Abstract
We assemble a versatile molecular scaffold from simple building blocks to create binary and multiplexed stable isotope reagents for quantitative mass spectrometry. Termed Protected Amine Labels (PAL), these reagents offer multiple analytical figures of merit including, (i) robust targeting of peptide N-termini and lysyl side chains, (ii) optimal mass spectrometry ionization efficiency through regeneration of primary amines on labeled peptides, (iii) an amino acid-based mass tag that incorporates heavy isotopes of carbon, nitrogen, and oxygen to ensure matched physicochemical and MS/MS fragmentation behavior among labeled peptides, and (iv) a molecularly efficient architecture, in which the majority of hetero-atom centers can be used to synthesize a variety of nominal mass and sub-Da isotopologue stable isotope reagents. We demonstrate the performance of these reagents in well-established strategies whereby up to four channels of peptide isotopomers, each separated by 4 Da are quantified in MS-level scans with accuracies comparable to current commercial reagents. In addition we utilize the PAL scaffold to create isotopologue reagents in which labeled peptide analogs differ in mass based on the binding energy in carbon and nitrogen nuclei, thereby allowing quantification based on MS or MS/MS spectra. We demonstrate accurate quantification for reagents that support 6-plex labeling and propose extension of this scheme to 9-channels based on a similar PAL scaffold. Finally we provide exemplar data that extends the application of isotopologe-based quantification reagents to medium resolution, quadrupole time-of-flight mass spectrometers.
INTRODUCTION
Collective developments in mass spectrometry, along with advances in sample processing, enrichment, separations, and data analysis have shifted the experimental focus of proteomics applications from simple protein catalogs to the construction of dynamic networks, where changes in protein expression and post-translational modification status are monitored as a function of biological state or perturbation. The ability to use these quantitative data in models that support predictions in the context of cellular physiology will have a profound impact on human health.
Over the past few years a number of isotope label-based and label-free approaches have been developed for relative protein quantification by LC-MS/MS [1–3]. Label-free methods do not require isotopically enriched reagents, although the variance in peptide ion current and chromatographic retention may increase significantly in experiments requiring complex sample handling or fractionation procedures. Spectral counting is an alternative label-free approach that utilizes the number of MS/MS scans acquired per peptide as a surrogate for relative abundance; this method has proven to be very reliable at the level of discrete protein complexes [4], although it has fared less well for quantification of high complexity biological mixtures. As a result of these limitations, techniques based on isotope dilution remain the gold-standard for quantitative mass spectrometry measurements across a broad range of biological samples and matrices.
Stable isotope-based labels for proteomic studies generally fall into one of three categories[1]: enzymatic, metabolic, and synthetic chemical reagents, with the latter two representing the majority of applications. Metabolic labels are best represented by SILAC [5], in which isotopically enriched essential amino acids are incorporated into cellular proteomes in vitro during serial passage, and SILAM [6–9] which relies on 15N-labeled feedstock to yield isotopically labeled proteins in vivo. Metabolic approaches minimize systematic sample processing errors by incorporating isotopes during cell culture or animal husbandry; however the reagents must be compatible with living systems, thereby limiting the chemical space for synthesis as well as the degree of sample multiplexing.
In contrast to metabolic approaches, introduction of isotopically enriched labels after protein purification from cells or tissues provides an opportunity to "fine-tune" reagents in terms of target site, impact on gas phase basicity, peptide fragmentation, and incorporation of affinity tags to provide dual-purpose enrichment and quantification reagents [10–13]. As a result, synthesis of reagents de novo provides a wide scope for creative molecular design compared to metabolic labeling, provided that care is taken to process samples reproducibly in parallel until the point at which proteins and/or peptides are isotopically encoded and mixed. Isobaric multiplexed labels (iTRAQ [11,14] and TMT [12]) represent the most widely used chemical-based reagents. Peptides derived from four (iTRAQ 4-plex [11]), six (TMT 6-plex [12]), or eight (iTRAQ 8-plex [14]) biological conditions are labeled and subsequently detected at the same nominal m/z in the MS-level spectrum. Fragmentation during MS/MS yields typical b- and y-type ions, along with a series of 4, 6, or 8 low-m/z "reporter" ions that are used for relative quantification. The inherent multiplexing capability of these reagents supports higher sample throughput, although simultaneous fragmentation of peptides having similar m/z can "compress" reporter ion ratios towards the mean value of all peptides in the sample [15–18].
Most quantification strategies in proteomics rely on comparison of isotopologues that contain a different number of neutrons (e.g., peptide precursors in the case of SILAC/SILAM or low-mass reporter ions in the case of iTRAQ/TMT). In contrast, the advent of mass spectrometers that operate with high duty cycle and resolving power provides an opportunity to quantify isotopologues that contain the same number of neutrons incorporated into different atoms. This approach has previously been utilized for metabolite identification [19], and demonstrated for quantitative proteomics by use of isotopologue-encoded: amino acid NHS-esters [20,21], TMT reagents [22,23], or small molecule protein biochemistry reagents [9,24]. Similarly, metabolic isotopologue reagents were recently described for use in peptide quantification [25,26]. In each of these methods, labeled peptides have the same nominal mass but differ in their exact mass based on the binding energies of carbon, nitrogen, or oxygen nuclei.
In this work we sought to develop a versatile molecular scaffold that would support synthesis of nominal mass and sub-Dalton stable isotope labels suitable for a variety of mass spectrometry platforms and proteomic applications. These reagents, which we term Protected Amine Labels (PAL), are created from readily available chemical building blocks and provide several desirable figures of merit: (i) robust targeting of peptide N-termini and lysyl side chains, (ii) regeneration of primary amines on labeled peptides to maintain facile ionization, (iii) matched physicochemical and MS/MS fragmentation behavior among peptide analogs, and (iv) efficient use of available hetero-atom centers to provide a variety of nominal mass and sub-Da isotopologue stable isotope reagents supporting both MS- and MS/MS-based quantification.
We use 3-channel PAL reagents in a phosphoproteomic workflow to derive accurate MS-level quantification of phosphopeptide isotopomers that comprise signaling cascades activated in primary hematopoietic progenitors stimulated with lineage-specific ligands. In addition we demonstrate the utility of PAL reagents for multiplexed, isotopologue-based quantification where peptide ratios are determined from MS or MS/MS spectra. Importantly, the latter mode of operation enables use of sub-Da isotopologue reagents on medium-resolution, quadrupole time-of-flight mass spectrometers, extending the range of instrumentation suitable for isotopologue quantification studies.
METHODS
Detailed methods describing the synthesis of each reagent, along with experimental procedures for cell culture, LC, mass spectrometry, and flow cytometry are available in Supplementary Experimental Procedures.
Synthesis of TFA-β-alanine N-hydroxysulfosuccinimide (sulfoNHS) esters
β-alanine or (13C-3, 15N-1)β-alanine was trifluoroacetylated with S-ethyl trifluorothioacetate. SulfoNHS was coupled by use of diisopropylcarbodiimide.
Synthesis of TFA-β-alanyl-β-alanine sulfoNHS esters
Preparation of these active esters involved reacting β-alanine, (13C-3, 15N-1)β-alanine, or (13C-3, 15N-1, 18O-1)β-alanine with TFA-β-alanine sulfoNHS, TFA-(13C-3, 15N-1)β-alanine sulfoNHS, or TFA-(13C-3, 15N-1, 18O-1)β-alanine sulfoNHS. Product was purified by C18, and sulfoNHS was coupled by use of diisopropylcarbodiimide.
18O incorporation
TFA-(13C-3, 15N-1)-β-alanine or TFA-(13C-3, 15N-1, 18O-2)-β-alanyl (13C-3, 15N-1)-β-alanine was dissolved in H2 18O, acidified with TFA, and heated. Water was removed, and the procedure repeated.
Synthesis of N-tert-butoxycarbonyl (boc)-glycyl-glycine pentafluorophenyl esters (Pfp)
(13C-1)glycine was treated with thionyl chloride in methanol, to afford the methyl ester. Boc-(13C-1)glycyl-(13C-1)glycine was prepared by coupling Boc-(13C-1)glycine and (13C-1)glycine methyl ester with EDC/HOBt. After saponification of the methyl ester, the Pfp ester was prepared by reacting N-boc-(13C-1)glycyl-(13C-1)glycine with pentafluorophenyltriflouroacetate. N-boc-(15N-1)glycyl-(15N-1)glycine Pfp was prepared in an analogous fashion, substituting (15N-1)glycine compounds for those containing (13C-1)glycine.
General protocol for labeling of peptides
Model peptides (1 nmol) or K562-derived trypsin or lys-C peptides (100 µg) were dissolved in 100 µL of 30% 500 mM TEAB/70% ethanol, treated with 1 µmol of the appropriate reagent, and incubated at room temperature for 1 h.
Removal of TFA protecting groups
TFA protecting groups were removed by adjusting the pH to ~11.5 with 1:5 ammonia/water and 250 mM sodium carbonate, pH 12.0. After incubation for 1 h at 56 °C, reactions were acidified with TFA, and evaporated to dryness by vacuum centrifugation.
Removal of N-boc protecting groups
Desalted, dried peptides were resuspended in 100 µL of 95% TFA, incubated at room temperature for 30 min, and dried by vacuum centrifugation.
Procedure for double labeling of peptides with N-boc-glycyl-glycine Pfp
Aliquots corresponding to 100 µg tryptic or lys-C peptides from K562 lysates were labeled with N-boc-(13C-1)glycyl-(13C-1)glycine Pfp or N-boc-(15N-1)glycyl-(15N-1)glycine Pfp according to the general protocol above. After deprotection and desalting, peptides were subjected to an additional round of labeling with the same reagent. Double label reactions were combined, dried by vacuum centrifugation, and deprotected.
Procedure for 6-plex labeling of K562 peptides with TFA-β-alanyl-alanine sulfoNHS and N-boc-glycyl-glycine Pfp
SulfoNHS esters of TFA-β-alanyl-β-alanine (“LL”), TFA-β-alanyl-(13C-3, 15N-1)β-alanine (“HL”), and TFA-(13C-3, 15N-1)β-alanyl-(13C-3, 15N-1)β-alanine (“HH”) (1 µmol) were used to label two aliquots of 100 µg lys-C peptides according to the general procedure. After drying and deprotection, one aliquot each of “LL”, “HL”, and “HH” labeled peptides was treated with N-boc-(15N-1)glycyl-(15N-1)glycine Pfp; the remaining aliquot of each was reacted with N-boc-(13C-1)glycyl-(13C-1)glycine Pfp. Reactions were combined, dried by vacuum centrifugation, and deprotected.
RESULTS AND DISCUSSION
Protected Amine Label (PAL) Architecture and Performance
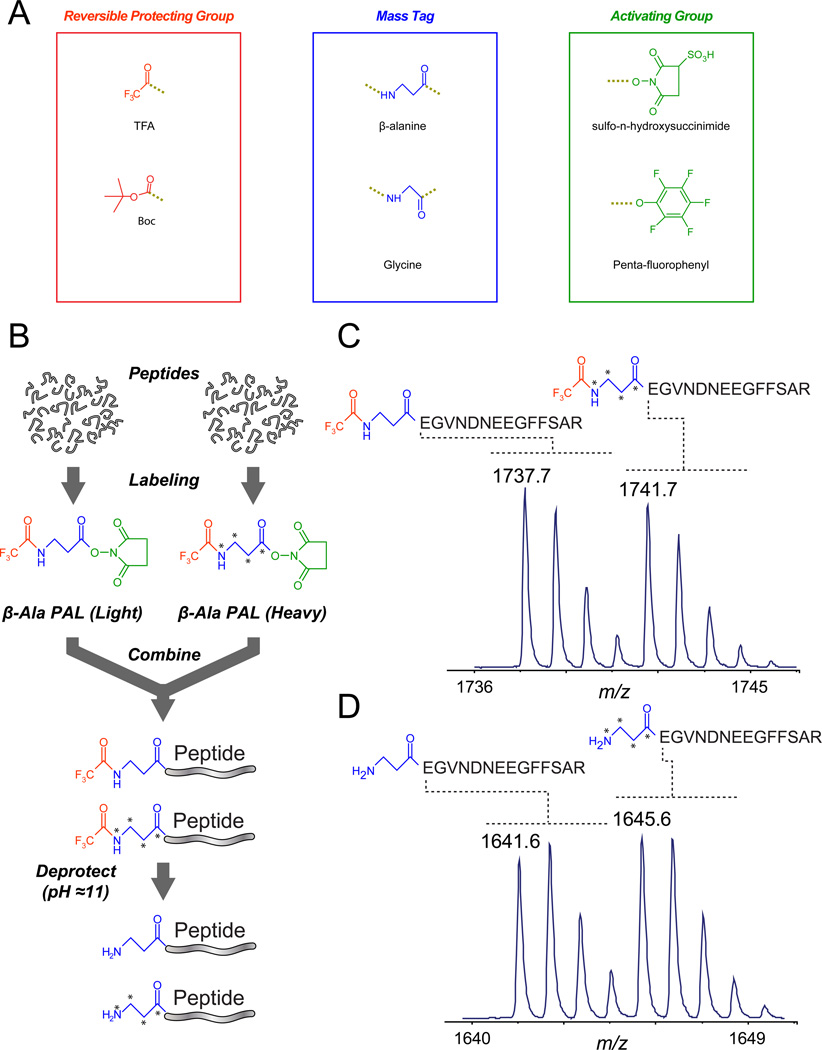
PAL reagents are synthesized from three building blocks (Figure 1A): (i) an amino acid-based mass tag carrying either light or heavy isotopes of carbon, nitrogen, or oxygen, (ii) an activating group that targets peptide primary amines, and (iii) a reversibly-bound protecting group that enables controlled introduction of multiple tags and allows regeneration of primary amines to facilitate ionization. As an initial evaluation of this technology toward traditional MS-based quantification, we synthesized a binary reagent based on β-alanine (3-aminopropanoic acid; β-Ala PAL) and labeled [glu-1]-fibrinopeptide B (Glu-Fib, Figure 1B). We observed a ~1:1 ratio of light/heavy protected (Figure 1C) and deprotected (Figure 1D) peptides; in addition, the labeling proceeded quantitatively with no appreciable polymerization. A small fraction (~15%) of Glu-Fib was deamidated during deprotection (note slight increase in carbon-13 peak in Figure 1D and Supplementary Figure S1), likely due to the N-D sequence motif which is known to have one of the highest rates of deamidation [8], second only to N-G. The standard peptide Substance P, which contains two glutamine residues but no N-D motif, exhibited virtually no deamidation under the deprotection conditions used herein (Supplementary Figure S2).
Figure 1. PAL architecture and binary labels.
(A) The PAL strategy utilizes amino acids (middle) as mass tags based on incorporation of 13C, 15N, or 18O. The primary amine is reversibly protected (left) whereas the C-terminus is activated by an amine-directed ester (right). (B) Labeling scheme for β-Ala PAL binary reagents. After protein purification and enzymatic digestion, peptides (top) are incubated at pH 8.5 at room temperature for 1 h with light and heavy reagents, and mixed. Incubation at pH 11 for 1 h regenerates primary amines for facile mass spectrometry ionization. A peptide standard (Glu-Fib, EGVNDNEEGFFSAR) was labeled with β-Ala PAL light and heavy reagents, mixed in a relative ratio of 1:1, and analyzed by MALDI-TOF mass spectrometry (C) before and (D) after deprotection.
MS/MS analysis by MALDI TOF/TOF revealed nearly identical fragmentation patterns between the labeled and unlabeled Glu-Fib peptides (Supplementary Figure S3). To explore the performance of β-Ala PAL across a wider range of peptides, we labeled tryptic digests of four standard proteins and mixed the light/heavy peptides in relative ratios ranging from 1:1 to 5:1. LC-MS/MS analysis demonstrated that lysine-containing peptides, labeled on both the amine terminus and side chain, yielded readily interpretable fragment ion spectra under typical low-energy collisional activated dissociation conditions. Finally, extracted ion chromatograms (XICs) for light/heavy peptide pairs overlapped in chromatographic elution time, with relative peak areas corresponding to the expected ratios (Supplementary Figure S4).
As noted above, the PAL protecting group prevents polymerization during the labeling reaction, but is easily removed to regenerate peptide N-termini and maintain optimal ionization efficiency. The trifluoroacetyl group imparted improved aqueous stability to β-Ala PAL reagents compared to the N-methyl-piperizine moiety of iTRAQ reagents (Supplementary Figure S5). We leveraged this characteristic to label tryptic peptides in situ after capture on reversed-phase resin (Supplementary Figure S6), affording the possibility of improved peptide recovery in sample-limited experiments [27].
PAL Multiplexed (PAL-MP) Labels
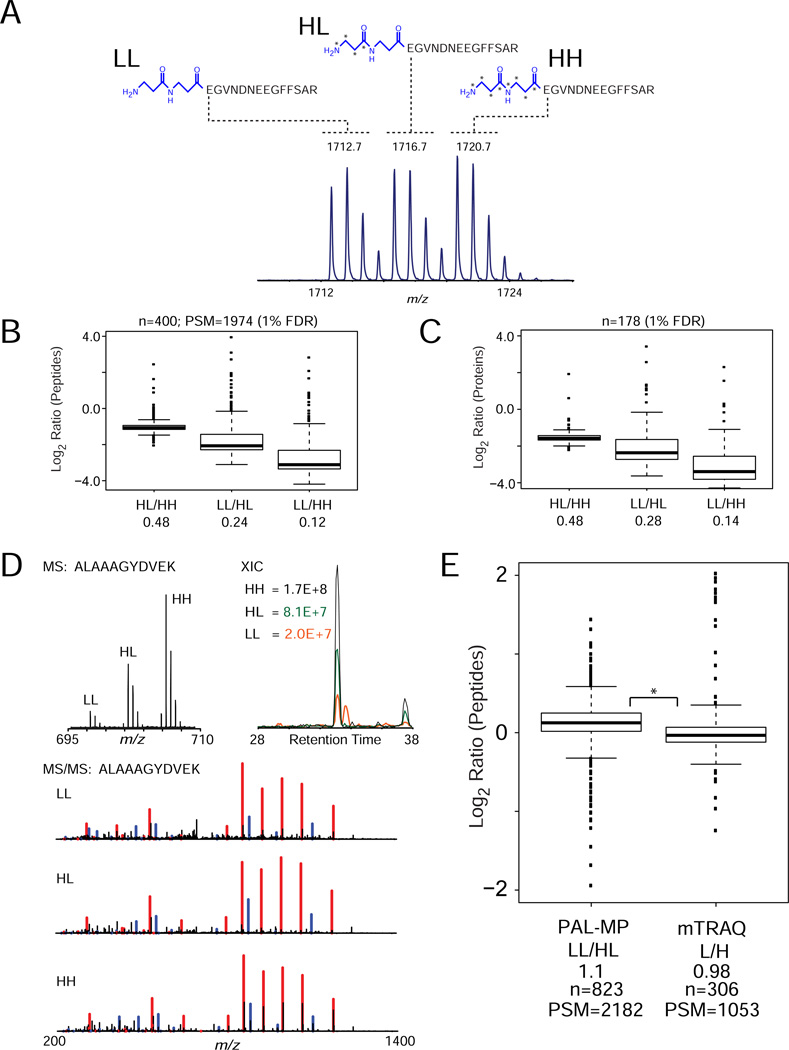
As noted above, there is significant interest in labeling strategies that enable multiplexed analysis of more than two biological states simultaneously. This approach improves sample throughput and supports more complex time-course or kinetic experiments [28]. The modular architecture of the PAL reagents enables polymerization of multiple, isotope-encoded amino acids to provide a 4-Da mass shift across multiple reagent channels. Combining light and heavy carbon and nitrogen isotopes within two β-alanine residues provides a 3-channel multiplexed reagent (β-Ala2 PAL, Figure 2A). As an initial demonstration, the model peptide Glu-Fib was derivatized with each reagent, mixed in a 1:1:1 ratio, deprotected, and analyzed by MALDI-TOF MS (Figure 2A). To further explore the performance of these multiplexed reagents, we labeled tryptic peptides derived from human myeloid K562 cells in relative ratios of 10:5:1 and analyzed the resulting mixture by LC-MS/MS. Peptides and proteins were accurately quantified (Figure 2B and C) across all three combinations. In addition, labeled peptides co-eluted and yielded readily interpretable MS/MS spectra (Figure 2D). Moreover, the distribution of peptide ratios was similar to that observed with mTRAQ labels (Figure 2E). Significantly, PAL reagents are "molecularly efficient" in that all heteroatom centers can be utilized to create higher-order multiplexed labels; for example, incorporation of 18O into each carbonyl of the β-alanine scaffold above yields a 4-plex reagent in which each channel is separated by 4 Da (Supplementary Figure S7). As in a recent report [29], this strategy could in principle be combined with commercially available isobaric labels (i.e., TMT/iTRAQ) to yield high-order (24- or 32-plex) multiplexed reagents.
Figure 2. PAL multiplexed labels.
(A) A peptide standard (Glu-Fib, EGVNDNEEGFFSAR) was labeled with β-Ala2 PAL multiplexed reagents (LL, HL, and HH), mixed in a relative ratio of 1:1:1 and analyzed by MALDI-TOF mass spectrometry. (B) Tryptic peptides derived from human cell lysate were labeled with β-Ala2 PAL multiplexed reagents, mixed in a relative ratio of 1:5:10 (LL:HL:HH), and analyzed by LC-MS/MS. Peptide ratios were calculated based on precursor ion peak heights, transformed to Log2 space, and displayed as box plots. Boxes encompass the interquartile range of all ratios with respective median values indicated with stripes; whiskers represent 1.5× the interquartile range with outliers shown as filled circles. Non-transformed, median values are listed below each ratio designation (bottom). Number of unique quantified peptides (n) and peptide spectral matches (PSM) are shown above the plot. (C) Peptide ratios were aggregated to yield protein-level ratios that were transformed to Log2 scale and displayed in box-plot format. Non-transformed, median values are listed below each ratio designation (bottom). Number of unique quantified proteins (n) is shown above the plot. (D) Mass spectrum (left) for the tryptic peptide (ALAAAGYDVEK) with isotopic distributions for the LL, HL, and HH analogs. Labeled peptides exhibit physicochemically equivalent behavior in terms of (right) chromatographic co-elution as evidenced by overlapping XICs and (bottom) nearly identical fragmentation spectra. Assigned y- and b-type fragment ions are indicated with red and blue lines in each MS/MS spectrum. (E) Tryptic peptides derived from human cell lysate were labeled separately with (i) two channels of β-Ala2 PAL multiplexed reagents (LL and HL) and (ii) commercial mTRAQ reagents (light iTRAQ and iTRAQ-117). Light and heavy peptides from each reagent were mixed in a relative ratio of 1:1 and analyzed in separate LC-MS/MS experiments. Ratios for peptides labeled with β-Ala2 PAL and mTRAQ were calculated based on precursor ion peak heights, transformed to Log2 space, and displayed in box-plot format. Non-transformed, median values are listed below each ratio designation (bottom). *, p-value for Levene’s test of peptide ratios (1st to 3rd quartiles of data) > 0.05.
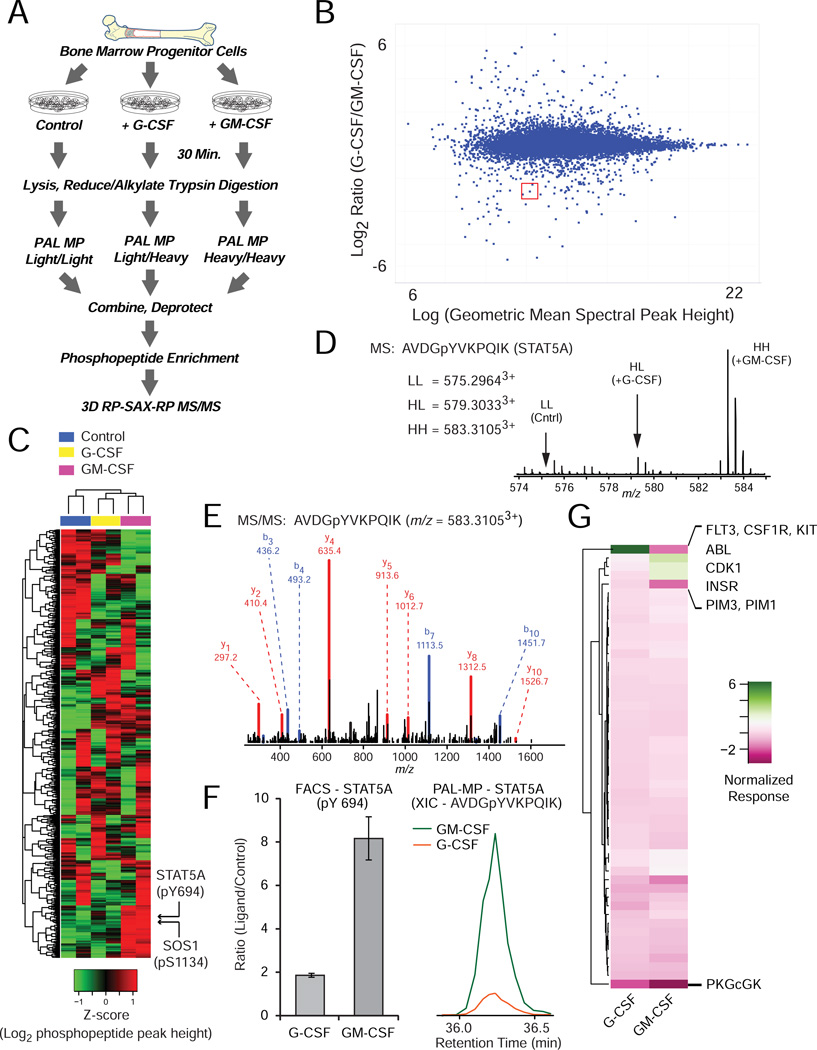
As an application of β-Ala2 PAL 3-plex reagents we interrogated signaling pathways mediated by Granulocyte-Macrophage Colony-Stimulating factor (GM-CSF) and Granulocyte Colony-Stimulating factor (G-CSF), which independently promote differentiation of bone marrow hematopoietic progenitors along eosinophil and neutrophil lineages [30–33]. Although these ligands target different receptors, both propagate phosphorylation through the JAK/STAT, RAS/ERK, and PI3K/AKT pathways [34–37], suggesting that recruitment of distinct mediators provides specificity and pleiotropy. To identify molecular determinants downstream of GM-CSF and G-CSF, we cultured CD34+ cells isolated from fresh, human bone marrow samples in the presence of vehicle or ligand (Figure 3A). After harvest and digestion, the tryptic peptides were labeled with β-Ala2 PAL 3-plex reagents, mixed, and deprotected. Phosphopeptides were enriched [38] and analyzed by RP-SAX-RP MS/MS analysis [39]. Across biological replicates we quantified 7,363 unique phosphopeptide sequences that mapped uniquely to 2,794 genes (Figure 3B), with each ligand inducing a unique phosphorylation profile (Figure 3C). Notably, peptide dehydration and deamidation rates were very similar to those observed in a previous study from our lab[39] that analyzed unlabeled phosphopeptides derived from K562 tryptic digests at a depth of 53 RP-SAX-RP fractions (similar to the conditions utilized here), providing further evidence that our PAL-MP labeling scheme does not promote a significant degree of undesirable side reactions and is compatible with large-scale proteomic workflows. As expected, we detected increased phosphorylation of STAT5 Y694 (Figure 3D and E) and SOS1 S1134 in the context of GM-CSF stimulation [40–42]. Biochemical analysis of STAT5 activation confirmed the mass spectrometry data (Figure 3F). We aggregated phosphopeptide ratios across replicate experiments and used these data as input to the NetworkKIN kinase prediction web resource [43]. This analysis suggested differential activation of kinases in multiple families, including FLT3/CSF1R/KIT, ABL, CDK1, INSR, PIM1/PIM3 and PKGcGK, in response to stimulation by G-CSF or GM-CSF (Figure 3G). Ultimately these ligands may promote expression of distinct hematopoietic, lineage-specific genes through phosphorylation of transcription regulatory factors identified in our proteomic analysis, such as IRF2BP2, FOXK1, SON, SPN and DDX21 (G-CSF), or ACIN1, TRAP3, BCLAF1 and PDCD4 (GM-CSF). Interestingly, each of these co-regulatory proteins has been functionally linked to myeloid differentiation, although the specific mechanisms are not yet fully defined. Collectively, these results demonstrate the compatibility of PAL reagents for large-scale multiplex profiling of cellular signaling pathways.
Figure 3. Quantification of hematopoietic signaling with β-Ala2 PAL 3-plex reagents.
(A) Primary CD34+ hematpoietic progenitors were isolated from fresh, human bone marrow samples and cultured in the presence of vehicle, G-CSF, or GM-CSF. After digestion, tryptic peptides were labeled with β-Ala2 PAL 3-plex reagents, mixed, and deprotected at pH 11. Labeled phosphopeptides were enriched and analyzed by 3D RP-SAX-RP LC-MS/MS. (B) Phosphopeptide ratios corresponding to (+G-CSF/+GM-CSF) were converted to Log2 scale and plotted versus the Log2 transformed geometric mean of the corresponding spectral peak heights. (C) The scheme in (A) was repeated for CD34+ progenitors derived from the bone marrow of a second donor. Hierarchical clustering of PAL-MP labeled phosphopeptide peak heights from these bioreplicate experiments revealed a distinct signaling pattern associated with each ligand, including phosphorylation of STAT5A and SOS1. (D) Mass spectrum for a tyrosine phosphorylated peptide (AVDGpYVKPQIK) from STAT5A with isotope distributions for the LL (control), HL (+G-CSF), and HH (+GM-CSF) conditions. (E) MS/MS spectrum for the HH-labeled phosphopeptide for STAT5A; assigned y- and b-type fragment ions are indicated with red and blue lines. Activation of STAT5A was verified in CD34+ progenitors by (F, left) flow cytometry analysis with a phospho-specific STAT5 (Y694), Alexa fluor 647-conjugated antibody following stimulation with G-CSF or GM-CSF. Relative phosphorylation was calculated from the mean fluorescence intensity (MFI) in G-CSF or GM-CSF stimulated versus control (unstimulated) cells. Data are presented as the mean ± SEM of two independent experiments with P=0.03. (F, right) Extracted ion chromatograms (XIC) for HL and HH labeled AVDGpY(694)VKPQIK peptides derived from STAT5A. (G) Aggregate phosphopeptide ratios were used as input to the NetworkKIN web resource to predict the relative activation state of kinase families as a result of stimulation with each ligand.
PAL High Resolution (PAL-HR) Stable Isotope Labels
Continued technological advances have resulted in mass spectrometers that provide high mass resolution and mass accuracy, along with fast acquisition rate; collectively, these analytical figures of merit enable quantification strategies whereby labeled peptides differ in mass based on the nuclear binding energy in carbon, nitrogen, or oxygen. In this scenario, analogous peptides from different biological conditions appear at the same nominal mass, similar to isobaric tags (e.g., iTRAQ and TMT). However, shifting neutrons from carbon to nitrogen in molecules that are otherwise identical creates a pair of isotopologues that differ in mass by ~6.3 mDa per atom. With sufficient mass resolution, peptide ratios can be measured for intact precursors in MS spectra as well as discrete N- and C-terminal fragment ions in MS/MS scans. Notably, isotopologue pairs will be simultaneously isolated for fragmentation, resulting in improved duty cycle compared to nominal mass labeling schemes, along with higher sensitivity when low-resolution MS/MS scans are acquired.
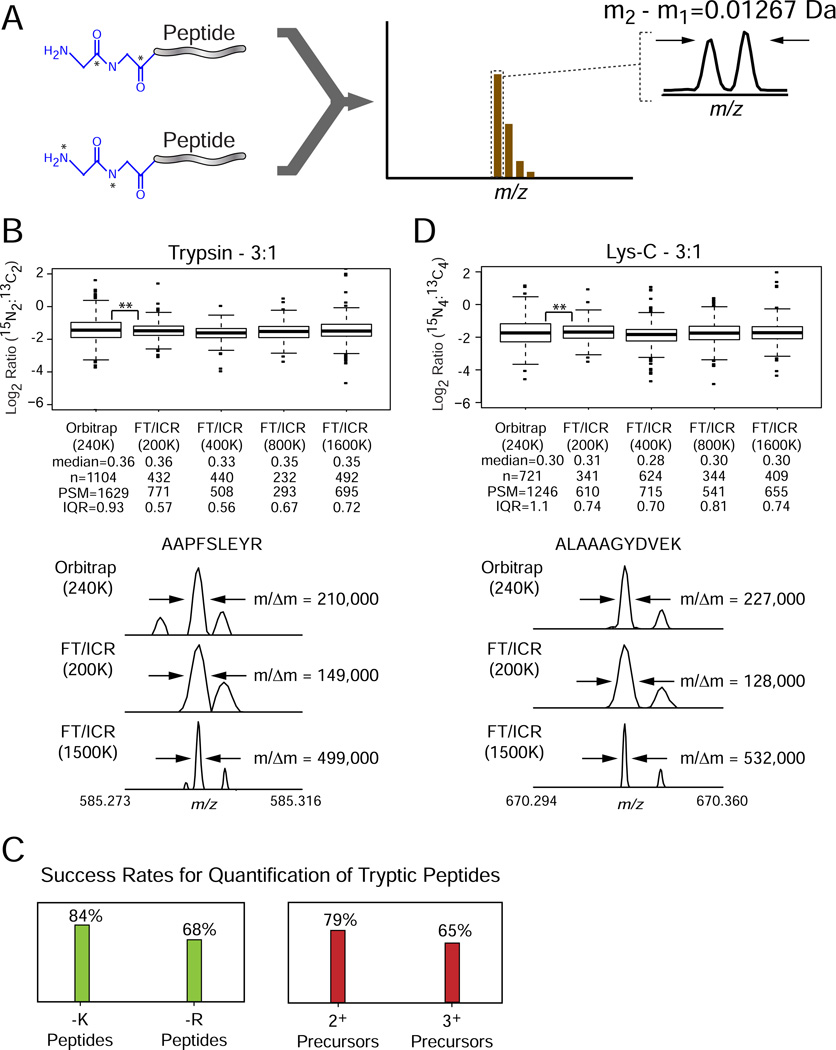
We used a PAL scaffold with two glycine residues to create an isotopologue reagent pair containing 13C2 or 15N2 that differ in mass by ~12.7 mDa (Gly2 PAL-HR, Figure 4A). To illustrate compatibility of PAL reagents with different protection schemes and amine-targeting active esters (Figure 1A), we installed an N-terminal Boc protecting group and a C-terminal amine reactive pentaflourophenyl ester. The Boc group is removed under acidic conditions, and may be preferred for analysis of peptides carrying modifications not amenable to high-pH deprotection (e.g., glycosylation). In addition, direct reaction of pentafluorophenyltrifluoroacetate with protected isotopologue acid intermediates in THF provides a simple and convenient synthetic route to obtain the final product. Tryptic peptides derived from human cell lysate were encoded with these reagents and mixed in a ratio of 3:1 for light (15N2) and heavy (13C2) channels, respectively. We acquired LC-MS/MS data with a hybrid linear ion trap (LTQ)/Orbitrap mass spectrometer, modified to allow acquisition of time-domain transients of up to ~3 sec, providing MS1 spectra with a mass resolving power of ~240,000 at m/z = 400. In addition we acquired LC-MS/MS data with a custom-built LTQ/FT-ICR mass spectrometer equipped with a 14.5 T magnet [44,45]. For the latter, we obtained MS1 spectra over a resolving power range of ~200,000 to ~1.6M (at m/z = 400). On both platforms, low resolution MS/MS spectra were acquired to facilitate peptide identification. Analysis of these data revealed a consistent median ratio across all conditions, with the LTQ/FT-ICR providing a narrower distribution than the Orbitrap (Figure 4B, top, with representative MS spectra for isotopologues below). As expected, increased acquisition period resulted in higher resolving power, although the precision of peak height ratios is likely limited by concomitant damping of time-domain transients [46,47]. In addition, we found that the success rate for quantification was higher for lysine-terminated peptides, expected to carry two labels, compared to those terminated with arginine (Figure 4C); similarly, the quantification rate was higher for doubly- as compared to triply-charge tryptic peptides. Based on these data we next digested human cell lysate with endoprotease Lys-C to ensure incorporation of multiple Gly2 PAL-HR labels on each peptide. After mixing these samples in a 3:1 ratio, we acquired LC-MS/MS data across the same resolving power range as above (Figure 4D, top). Analysis of the resulting peptide ratios revealed a nearly uniform success rate of quantification across all experiments. Although Lys-C, compared to trypsin, is expected to generate larger peptides with higher basicity, we nevertheless observed that most of the precursors were doubly-charged, with a concomitant increase in m/z separation between isotopologues (Figure 4D, bottom).
Figure 4. PAL Gly2 high resolution binary labels.
(A) Peptides labeled with binary PAL reagents based on di-glycine encoded with either 15N2 or 13C2 differ in mass by ~12.7 mDa per primary amine. (B, top) Tryptic peptides derived from human cell lysate were labeled with Gly2 PAL-HR reagents, mixed in a relative ratio of 3:1 (15N2:13C2) and analyzed by LC-MS/MS with LTQ/Orbitrap and LTQ/FT-ICR mass spectrometers, across a range of resolving power (specified at m/z = 400) as indicated in parentheses below each instrument designation. Peptide ratios were calculated based on precursor ion peak heights, transformed to Log2 space, and displayed in box-plot format. Non-transformed, median ratios, number of unique quantified peptides (n), number of peptide spectral matches (PSM), and interquartile ranges (IQR) are listed below each instrument designation. **, p-value for Levene’s test on peptide ratios (1st to 3rd quartiles of data) < 0.0001. (B, bottom) Representative mass spectral segments for the peptide, AAPFSLEYR, illustrating baseline resolution of isotopologues under different instrument/resolving power combinations. Resolving power for an undamped time-domain signal is listed in parentheses; actual experimental resolving power is shown at the right of each m/z segment. (C) Success rate for quantification of tryptic peptide isotopologue pairs as a function of (left) lysine- versus arginine-terminated peptides and (right) doubly- versus triply-charged precursors. (D, top) Lys-C peptides derived from human cell lysate were labeled with Gly2 PAL-HR reagents, mixed in a relative ratio of 3:1 (15N4:13C4) and analyzed by LC-MS/MS with LTQ/Orbitrap and LTQ/FT-ICR mass spectrometers, across a range of resolving power (specified at m/z = 400) as indicated in parentheses below each instrument designation. Peptide ratios were calculated based on precursor ion peak heights, transformed to Log2 space, and displayed in box-plot format. Non-transformed, median ratios, number of unique quantified peptides (n), number of peptide spectral matches (PSM), and interquartile ranges (IQR) are listed below each instrument designation. **, p-value for Levene’s test of peptide ratios (1st to 3rd quartiles of data) < 0.0001. (D, bottom) Representative mass spectral segments for the peptide, ALAAAGYDVEK, illustrate improved resolution of isotopologues, under each instrument/resolving power combination, compared to that for tryptic peptides (B, bottom) acquired under identical LC-MS/MS conditions.
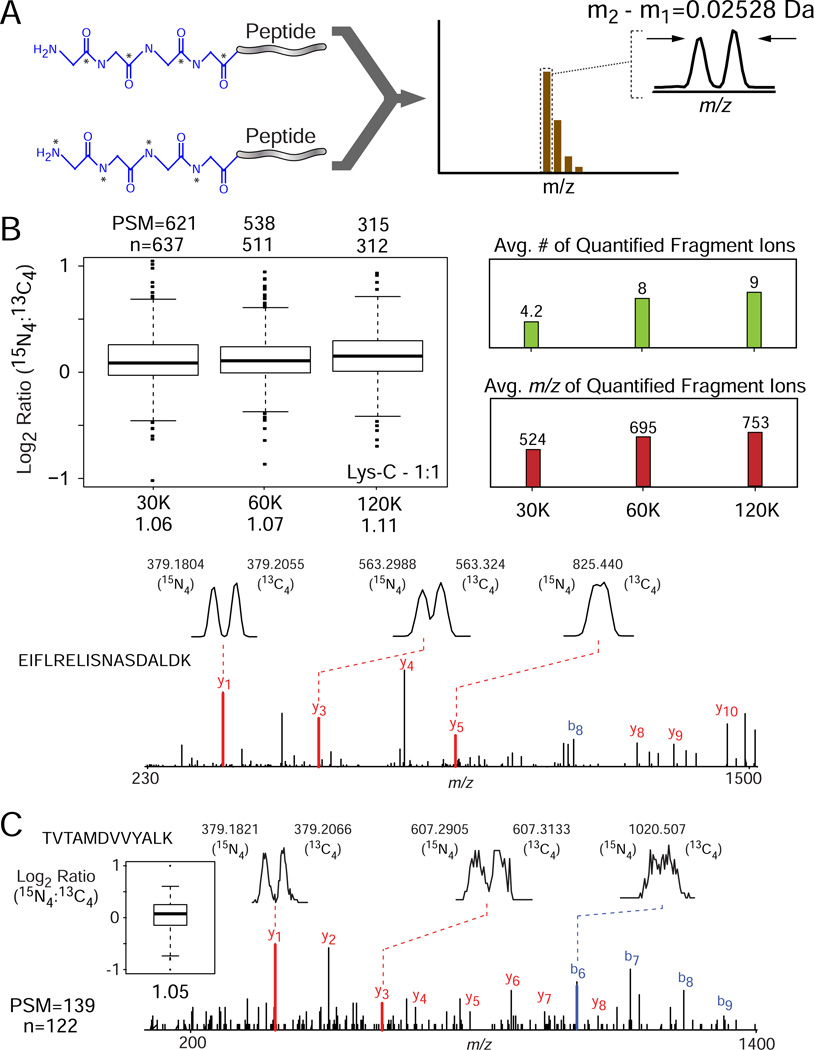
As noted above, the PAL architecture affords an opportunity to "tune" the encoded mass difference between isotopologues through polymerization of additional labeling groups after deprotection. For example, introduction of two tandem Gly2 PAL-HR labels (label-deprotect-label-deprotect) would increase the mass difference from ~12.7 mDa (13C2 or 15N2) to ~25 mDa (13C4 or 15N4) per double-labeled amine, or ~50 mDa (13C8 or 15N8) in total for lysine-terminated peptides. We reasoned that this approach would extend the m/z range over which precursors are successfully quantified in MS1 scans and extend the use of PAL chemistry to MS/MS level quantification, where b- and y-type fragments are predominantly singly-charged, in addition to other mass spectrometry platforms (e.g., TOF). Pursuant to this hypothesis, we labeled Lys-C peptides derived from human cell lysate with Gly2 PAL-HR reagents, and then repeated the labeling procedure after the deprotection step, effectively creating Gly4 PAL-HR peptides (Figure 5A). We mixed labeled peptides in a 1:1 ratio and acquired LC-MS/MS data with a LTQ/Orbitrap; fragment ions detected at high-resolution in the Orbitrap mass analyzer. Peptides were accurately quantified in MS/MS scans, across a resolving power range of ~30,000 to ~120,000 (Figure 5B, left), with both the average number of quantified fragments and their corresponding mass-to-charge ratio increasing as a function of resolving power (Figure 5B, right panels). We next analyzed the same sample by LC-MS/MS with a tandem quadrupole time-of-flight mass spectrometer and resolved fragment ion isotopologues up to m/z ≈ 600 (Figure 5C). Although this mode is similar to iTRAQ/TMT strategies in that quantitation is derived from MS/MS spectra, the PAL approach offers several advantages. First, relative quantitation is derived from b- and y-type fragments that are sequence-specific and hence in principle devoid of contamination and associated ratio compression[16–18]. In addition, quantification is based on multiple fragment ions derived from the same peptide, rather than a single reporter ion as for iTRAQ/TMT isobaric reagents. Finally, the collision energy for MS/MS is not compromised by the need to balance b-/y-fragment and reporter ion relative abundances as is often the case with commercially available isobaric labels [48].
Figure 5. PAL Gly4 high resolution binary labels.
(A) Peptides labeled with binary PAL reagents based on 2 × di-glycine encoded with either 15N4 or 13C4 differ in mass by ~25.0 mDa per primary amine. (B, top) Lys-C peptides derived from human cell lysate were labeled with Gly4 PAL-HR reagents, mixed in a relative ratio of 1:1 (15N4:13C4) and analyzed by LC-MS/MS with a LTQ/Orbitrap mass spectrometer with fragment ions detected over a range of resolving power from 30,000 to 120,000 (specified at m/z = 400). Peptide ratios were calculated based on peak heights for fragment ion isotopologue pairs, transformed to Log2 space, and displayed in box-plot format. Non-transformed, median values are listed below each resolving power designation. Number of unique quantified peptides (n) and peptide spectral matches (PSM) are shown above the plot. (B, right) Average number and m/z of quantified isotopologues as a function of MS/MS resolving power for the LTQ/Orbitrap. (B, bottom) Representative MS/MS spectrum for the peptide, EIFLRELISNASDALDK, acquired at 30,000 resolving power. Selected y- and b-type fragment ions are indicated in red and blue, with insets illustrating resolution of isotopologue pairs as a function of fragment m/z. (C) Representative fragment ion spectrum for the peptide, TVTAMDVVYALK, derived from the Lys-C digest described in (A) and acquired by LC-MS/MS with a tandem quadrupole time-of-flight mass spectrometer. Selected y- and b-type fragment ions are indicated in red and blue, with insets illustrating (top) the resolution of isotopologue pairs as a function of fragment m/z, and (left) distribution of ratios calculated from an average of fragment ion isotopologues pairs, transformed to Log2 scale, and displayed in box-plot format. Number of unique quantified peptides (n) and peptide spectral matches (PSM) are listed on the bottom left.
Controlled polymerization inherent to PAL chemistry also enables a "mix-and-match" of low- and high-resolution labels to achieve improved multiplexing. As an example, we labeled Lys-C peptides derived from human cell lysate first with β-Ala2 PAL 3-plex reagents and then with Gly2 PAL-HR binary reagents to create a 6-channel multiplex scheme in which isotopologue pairs are separated by ~25 mDa (13C4 vs. 15N4; m2 − m1 = 0.02534) for peptides labeled on both N-termini and C-terminal lysine residues. We combined labeled peptides to create mixtures with uniform (Figure 6A) and variable (Figure 6B) relative ratios, and acquired LC-MS/MS data with a LTQ/Orbitrap with mass spectral resolving power of ~240,000. Low resolution MS/MS spectra were acquired to facilitate peptide identification. Isotopologues within each 3-plex isotope cluster were readily resolved and identified by MS/MS, with median ratios that agreed well with the expected values (Figure 6C).
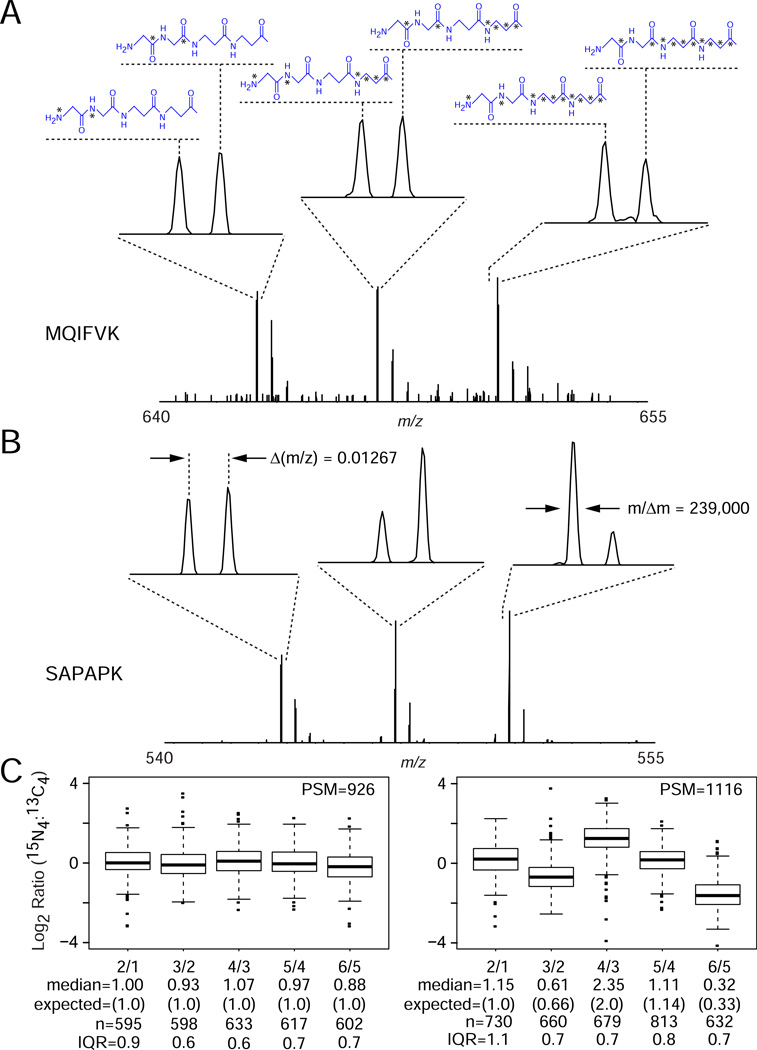
Figure 6. PAL high resolution multiplexed labels.
Lys-C peptides derived from human cell lysate were iteratively labeled, first with β-Ala2 PAL 3-plex reagents and then with Gly2 PAL-HR binary reagents to provide a 6-channel labeling scheme in which isotopologues within each 3-plex isotope cluster were separated by m2 − m1 = 0.01267 (13C2 or 15N2) per labeled amine group. Peptides were mixed in relative ratios of (A) [(1):(1):(1):(1):(1):(1)] or (B) [(1):(1):(0.66):(1.32):(1.5):(0.5)] followed by LC-MS/MS analysis with an LTQ/Orbitrap mass spectrometer with MS spectra acquired at a resolving power of ~240,000 (specified at m/z = 400). Representative mass spectral segments shown for peptides (A) MQIFVK and (B) SAPAPK. (C) Peptide ratios were calculated based on precursor ion peak heights, transformed to Log2 scale, and displayed in box-plot format. Non-transformed median ratios, expected values, number of unique quantified peptides (n), and IQR values are listed below each channel designation.
Direct Comparison of Labeling Reagents
In an effort to assess the general performance of PAL labeling strategies, we directly compared PAL-MP, PAL-HR, and mTRAQ approaches. Equal aliquots of K562-derived tryptic peptides were labeled to yield binary mixtures: PAL-MP (LL and LH), PAL-HR, or mTRAQ (light iTRAQ and iTRAQ-117). After deprotection of PAL labeled peptides, equivalent amounts of each peptide mixture (250 ng) were analyzed by the same LC-MS/MS method to allow direct comparison of labeling efficiency, as well as numbers of PSM and unique peptide sequences afforded by each reagent. Unlabeled peptides were included in this analysis as a control (Table 1). In all experiments, we observed labeling efficiencies of ≥90% (Supplementary Experimental Procedures). Although we observed no significant impact on Mascot scores for PAL-labeled peptides (Supplemental Figure S08), it is possible that label-specific fragmentation, such as amide bond cleavage within larger PAL reagents, could negatively impact peptide identification rates. However it is likely that the search algorithm or its associated parameters could be modified to account for these fragments. Not surprisingly, each labeling scheme reduced the number of PSM and unique peptides vs. the equivalent analysis of unlabeled peptides. Although these data provide proof-of-principle for the use of PAL reagents in quantitative proteomic studies, further investigation is needed to determine the relative impact of production variables such as reagent purity, which can presumably be addressed with commercial-grade synthesis capabilities, versus fundamental limitations in fragmentation efficiency, on the overall success of peptide identification relative to mTRAQ labels.
Table 1.
Performance evaluation of mTRAQ, PAL-MP, and PAL-HR labeling schemes.
| Labeling Scheme |
PSM | Labeling Efficiency |
Unique Peptides |
Unique Proteins |
Number of MS2 Spectra |
% Identified MS2 Spectra |
|---|---|---|---|---|---|---|
| mTRAQ | 3151 | 98.7% | 1645 | 444 | 6930 | 45.5% |
| PAL-MP | 2541 | 98.4% | 1367 | 411 | 6878 | 36.9% |
| PAL-HR | 1521 | 90.2% | 1226 | 411 | 5423 | 28.0% |
| Unlabeled | 4592 | NA | 3408 | 857 | 7614 | 60.3% |
CONCLUSIONS
Recent advances in DNA sequencing technologies [49–51] illustrate the potential to derive biological insight from systematic, genome-scale measurements. Quantitative proteomic data complement those from high-throughput Next-Gen sequencing and are critical components of system-level studies designed to monitor biological response to perturbation or build predictive models of cellular physiology. As a result, there is significant motivation for continued technological advances in mass spectrometry-based proteomics. Herein we provide proof-of-principle data for a new class of stable isotope labels to support quantitative proteomic applications. Termed Protected Amine Labels (PAL), these reagents offer several desirable figures of merit for mass spectrometry-based protein characterization. First, the use of amino acids encoded with 13C, 15N, and 18O ensures physicochemical equivalence with respect to chromatographic and fragmentation behavior among labeled peptide analogs. This attribute is particularly relevant given the recent advances in multi-dimensional peptide fractionation [52,53] along with the introduction of novel mass spectrometry fragmentation schemes [54,55]. Moreover, PAL reagents achieve broad, quantitative labeling of peptide primary amines via activated esters, but also include a reversible protecting group to enable regeneration of free amines, and hence facile ionization during mass spectrometry analysis. Importantly, these reactions go to completion under mild conditions, ensuring compatibility with efforts directed at quantitative analysis of post-translational modifications. We demonstrated the latter observation through a large-scale, quantitative analysis of phosphorylation cascades mediated by GM-CSF or G-CSF ligands in primary, human CD34+ hematopoietic progenitor cells.
Building on previous reports [9,20–26], we used the PAL scaffold to develop stable isotope labels in which peptides are mass-encoded based on the difference in binding energy between carbon and nitrogen nuclei. The Gly2 PAL-HR binary reagent affords a mass difference of 0.01267 Da/label (13C2 vs. 15N2; m2 − m1 = 0.01267). We observed fully resolved peptide precursors and fragment ion isotopologues in MS and MS/MS scans for both Orbitrap and FT-ICR mass analyzers. The Orbitrap exhibited a higher susceptibility to space charge-induced coalescence of isotopes, which may diminish the overall success rate of quantification depending on the specific sample type and acquisition parameters. However, recent commercial releases of this instrument operate at higher resolving power and hence may fare better in this regard [56–58]. Although the Gly2 PAL-HR reagents described herein enable binary quantification schemes, it is possible to extend the multiplexing capabilities by use of the same scaffold (for example, Supplemental Figure S9A). In an analogous fashion, strategic incorporation of 13C, 15N, and 18O into the Gly4 PAL-HR scaffold could be used to implement a 9-plex quantification scheme (Supplemental Figure S9B).
Importantly, the PAL workflow supports iterative steps of "label-deprotect-label-deprotect," allowing incorporation of multiple labels, and hence a greater mass separation between corresponding isotopologues. We leveraged this aspect of PAL chemistry to achieve accurate quantification results for Lys-C peptides with an Orbitrap mass analyzer; in fact, we detected resolved isotopologue fragment ions, up to m/z ≈ 600 with a tandem quadrupole time-of-flight instrument, highlighting the versatility of PAL reagents and their capacity to accommodate quantitative proteomic analysis across a wide range of mass spectrometry platforms. Finally, we leveraged the versatility of PAL chemistry to create a 6-plex high-resolution, multiplexed reagent (β-Ala2-Gly2 PAL HR MP).
Although we successfully applied PAL chemistry in multiple contexts, we also observed several limitations at this early stage of reagent development. First, regeneration of peptide free amines requires an extra step of sample handling that may negatively impact the number of identified peptides; this effect would presumably be amplified during iterative stages of "label-deprotect-label-deprotect." However, the more complex labels described herein (Gly4 PAL HR and β-Ala2-Gly2 PAL HR MP labels) can be readily produced by standard techniques in commercial peptide synthesis, thereby simplifying the overall method to a single labeling/deprotection step. In addition, PAL-HR labels require instruments that can provide high resolution mass measurements of precursor or product ions, leading to a potentially deleterious compromise between peptide identification and quantification. Fortunately, as noted above, developments in instrument hardware and data processing are clearly moving in the direction of improved scan rate concurrent with higher resolution. Moreover, current time-of-flight instruments provide fast acquisition rate at sufficient resolution to enable MS/MS-based quantitation with PAL HR labels. Finally we note that quantification accuracy may be comprosmised for extreme ratios if PAL isotopologues are not fully baseline resolved.
PAL regents are readily synthesized from simple building blocks, making them widely accessible for proteomic applications and further technical development throughout the research community. Their versatility with respect to sample type, experimental workflow, and mass spectrometry instrumentation will make PAL reagents an important addition to the quantitative proteomics toolbox.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Michael Senko at ThermoFisher Scientific for providing modified ITCL code and guidance to enable acquisition of extended length time domain transients with the Orbitrap XL instrument. Generous financial support for this work was provided by the Susan Smith Center for Women’s Cancers and the Strategic Research Initiative at the Dana-Farber Cancer Institute, in addition to the National Institutes of Health, P01NS047572 (to J.A.M.) and HG006097-03 (to N.S.G). This work was also supported by the National Science Foundation Division of Materials Research through DMR-11-57490 (to A.G.M) and the State of Florida.
REFERENCES
- 1.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative Mass Spectrometry in Proteomics: A Critical Review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 2.Pan S, Aebersold R. Quantitative Proteomics by Stable Isotope Labeling and Mass Spectrometry. Methods Mol Biol. 2007;367:209–218. doi: 10.1385/1-59745-275-0:209. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber TB, Mausbacher N, Breitkopf SB, Grundner-Culemann K, Daub H. Quantitative Phosphoproteomics--an Emerging Key Technology in Signal-Transduction Research. Proteomics. 2008;8:4416–4432. doi: 10.1002/pmic.200800132. [DOI] [PubMed] [Google Scholar]
- 4.Ramisetty SR, Washburn MP. Unraveling the Dynamics of Protein Interactions with Quantitative Mass Spectrometry. Crit Rev Biochem Mol Biol. 2011;46:216–228. doi: 10.3109/10409238.2011.567244. [DOI] [PubMed] [Google Scholar]
- 5.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, Silac, as a Simple and Accurate Approach to Expression Proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., 3rd 15n Metabolic Labeling of Mammalian Tissue with Slow Protein Turnover. J Proteome Res. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the Synaptosomal Proteome of the Rat Cerebellum During Post-Natal Development. Genome Res. 2007;17:1378–1388. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao P, Ren Y, Alpert AJ, Sze SK. Detection, Evaluation and Minimization of Nonenzymatic Deamidation in Proteomic Sample Preparation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O111.009381. O111 009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulbrich A, Merrill AE, Hebert AS, Westphall MS, Keller MP, Attie AD, Coon JJ. Neutron-Encoded Protein Quantification by Peptide Carbamylation. J Am Soc Mass Spectrom. 2013 doi: 10.1007/s13361-013-0765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative Analysis of Complex Protein Mixtures Using Isotope-Coded Affinity Tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 11.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed Protein Quantitation in Saccharomyces Cerevisiae Using Amine-Reactive Isobaric Tagging Reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by Ms/Ms. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 13.Palaniappan KK, Pitcher AA, Smart BP, Spiciarich DR, Iavarone AT, Bertozzi CR. Isotopic Signature Transfer and Mass Pattern Prediction (Isostamp): An Enabling Technique for Chemically-Directed Proteomics. ACS Chem Biol. 2011;6:829–836. doi: 10.1021/cb100338x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-Plex Quantitation of Changes in Cerebrospinal Fluid Protein Expression in Subjects Undergoing Intravenous Immunoglobulin Treatment for Alzheimer's Disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mertins P, Udeshi ND, Clauser KR, Mani DR, Patel J, Ong SE, Jaffe JD, Carr SA. Itraq Labeling Is Superior to Mtraq for Quantitative Global Proteomics and Phosphoproteomics. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014423. M111 014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ting L, Rad R, Gygi SP, Haas W. Ms3 Eliminates Ratio Distortion in Isobaric Multiplexed Quantitative Proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. Itraq Underestimation in Simple and Complex Mixtures:"The Good, the Bad and the Ugly". J Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 18.Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Gas-Phase Purification Enables Accurate, Multiplexed Proteome Quantification with Isobaric Tagging. Nat Methods. 2011;8:933–935. doi: 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blank LM, Desphande RR, Schmid A, Hayen H. Analysis of Carbon and Nitrogen Co-Metabolism in Yeast by Ultrahigh-Resolution Mass Spectrometry Applying 13c- and 15n-Labeled Substrates Simultaneously. Anal Bioanal Chem. 2012;403:2291–2305. doi: 10.1007/s00216-012-6009-4. [DOI] [PubMed] [Google Scholar]
- 20.Marto JA. Protected Amine Labels and Use in Detecting Analytes. 2011 [Google Scholar]
- 21.Hebert AS, Merrill AE, Stefely JA, Bailey DJ, Wenger CD, Westphall MS, Pagliarini DJ, Coon JJ. Amine-Reactive Neutron-Encoded Labels for Highly Plexed Proteomic Quantitation. Mol Cell Proteomics. 2013;12:3360–3369. doi: 10.1074/mcp.M113.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M. High-Resolution Enabled Tmt 8-Plexing. Anal Chem. 2012;84:7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- 23.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Increasing the Multiplexing Capacity of Tmts Using Reporter Ion Isotopologues with Isobaric Masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Shan Y, Wu Q, Zhang S, Zhang L, Zhang Y. Mass Defect-Based Pseudo-Isobaric Dimethyl Labeling for Proteome Quantification. Anal Chem. 2013 doi: 10.1021/ac402834w. [DOI] [PubMed] [Google Scholar]
- 25.Hebert AS, Merrill AE, Bailey DJ, Still AJ, Westphall MS, Strieter ER, Pagliarini DJ, Coon JJ. Neutron-Encoded Mass Signatures for Multiplexed Proteome Quantification. Nat Methods. 2013;10:332–334. doi: 10.1038/nmeth.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose CM, Merrill AE, Bailey DJ, Hebert AS, Westphall MS, Coon JJ. Neutron Encoded Labeling for Peptide Identification. Anal Chem. 2013;85:5129–5137. doi: 10.1021/ac400476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiring HD, van der Heeft E, ten Hove GJ, de Jong A. Nanoscale Lc-Ms(N): Technical Design and Applications to Peptide and Protein Analysis. J Sep Sci. 2002;25:557–568. [Google Scholar]
- 28.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-Resolved Mass Spectrometry of Tyrosine Phosphorylation Sites in the Epidermal Growth Factor Receptor Signaling Network Reveals Dynamic Modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Everley RA, Kunz RC, McAllister FE, Gygi SP. Increasing Throughput in Targeted Proteomics Assays: 54-Plex Quantitation in a Single Mass Spectrometry Run. Anal Chem. 2013;85:5340–5346. doi: 10.1021/ac400845e. [DOI] [PubMed] [Google Scholar]
- 30.Dong F, van Buitenen C, Pouwels K, Hoefsloot LH, Lowenberg B, Touw IP. Distinct Cytoplasmic Regions of the Human Granulocyte Colony-Stimulating Factor Receptor Involved in Induction of Proliferation and Maturation. Mol Cell Biol. 1993;13:7774–7781. doi: 10.1128/mcb.13.12.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermans MH, Ward AC, Antonissen C, Karis A, Lowenberg B, Touw IP. Perturbed Granulopoiesis in Mice with a Targeted Mutation in the Granulocyte Colony-Stimulating Factor Receptor Gene Associated with Severe Chronic Neutropenia. Blood. 1998;92:32–39. [PubMed] [Google Scholar]
- 32.Maun NA, Gaines P, Khanna-Gupta A, Zibello T, Enriquez L, Goldberg L, Berliner N. G-Csf Signaling Can Differentiate Promyelocytes Expressing a Defective Retinoic Acid Receptor: Evidence for Divergent Pathways Regulating Neutrophil Differentiation. Blood. 2004;103:1693–1701. doi: 10.1182/blood-2002-10-3247. [DOI] [PubMed] [Google Scholar]
- 33.Metcalf D. Hematopoietic Cytokines. Blood. 2008;111:485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hercus TR, Thomas D, Guthridge MA, Ekert PG, King-Scott J, Parker MW, Lopez AF. The Granulocyte-Macrophage Colony-Stimulating Factor Receptor: Linking Its Structure to Cell Signaling and Its Role in Disease. Blood. 2009;114:1289–1298. doi: 10.1182/blood-2008-12-164004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pazdrak K, Young TW, Stafford S, Olszewska-Pazdrak B, Straub C, Starosta V, Brasier A, Kurosky A. Cross-Talk between Icam-1 and Granulocyte-Macrophage Colony-Stimulating Factor Receptor Signaling Modulates Eosinophil Survival and Activation. J Immunol. 2008;180:4182–4190. doi: 10.4049/jimmunol.180.6.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Touw IP, De Koning JP, Ward AC, Hermans MH. Signaling Mechanisms of Cytokine Receptors and Their Perturbances in Disease. Mol Cell Endocrinol. 2000;160:1–9. doi: 10.1016/s0303-7207(99)00206-3. [DOI] [PubMed] [Google Scholar]
- 37.Ward AC, Loeb DM, Soede-Bobok AA, Touw IP, Friedman AD. Regulation of Granulopoiesis by Transcription Factors and Cytokine Signals. Leukemia. 2000;14:973–990. doi: 10.1038/sj.leu.2401808. [DOI] [PubMed] [Google Scholar]
- 38.Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic Bead Processor for Rapid Evaluation and Optimization of Parameters for Phosphopeptide Enrichment. Anal Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, Garg B, Adelmant G, Webber JT, Luckey CJ, Marto JA. Online Nanoflow Multidimensional Fractionation for High Efficiency Phosphopeptide Analysis. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.O111.011064. O111 011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe'er D, Tanner SD, Nolan GP. Single-Cell Mass Cytometry of Differential Immune and Drug Responses across a Human Hematopoietic Continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka S, Saito Y, Kunisawa J, Kurashima Y, Wake T, Suzuki N, Shultz LD, Kiyono H, Ishikawa F. Development of Mature and Functional Human Myeloid Subsets in Hematopoietic Stem Cell-Engrafted Nod/Scid/Il2rgammako Mice. J Immunol. 2012;188:6145–6155. doi: 10.4049/jimmunol.1103660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piu F, Magnani M, Ader ME. Dissection of the Cytoplasmic Domains of Cytokine Receptors Involved in Stat and Ras Dependent Proliferation. Oncogene. 2002;21:3579–3591. doi: 10.1038/sj.onc.1205444. [DOI] [PubMed] [Google Scholar]
- 43.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic Discovery of in Vivo Phosphorylation Networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaub TM, Hendrickson CL, Horning S, Quinn JP, Senko MW, Marshall AG. High-Performance Mass Spectrometry: Fourier Transform Ion Cyclotron Resonance at 14.5 Tesla. Anal Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 45.Tipton JD, Tran JC, Catherman AD, Ahlf DR, Durbin KR, Lee JE, Kellie JF, Kelleher NL, Hendrickson CL, Marshall AG. Nano-Lc Fticr Tandem Mass Spectrometry for Top-Down Proteomics: Routine Baseline Unit Mass Resolution of Whole Cell Lysate Proteins up to 72 Kda. Anal Chem. 2012;84:2111–2117. doi: 10.1021/ac202651v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Cottrell CE, Marshall AG. Effect of Signal-to-Noise Ratio and Number of Data Points Upon Precision in Measurement of Peak Amplitude, Position and Width in Fourier Transform Spectrometry. Chemometr Intell Lab. 1986;1:51–58. [Google Scholar]
- 47.Marshall AG. Theoretical Signal-to-Noise Ratio and Mass Resolution in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal Chem. 1979;51:1710–1714. [Google Scholar]
- 48.Zhang Y, Ficarro SB, Li S, Marto JA. Optimized Orbitrap Hcd for Quantitative Analysis of Phosphopeptides. J Am Soc Mass Spectrom. 2009;20:1425–1434. doi: 10.1016/j.jasms.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Rizzo JM, Buck MJ. Key Principles and Clinical Applications of "Next-Generation" DNA Sequencing. Cancer Prev Res (Phila) 2012;5:887–900. doi: 10.1158/1940-6207.CAPR-11-0432. [DOI] [PubMed] [Google Scholar]
- 50.Ecker JR. Forum: Genomics Encode Explained. Nature. 2012;489:52–53. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 51.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Link AJ. Multidimensional Peptide Separations in Proteomics. Trends Biotechnol. 2002;20:S8–S13. doi: 10.1016/s1471-1931(02)00202-1. [DOI] [PubMed] [Google Scholar]
- 53.Di Palma S, Hennrich ML, Heck AJ, Mohammed S. Recent Advances in Peptide Separation by Multidimensional Liquid Chromatography for Proteome Analysis. J Proteomics. 2012;75:3791–3813. doi: 10.1016/j.jprot.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 54.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 56.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach JJ, Cox J, Horning S, Mann M, Makarov A. Ultra High Resolution Linear Ion Trap Orbitrap Mass Spectrometer (Orbitrap Elite) Facilitates Top Down Lc Ms/Ms and Versatile Peptide Fragmentation Modes. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. O111 013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makarov A, Denisov E, Lange O. Performance Evaluation of a High-Field Orbitrap Mass Analyzer. J Am Soc Mass Spectrom. 2009;20:1391–1396. doi: 10.1016/j.jasms.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass Spectrometry-Based Proteomics Using Q Exactive, a High-Performance Benchtop Quadrupole Orbitrap Mass Spectrometer. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. M111 011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.