In the reported X-ray cocrystal structure of the DDR1 kinase domain in complex with the inhibitor DDR1-IN-1 (PDB code 4BKI), the indolin-2-one moiety was modeled with two hydrogen bonds to the kinase hinge residues Met704 and Asp702. Subsequent analysis of the electron density has revealed that the indolin-2-one group is flipped allowing only a single hydrogen bond to Met704. The amended coordinates have been released with the new PDB code 4CKR. The corrected Figure 2A is shown, together with the updated refinement statistics (Table 5) reported in Supporting Information Table 5 of the original paper. This change does not otherwise affect the scientific integrity of the article. We thank Oliver Smart, Global Phasing Ltd., for drawing our attention to the error.
Figure 2.
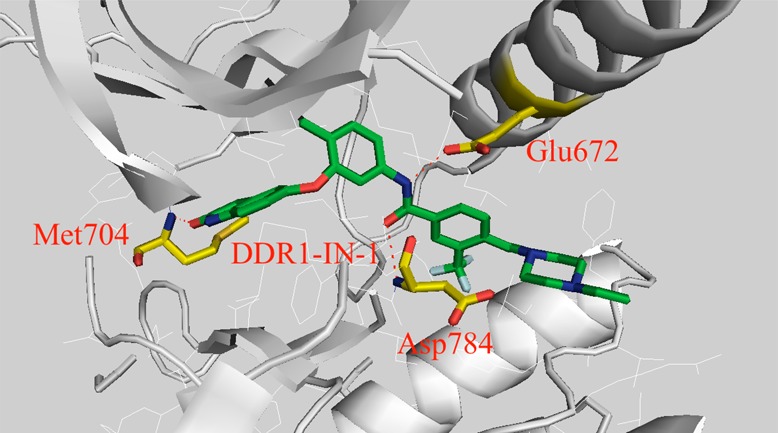
Binding information of DDR1-IN-1/2 against DDR1. (A) X-ray cocrystal structure of DDR1-IN-1 with DDR1 kinase.
Table 5. Data Processing and Refinement Statistics. Values in Parentheses Refer to the Highest Resolution Shell.
| DDR1 with DDR1-IN-1 | |
|---|---|
| Data | |
| wavelength (Å) | 0.9795 |
| resolution range (Å) | 49.44–2.2 (2.278–2.199) |
| space group | P41212 |
| unit cell (Å) | 59.3, 59.3, 178.5 |
| unit cell (deg) | 90, 90, 90 |
| total reflections | 183798 (14101) |
| unique reflections | 17062 (1526) |
| multiplicity | 11 (9.2) |
| completeness (%) | 100 (100.00) |
| I/σ(I) | 17.5 (2.4) |
| Wilson B-factor (Å2) | 36.2 |
| R-merge | 0.1 (0.956) |
| R-meas | 0.109 (1.071) |
| CC1/2 | 0.999 (0.720) |
| Refinement | |
| R-work/R-free | 0.1982/0.2470 |
| no. of atoms | 2422 |
| macromolecules | 2324 |
| ligands | 60 |
| water | 38 |
| protein residues | 301 |
| RMS(bonds) (Å) | 0.01 |
| RMS(angles) (°) | 1.3 |
| Ramachandran favored (%) | 96.2 |
| Ramachandran outliers (%) | 0.34 |
| Clashscore | 1.49 |
| avg B-factor (Å2) | 48.3 |
| macromolecules | 48.8 |
| ligands | 33.1 |
| solvent | 37.3 |
| PDB ID | 4CKR |
Funding Statement
National Institutes of Health, United States


