antimicrobial combinations should be developed in order to reduce the burden of pathogenic microorganisms, to increase a wide variety of host-compatible species and to achieve long-term clinical stability in individuals presenting RP.
Supplementary Material
Summary of findings.
Periodontitis patients presented a highly diverse subgingival microbiota. Putative and novel periodontal pathogens persisted in higher frequency in sites/subjects with “refractory” periodontitis as compared to good responders, whereas presumptive beneficial species were detected in lower prevalence in RP than GR after therapy.
Acknowledgments
Sources of support: Supported by NIDCR grants DE11443 and RR025771; CAPES, Brazil.
This work was supported by NIDCR grants DE11443 and RR025771; and by National Council for Scientific and Technological Development (CAPES), Brazil.
Footnotes
Hu-Friedy, Chicago, IL
Sigma-Aldrich, Inc., St. Louis, MO
Promega, Madison, WI
Sigma Genosys, The Woodlands, TX
Schott-Nexterion® Slide AL, Louisville, KY
QIAquick® PCR Purification Kit, Qiagen Inc., Valencia, CA
GE Healthcare Bio-Sciences Corp., Piscataway, NJ
Axon GenePix® 4000B, MDS Analytical Technologies, Sunnyvale, CA
Microarray Image Analysis GenePix Pro 6.0, MDS Analytical Technologies, Sunnyvale, CA
Statistical Package for the Social Sciences - SPSS Inc®.v.16 Chicago, IL, USA
The authors report no conflicts of interest related to this study.
References
- 1.Haffajee AD, Teles RP, Socransky SS. The effect of periodontal therapy on the composition of the subgingival microbiota. Periodontol 2000. 2006;42:219–258. doi: 10.1111/j.1600-0757.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 2.Teles RP, Haffajee AD, Socransky SS. Microbiological goals of periodontal therapy. Periodontol 2000. 2006;42:180–218. doi: 10.1111/j.1600-0757.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindhe J, Westfelt E, Nyman S, Socransky SS, Haffajee AD. Long-term effect of surgical/non-surgical treatment of periodontal disease. J Clin Periodontol. 1984;11:448–458. doi: 10.1111/j.1600-051x.1984.tb01344.x. [DOI] [PubMed] [Google Scholar]
- 4.Hirschfeld L, Wasserman B. A long-term survey of tooth loss in 600 treated periodontal patients. J Periodontol. 1978;49:225–237. doi: 10.1902/jop.1978.49.5.225. [DOI] [PubMed] [Google Scholar]
- 5.MacFall WT., Jr Tooth loss in 100 treated patients with periodontal disease. A long-term study. J Periodontol. 1982;53:539–549. doi: 10.1902/jop.1982.53.9.539. [DOI] [PubMed] [Google Scholar]
- 6.Haffajee AD, Socransky SS, Dzink JL, et al. Clinical, microbiological and immunological features of subjects with refractory periodontal diseases. J Clin Periodontol. 1988;15:390–398. doi: 10.1111/j.1600-051x.1988.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 7.Choi JI, Nakagawa T, Yamada S, Takazoe I, Okuda K. Clinical, microbiological and immunological studies in recurrent periodontal disease. J Clin Periodontol. 1990;17:426–434. [PubMed] [Google Scholar]
- 8.Collins JG, Offenbacher S, Arnold RR. Effects of a combination therapy to eliminate Porphyromonas gingivalis in refractory periodontitis. J Periodontol. 1993;64:998–1007. doi: 10.1902/jop.1993.64.10.998. [DOI] [PubMed] [Google Scholar]
- 9.Magnusson I, Marks RG, Clark WB, Walker CB, Low SB, MacArthur WP. Clinical, microbiological and immunological characteristics of subjects with “refractory” periodontal disease. J Clin Periodontol. 1991;18:291–299. doi: 10.1111/j.1600-051x.1991.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 10.Colombo AP, Haffajee AD, Dewhirst FE, et al. Clinical and microbiological features of refractory periodontitis subjects. J Clin Periodontol. 1998;25:169– 180. doi: 10.1111/j.1600-051x.1998.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 11.Colombo AP, Eftimiadi C, Haffajee AD, Cugini MA, Socransky SS. Serum IgG2 level, Gm(23) allotype and FcgammaRIIa and FcgammaRIIIb receptors in refractory periodontal disease. J Clin Periodontol. 1998;25:465–474. doi: 10.1111/j.1600-051x.1998.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 12.Colombo AP, Sakellari D, Haffajee AD, Tanner A, Cugini MA, Socransky SS. Serum antibodies reacting with subgingival species in refractory periodontitis subjects. J Clin Periodontol. 1998;25:596–604. doi: 10.1111/j.1600-051x.1998.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 13.Colombo AP, Haffajee AD, Smith CM, Cugini MA, Socransky SS. Discrimination of refractory periodontitis subjects using clinical and laboratory parameters alone and in combination. J Clin Periodontol. 1999;26:569–576. doi: 10.1034/j.1600-051x.1999.260902.x. [DOI] [PubMed] [Google Scholar]
- 14.Socransky SS, Smith C, Haffajee AD. Subgingival microbial profiles in refractory periodontal disease. J Clin Periodontol. 2002;29:206–268. doi: 10.1034/j.1600-051x.2002.290313.x. [DOI] [PubMed] [Google Scholar]
- 15.Haffajee AD, Uzel NG, Arguello EI, Torresyap G, Guerrero DM, Socransky SS. Clinical and microbiological changes associated with the use of combined antimicrobial therapies to treat refractory periodontitis. J Clin Periodontol. 2004;31:869–877. doi: 10.1111/j.1600-051X.2004.00573.x. [DOI] [PubMed] [Google Scholar]
- 16.Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkel EG, vanWinkelhoff AJ, Timmerman MF, Vansted T, van der Velden U. Effects of metronidazole in patients with refractory periodontitis associated with Bacteroides forsythus. J Clin Periodontol. 1997;24:573–579. doi: 10.1111/j.1600-051x.1997.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 18.Haffajee AD, Cugini MA, Dibart S, Smith C, Kent RL, Jr, Socransky SS. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324–334. doi: 10.1111/j.1600-051x.1997.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 19.Haffajee AD, Socransky SS, Goodson JM. Comparison of different data analyses for detecting changes in attachment level. J Clin Periodontol. 1983;10:298–310. doi: 10.1111/j.1600-051x.1983.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 20. [Accessed January 2009.];Human Oral Microbe Identification Microarray (HOMIM) Available at: http://mim.forsyth.org/homim.html.
- 21.Human Oral Microbiome Database (HOMD) The Forsyth Institute; [Accessed January 2009.]. Available at: http://www.homd.org/ [Google Scholar]
- 22. [Accessed March–December 2008.];Human Oral Microbe Identification Microarray (HOMIM) Available at: http://bioinformatics.forsyth.org/homim/
- 23.Berglundh T, Krok L, Liljenberg B, Westfelt E, Serino G, Lindhe J. The use of metronidazole and amoxicillin in the treatment of advanced periodontal disease. A prospective, controlledclinical trial. J Clin Periodontol. 1998;25:354–362. doi: 10.1111/j.1600-051x.1998.tb02455.x. [DOI] [PubMed] [Google Scholar]
- 24.Cugini MA, Haffajee AD, Smith C, Kent RL, Jr, Socransky SS. The effect of scaling and root planing on the clinical and microbiological parameters of periodontal diseases: 12-month results. J Clin Periodontol. 2000;27:30–36. doi: 10.1034/j.1600-051x.2000.027001030.x. [DOI] [PubMed] [Google Scholar]
- 25.Feres M, Haffajee AD, Allard KA, Som S, Socransky SS. Change in subgingival microbial profiles in adult periodontitis subjects receiving either systemically administered amoxicillin or metronidazole. J Clin Periodontol. 2001;28:597–609. doi: 10.1034/j.1600-051x.2001.028007597.x. [DOI] [PubMed] [Google Scholar]
- 26.Serino G, Rosling B, Ramberg P, Hellstrom M-K, Socransky SS, Lindhe J. The effect of systemic antibiotics in the treatment of patients with recurrent periodontitis. J Clin Periodontol. 2001;28:411–418. doi: 10.1034/j.1600-051x.2001.028005411.x. [DOI] [PubMed] [Google Scholar]
- 27.Ehmke B, Moter A, Beikler T, Milian E, Flemmig TF. Adjunctive antimicrobial therapy of periodontitis: long term effects on disease progression and oral colonization. J Periodontol. 2005;76:749–759. doi: 10.1902/jop.2005.76.5.749. [DOI] [PubMed] [Google Scholar]
- 28.Sigusch B, Beier M, Klinger G, Pfister W, Glockman E. A 2-step non-surgical procedure and systemic antibiotics in the treatment of rapidly progressive periodontitis. J Periodontol. 2001;72:275–283. doi: 10.1902/jop.2001.72.3.275. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane GD, Herzberg M, Wolff L, Hardie N. Refractory periodontitis associated with abnormal polymorphonuclear leukocyte phagocytosis and cigarette smoking. J Periodontol. 1992;63:908–913. doi: 10.1902/jop.1992.63.11.908. [DOI] [PubMed] [Google Scholar]
- 30.Carvalho RPM, Mesquita JS, Bonomo A, Elsas PX, Colombo APV. Relationship of neutrophil phagocytosis and oxidative burst with the subgingival microbiota of generalized aggressive periodontitis. Oral Microbiol Immunol. 2009;24:124–132. doi: 10.1111/j.1399-302X.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- 31.Dahlén G, Lindhe J, Sato K, Hanamura H, Okamoto H. The effect of supragingival plaque control on the subgingival microbiota in subjects with periodontal disease. J Clin Periodontol. 1992;19:802–809. doi: 10.1111/j.1600-051x.1992.tb02174.x. [DOI] [PubMed] [Google Scholar]
- 32.McNabb H, Mombelli A, Lang NP. Supragingival cleaning 3 times a week. The microbiological effects in moderately deep pockets. J Clin Periodontol. 1992;19:348–356. doi: 10.1111/j.1600-051x.1992.tb00658.x. [DOI] [PubMed] [Google Scholar]
- 33.Westfelt E, Rylander H, Dahlen G, Lindhe J. The effect of supragingival plaque control on the progression of advanced periodontal disease. J Clin Periodontol. 1998;25:536–541. doi: 10.1111/j.1600-051x.1998.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 34.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 35.Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colombo AP, Teles R, Torres MC, et al. Subgingival microbiota of Brazilian Subjects with untreated chronic periodontitis. J Periodontol. 2002;73:360–369. 24. doi: 10.1902/jop.2002.73.4.360. [DOI] [PubMed] [Google Scholar]
- 39.Souto R, Colombo AP. Detection of Helicobacter pylori by polymerase chain reaction in the subgingival biofilm and saliva of non-dyspeptic periodontal patients. J Periodontol. 2008;79:97–103. doi: 10.1902/jop.2008.070241. [DOI] [PubMed] [Google Scholar]
- 40.Heller D, Varela VM, Silva-Senem MX, Torres MCB, Feres-Filho EJ, Colombo APV. Impact of systemic antimicrobials combined to anti-infective mechanical debridement on the microbiota of generalized aggressive periodontitis: a 6-month RCT. J Clin Periodontol. 2011;38:355–364. doi: 10.1111/j.1600-051X.2011.01707.x. [DOI] [PubMed] [Google Scholar]
- 41.Moore WEC, Moore LH. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 42.Leys EJ, Lyons SR, Moeschberger ML, Rumpf RW, Griffen AL. Association of Bacteroides forsythus and a novel Bacteroides phylotype with periodontitis. J Clin Microbiol. 2002;40:821–825. doi: 10.1128/JCM.40.3.821-825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferraro CT, Gornic C, Barbosa AS, et al. Detection of Dialister pneumosintes in the subgingival biofilm of subjects with periodontal disease. Anaerobe. 2007;13:244–248. doi: 10.1016/j.anaerobe.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 45.Wormser GP, Bottone EJ. Cardiobacterium hominis: review of microbiologic and clinical features. Rev Infect Dis. 1993;5:680–691. doi: 10.1093/clinids/5.4.680. [DOI] [PubMed] [Google Scholar]
- 46.Rossmann SN, Wilson PH, Hicks J, et al. Isolation of Lautropia mirabilis from Oral Cavities of Human Immunodeficiency Virus-Infected Children. J Clin Microbiol. 1998;36:1756–1760. doi: 10.1128/jcm.36.6.1756-1760.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR. Infective endocarditis caused by HACEK microorganisms. Annu Rev Med. 1997;48:25–33. doi: 10.1146/annurev.med.48.1.25. [DOI] [PubMed] [Google Scholar]
- 48.Chen C. Distribution of a newly described species, Kingella oralis, in the human oral cavity. Oral Microbiol Immunol. 1996;11(6):425–427. doi: 10.1111/j.1399-302x.1996.tb00206.x. [DOI] [PubMed] [Google Scholar]
- 49.Takeshita T, Nakano Y, Kumagai T, et al. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME Journal. 2009;3:65–78. doi: 10.1038/ismej.2008.91. [DOI] [PubMed] [Google Scholar]
- 50.Woo PC, Fung AM, Lau SK, Chan BY, Chiu SK, Teng JL, Que TL, Yung RW, Yuen KY. Granulicatella adiacens and Abiotrophia defectiva bacteraemia characterized by 16S rRNA gene sequencing. J Med Microbiol. 2003;52:137–140. doi: 10.1099/jmm.0.04950-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.