Abstract
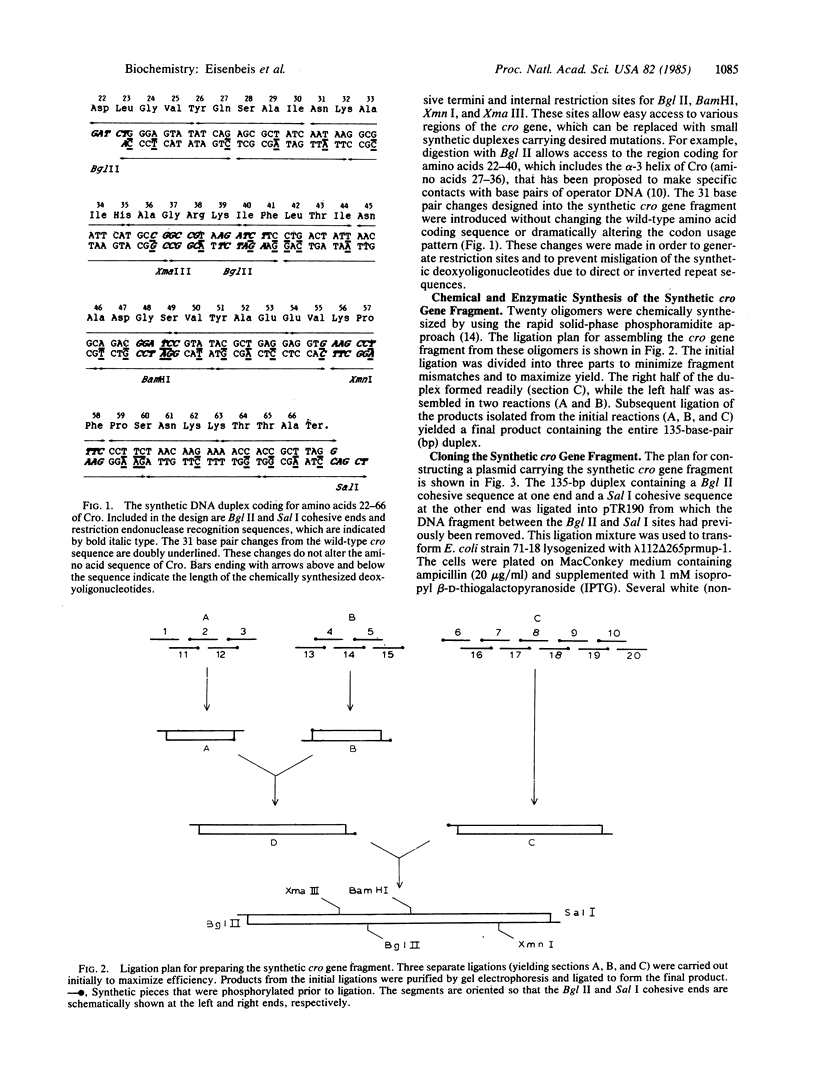
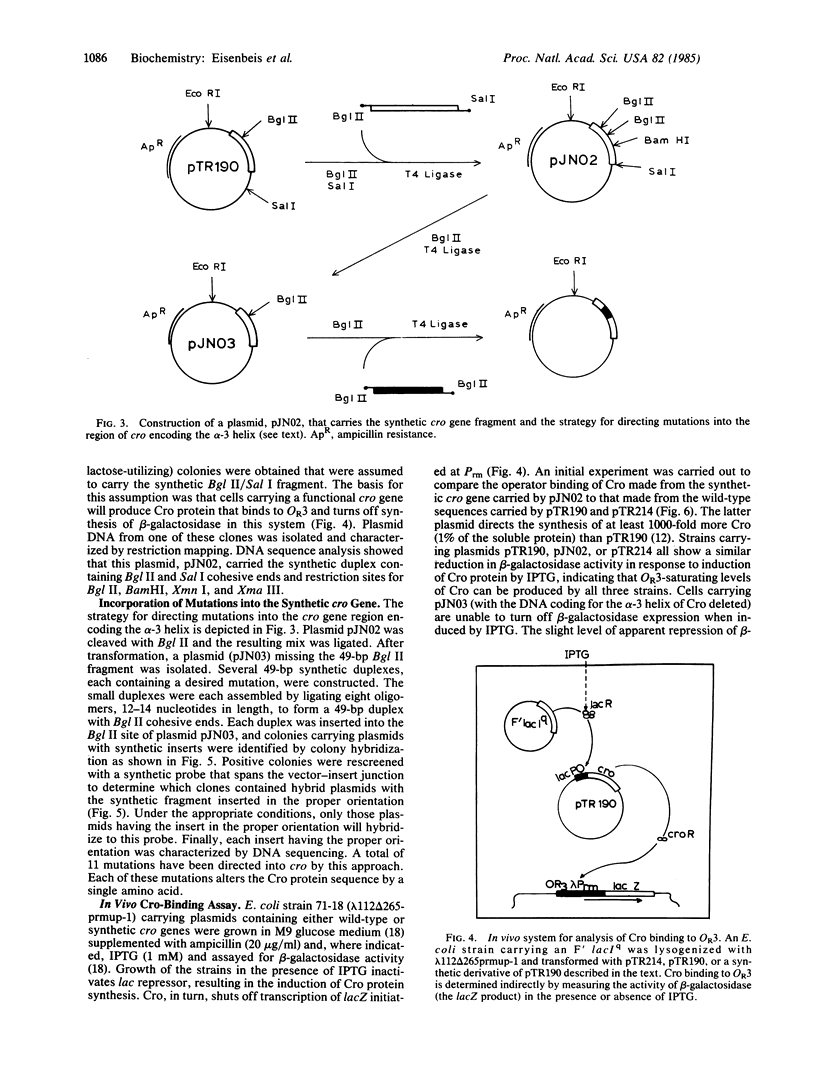
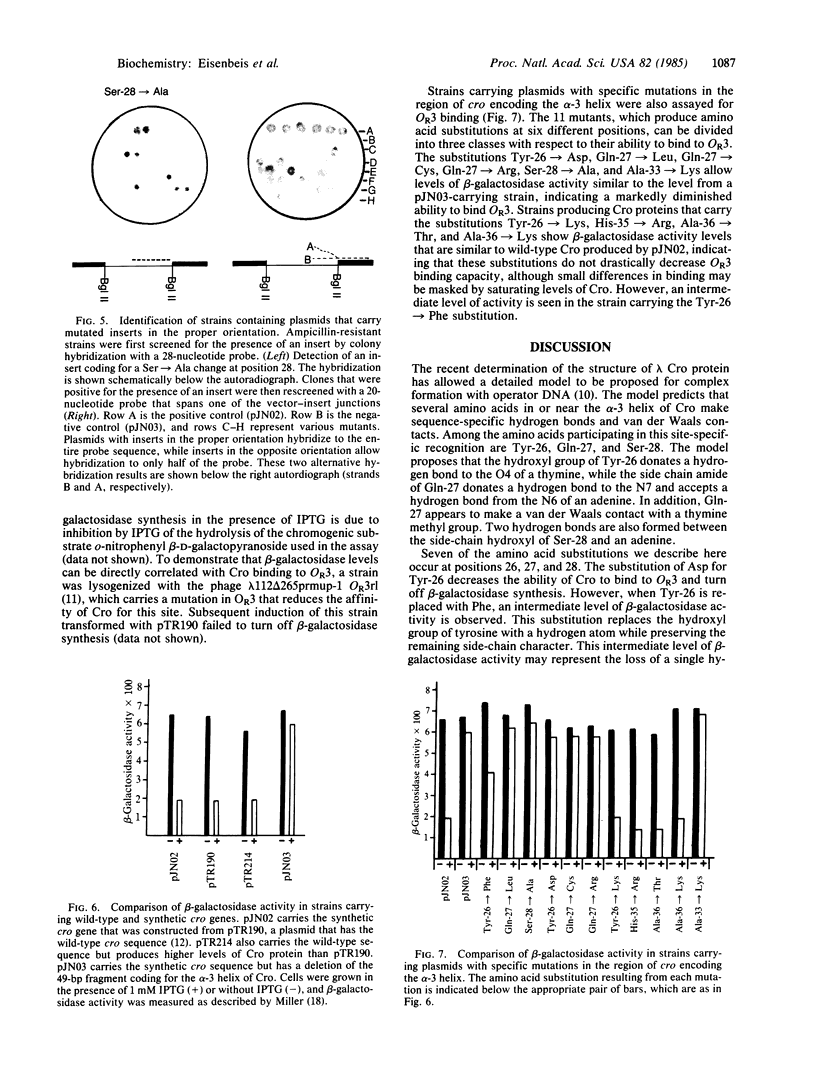
A portion of the gene coding for the Cro repressor protein of bacteriophage lambda has been chemically synthesized, incorporating base pair changes that generate restriction endonuclease sites without altering the amino acid coding sequence. These restriction endonuclease sites were used to remove small segments of the synthetic cro gene and the segments were replaced with duplexes carrying desired mutations. Altered Cro proteins produced by mutants constructed in this manner were then assayed for binding to lambda operator OR3 in vivo. Mutations directed into the region of the cro gene encoding the alpha-3 helix produced altered Cro proteins with a range of affinities for operator DNA. These changes suggest which amino acids play an important role in Cro-OR3 complex formation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F. Proposed alpha-helical super-secondary structure associated with protein-dna recognition. J Mol Biol. 1982 Aug 25;159(4):745–751. doi: 10.1016/0022-2836(82)90111-5. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M., Jeffrey A., Wang J., Ladner R., Ptashne M., Pabo C. O. Structure of the operator-binding domain of bacteriophage lambda repressor: implications for DNA recognition and gene regulation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):435–440. doi: 10.1101/sqb.1983.047.01.051. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R., Meyer B., Ptashne M. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980 May 15;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Hecht M. H., Sauer R. T. Mutations defining the operator-binding sites of bacteriophage lambda repressor. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):441–449. doi: 10.1101/sqb.1983.047.01.052. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Kacich R., Ptashne M. A general method for maximizing the expression of a cloned gene. Proc Natl Acad Sci U S A. 1979 Feb;76(2):760–764. doi: 10.1073/pnas.76.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Khorana H. G. CXII. Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J Mol Biol. 1972 Dec 28;72(2):427–444. doi: 10.1016/0022-2836(72)90155-6. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., McKay D. B., Steitz T. A. Two helix DNA binding motif of CAP found in lac repressor and gal repressor. Nucleic Acids Res. 1982 Aug 25;10(16):5085–5102. doi: 10.1093/nar/10.16.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]