Abstract
Signal transduction pathways downstream of receptor tyrosine kinases (RTKs) are often deregulated during oncogenesis, tumor progression, and metastasis. In particular, the peptide growth factor hormone, hepatocyte growth factor (HGF), and its specific receptor, Met tyrosine kinase, regulate cancer cell migration, thereby conferring an aggressive phenotype (Nakamura et al., J Clin Invest 106(12):1511–1519, 2000; Huh et al., Proc Natl Acad Sci U S A 101:4477–4482, 2004). Additionally, overexpression of Met is associated with enhanced invasiveness of breast cancer cells (Edakuni et al., Pathol Int 51(3):172–178, 2001; Jin et al., Cancer 79(4):749–760, 1997; Tuck et al., Am J Pathol 148(1):225–232, 1996). Here, we review the regulation of recently identified novel downstream mediators of HGF/Met signaling, Breast tumor kinase (Brk/PTK6), and Src-associated substrate during mitosis of 68 kDa (Sam68), and discuss their relevance to mechanisms of breast cancer progression.
Keywords: HGF, Met, Brk, PTK6, Erk5, Sam68, Cell migration, Breast cancer
Introduction
Met receptor activity is critical to cell migration in both normal physiological and pathological invasive growth processes, such as during wound healing or as part of cancer cell metastasis. The ability of cells to migrate, invade neighboring tissues, survive in a foreign microenvironment, and settle/proliferate at distant sites defines a biological event known as the “invasive growth program.” This program is appropriately utilized during normal embryonic development, organ formation, as part of inflammatory responses, and for injury repair. A similar process is also prevalent during early cancer cell transformation, such as during epithelial-to-mesenchymal transition (EMT) that precedes metastasis. EMT is characterized by release of epithelial cell junctions, changes in cell polarity, cytoskeletal rearrangement, and an increased ability to move through the extracellular matrix (ECM). Met-driven signal transduction (primarily PI3K/Akt, STAT, and Ras/ERK1/2 MAP kinase driven) is now well studied in both normal and neoplastic models of cell motility and invasive cellular behavior. Herein, we review recent studies that suggest the addition of a novel signaling pathway characterized by regulation of breast tumor kinase (Brk/PTK6), Erk5, and Sam68 molecules that function as mediators of cellular migration downstream of Met receptor activation.
HGF/MET Tyrosine Kinase Growth Factor Receptor Signaling: Structure and Function
HGF
The Met receptor ligand, hepatocyte growth factor (HGF), also named scatter factor (SF), is a pleiotropic growth factor initially isolated from the serum of hepatectomized rats [6, 7] and later from fibroblasts [8]. HGF belongs to the family of plasminogen-related growth factors (PRGFs) and is also called PRGF-1. Human HGF proteins are transcribed as inactive 90 kDa, single-chain precursors (pro-HGF) from a single gene located on chromosome 7 q 21.1 [9, 10]. Pro-HGF is then converted into an active, mature heterodimeric form, consisting of a disulfide-linked 69 kDa α chain and 34 kDa β chain [11, 12].
HGF is widely expressed in different tissues and mainly produced by mesenchymal and stromal cells. Biological activation of pro-HGF is mediated by a single cleavage event performed by several tightly regulated serine proteases [13–18]. These enzymes require proteolytic activation by other proteases which are also regulated by specific endogenous inhibitor proteins. The proteolytic cleavage of pro-HGF occurs in the extracellular environment in response to tissue damage or during tumor progression [13]. HGF is an essential regulator of diverse cellular functions including cell proliferation, motility, morphogenesis, and angiogenesis [19, 20] and has been called a mitogen, motogen, and morphogen [21]. HGF acts in a paracrine manner [22, 23] on epithelial cells [24], endothelial cells [25], and cells of the macrophage/monocyte lineage [26], through binding to an HGF-specific receptor tyrosine kinase (RTK) termed Met.
Met
The Met receptor is a transmembrane RTK encoded by the c-Met proto-oncogene located on chromosome 7q 21–31 [27]. c-Met was originally identified as a fusion protein (Tpr/Met) generated from chromosomal translocation in human osteogenic sarcoma (HOS) cells treated with the carcinogen, N-methyl-N′-nitro-N-nitrosoguanidine [28]. In these cells, the translocated promoter region locus (TPR) on chromosome 1 is placed upstream of a portion of the c-Met oncogene [29], generating the Trp/Met fusion protein characterized by a constitutively phosphorylated and thus active Met kinase domain.
Met is expressed in both epithelial and endothelial cells [20] and is synthesized as a 170-kDa single-chain precursor which is proteolytically converted into a disulfide-linked, highly glycosylated extracellular 50 kDa α subunit and a transmembrane 145-kDa β chain. The extracellular segment of the Met receptor, which includes the full α subunit, contains an atypical protein–protein interaction motif, the Sema domain, characterized by low ligand-binding affinity. The extracellular portion of Met receptor also features a cysteine-rich domain (Cys-domain) known as Met-related sequence (MRS) and a classical protein–protein interaction region made of four immunoglobulin-like motifs (IPT domain).
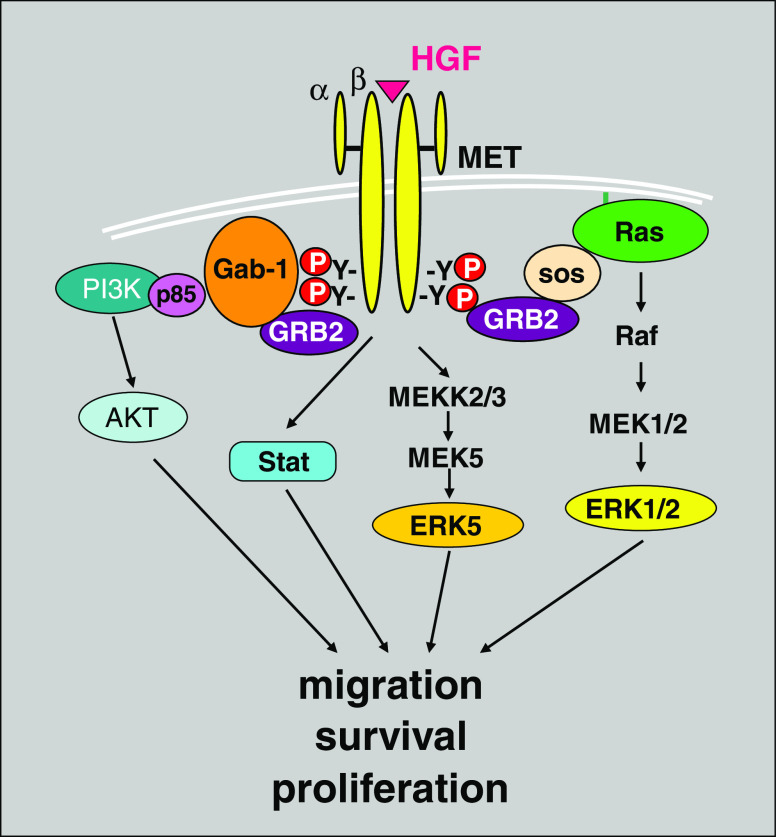
The intracellular portion of the Met receptor contains a juxtamembrane region, a tyrosine kinase domain, and a C-terminal regulatory tail. The juxtamembrane domain plays an essential role in receptor downregulation [20]. This domain contains a serine residue (Ser985) that, upon phosphorylation by PKC, is responsible for inhibition of kinase activity and a tyrosine residue (Tyr 1003) that mediates binding of the ubiquitin ligase Cbl; polyubiquitination of Met receptor by Cbl subsequently results in Met degradation [30, 31]. The Met kinase domain contains key tyrosine residues (Tyr 1230, 1234, and 1235) within the activation loop responsible for the regulation of Met kinase activity [32]. Upon HGF binding, Met receptors undergo dimerization and autophosphorylation of Tyr1234 and Tyr1235, leading to phosphorylation of Tyr1349 and Tyr1356 of the C-terminal regulatory tail. When phosphorylated, the C-terminal regulatory tail acts as a docking site for intracellular adaptor molecules, such as Shc. Binding of downstream kinases to Met can be direct, as it is for c-Src and the p85 subunit of PI3K, or mediated by the scaffolding protein Gab1 [33]. Recruitment of Src homology 2 (SH2) domain-containing proteins favors the activation of downstream signal transduction cascades. These include mitogen-activated protein kinase (MAPK), which is important for cell proliferation and transformation [33], PI3K, which is essential for cell motility [34] and preventing apoptosis via Akt-activation [35], and JAK/STAT pathways members [36], which are required for epithelial tubulogenesis [37] (Fig. 1).
Fig. 1.
HGF/Met receptor signaling pathway. Stimulation of Met receptor by its specific ligand HGF leads to activation of a variety of downstream signaling molecules, targeting cell proliferation, migration, and survival
As described above, Met is a single transmembrane receptor extracellularly exposed on the phospholipidic bi-layer. The combination of ligand activation and dynamic interaction with other cell surface receptors results in synergetic integration of downstream signaling. For example, Met interacts with additional tyrosine kinase receptor family members. While the recepteur d'origine nantais (RON) shares significant homology with the Met receptor, it is specifically activated by binding to macrophage-stimulating protein (MSP) [38]. However, when overexpressed, Met and RON interact in the absence of ligand; activation of each receptor can induce the transphosphorylation of the other family member. This cross talk appears to occur independently of the C-terminal docking site on either receptor but requires RON kinase activity, as kinase-dead mutant RON is able to suppress the transforming actions of Met receptor [39]. Additionally, Met shares structural similarities with the receptor members of the semaphorin family, such as plexins. Met interacts constitutively with plexin B, and clustering of Met with activated plexin BI causes HGF-independent stimulation of Met and invasive growth [40].
Met is known to interact with multiple membrane-associated proteins which function to modulate cell signaling in response to HGF. For example, Met constitutively associates with integrins, generating a platform for downstream signaling effectors necessary to promote cell migration and invasion [20]. Met can also interact with the death receptor Fas to protect cells from apoptosis [41]. Finally, Met receptors associate with CD44, a transmembrane glycoprotein that functions as a receptor for the major ECM component, hyaluronic acid. Specific CD44 isoforms generated by alternative splicing are able to trigger or enhance Met activation. For example, the CD44 isoform containing the variant exon 3 (CD44v3) strongly binds HGF and is involved in concentrating HGF at the cell surface and thus presents HGF to the Met receptor [42]. In addition, CD44v6 is required for ligand-dependent activation of Met [43]. Recently, Locatelli and Lange [44] showed that activated Met receptors regulate keratinocyte cell migration through expression of CD44v5, an isoform that correlates with the invasiveness of renal cell carcinoma [45].
Met Signaling Drives an Invasive Growth Program in Normal and Cancerous Tissues
The Met pathway is essential for normal development. Lack of either HGF or Met in knockout mice leads to lethal morphogenetic defects during embryogenesis [36]; HGF/Met complexes control proliferation and cell survival of hepatocytes, placental formation, nervous system organization, bone remodeling, angiogenesis, and cell migration from the dermomyotome to targets where cells form skeletal muscle [46–48]. In adult conditional mutant mice, deregulated HGF or Met signaling contributes to compromised tissue repair after injury [49]. HGF is upregulated during liver regeneration [50–52] and repair of other tissues like lung, kidney, heart, and skin [1, 53–56]. In wound healing models, the HGF/Met pathway promotes motility [21, 57, 58] and rapid migration [24, 59] of keratinocyte (skin) cells. Birchmeier and colleagues have shown that Met is specifically localized to the leading edge of migrating keratinocytes and acts in an autocrine fashion to promote wound healing; skin cells expressing mutant Met receptors (loss of function) are no longer able to proliferate and migrate into the wound area [49]. In the normal mammary gland, HGF, primarily produced by stromal fibroblasts [60], promotes bud elongation and infiltrating branching tubulogenesis via paracrine stimulation of Met receptors expressed in normal ductal and lobular breast epithelial cells [61].
In addition to normal processes, aberrant activation of the HGF/Met signaling pathway leads to oncogenesis, tumor progression, and metastasis [1, 2]. Constitutive activation of the Met receptor occurs under the following conditions: establishment of ligand–receptor activation loops that induce sustained activation of the receptor in a paracrine or autocrine manner [62], receptor overexpression triggering oligomerization and reciprocal activation, a condition that sensitizes cells to lower levels of HGF, activating point mutations within the kinase domain, pathway activation via hypoxic conditions (i.e., that increase Met expression), transactivation by other membrane proteins, and loss of negative regulators [63].
Mutations of Met proteins are not particularly frequent in tumors, whereas enhanced Met expression has been reported in a variety of solid tumors such as osteosarcomas [62], renal [64], ovarian [65], hepatocellular [66], non-small cell lung [67], gastric, pancreatic [68], prostatic [69], and invasive breast carcinomas [4]. Overexpression of Met receptor is associated with more aggressive and invasive phenotypes in these tumors and poor prognosis. In the absence of gene amplification [65] or mutation, overexpression of Met can be driven by activation of oncogenes including Ras, RET (rearranged during transfection), or Ets [70]. Interestingly, the Met promoter contains four putative binding sites for members of the Ets family of transcription factors, which are involved in promoting invasive growth programs. Ets I induces increased Met mRNA and protein levels which, in turn, can mediate enhanced Ets I mRNA production, suggesting that Ets protein family members act both upstream and downstream of the HGF/Met signaling pathway [71] in a “feed-forward” manner. In colorectal cancers, the WNT/β catenin cascade leads to overexpression and constitutive activation of Met receptors [72]. In the mammary gland, Met shows a gradient of increasing expression from normal breast/benign mammary hyperplasia (lowest expression) to ductal carcinoma in situ (DCIS), and is overexpressed in invasive carcinoma [4]; Met mRNA shows a similar gradient of strong presence at the stromal–epithelial cell interface to weak expression in the inner part of the tumor [5]. Met and HGF are expressed at the active invading front of breast tumors [3, 5]; deregulated expression of HGF/Met promotes the invasive growth program, leading to breast tumor progression and metastasis.
Recent studies conducted in knock-in mice expressing an activated Met receptor mutant revealed its tumorigenic effect on mammary epithelium: Met mutant mice develop diverse mammary tumors with high incidence, and these adenocarcinomas show basal characteristics [73]. Met RNA and protein expression is associated with the human basal-like breast cancer subtype and poor outcome [73, 74]; moreover, Met expression correlates with increased levels of EMT markers in breast tumors [74]. In accordance with Met association with the basal-like breast tumor subtype, there is an inverse correlation between Met and estrogen receptor (ER), progesterone receptor (PR), and ErbB2/HER2 expression in tumor hyperplastic regions, suggesting a compensatory effect of Met signaling in the presence of decreased or negligible detection of these other receptors [73].
Activation of Brk/PTK6 Downstream of MET
Similar to the pattern of Met receptor overexpression in invasive breast carcinomas, the expression of the non-RTK, breast tumor kinase/protein tyrosine kinase 6 (Brk/PTK6), increases in association with increasing histological grade of breast tumors [75] and invasiveness of breast cancer cell lines [76], while this kinase is notably absent from normal mammary tissue [77]. We recently found that HGF robustly activates Brk kinase activity in Met+ breast cancer cells [78]. Brk was initially cloned in a screen for tyrosine kinases expressed in metastatic breast cancers [79] and has since been shown to be expressed in up to 86% of invasive ductal breast carcinomas [75], as well as prostate and colon carcinomas [77, 80, 81], 70% of serous ovarian carcinomas [82], 37.5% of a small sample of head and neck squamous cell carcinoma [83], and a small percentage of metastatic melanomas [84].
Brk belongs to a family of soluble kinases similar to c-Src; as such, Brk phosphorylation and kinase activity can be induced via activation of growth factor receptor signaling cascades, such as EGF, heregulin [75], or HGF [78]. The Brk protein consists of an N-terminal SH2 domain, a Src homology 3 (SH3) domain, and a C-terminal kinase domain that is subject to autophosphorylation and autoinhibition [85] (Fig. 2). However, unlike c-Src, Brk lacks an SH4 domain required for myristoylation, thereby rendering it soluble (i.e., located in both cytoplasmic and nuclear compartments). Similar to c-Src, Brk kinase activity is inhibited through interactions with its own SH2 and SH3 domains [85], but the Brk kinase domain shares higher homology to other family members (Srm, Frk, and Src42A) than it does to the prototypical c-Src kinase domain [86]. Proteins that can associate with Brk (shown by immunoprecipitation) include the RTKs EGFR/ErbB1, ErbB2, ErbB3 [87–89], adaptor proteins, such as insulin receptor substrate-4 and GAP-A.p65 [90], RNA-binding proteins (Sam68; discussed below), and Erk5 MAPK [78] (Fig. 2). Similarly, Brk substrates include a number of RNA-binding proteins and signaling molecules, such as Sam68 [91], BKS/STAP-2 [92, 93], SLM-1 and -2 [94], paxillin [76], KAP3A [95], p190RhoGAP [96], and PSF [97] (Fig. 2); most recently, Brk has been reported to directly phosphorylate beta-catenin [98] and Akt/pkB [99]. Notably, signal transducers and activators of transcription (STAT) 3 and STAT5b are direct substrates of Brk in vitro [100, 101] and critical regulators of mammary gland function [102, 103], in particular lactogenic differentiation (STAT5 [102]) and regression (STAT3 [104]) [105].
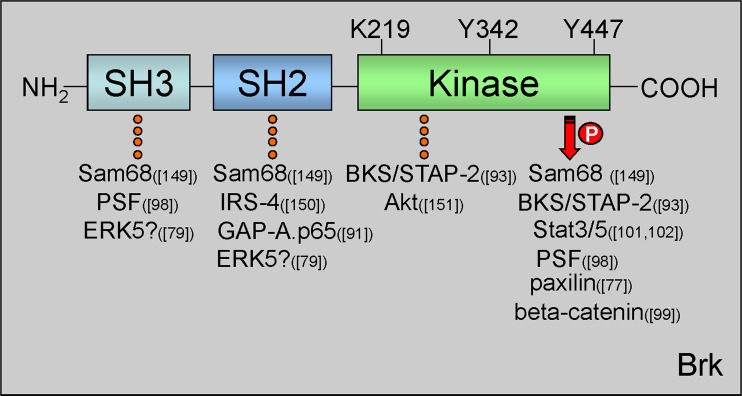
Fig. 2.
Brk protein domains and important regulatory residues. Brk protein structure consists of one N-terminal SH3 domain, one SH2 domain, and a C-terminal kinase domain. Within the kinase domain are three residues important for Brk kinase activity. Lys219 (K219) in the ATP binding pocket is required for kinase activation. Mutation of this site to Met (M) renders Brk kinase-inactive. Tyr342 (Y342) is autophosphorylated upon Brk activation. Tyr447 (Y447) is required for Brk autoinhibition. Substitution of Y to Phe (F) at this site mimics phosphorylation and results in a constitutively active Brk molecule. Brk domain-specific interacting proteins are indicated with dotted line and substrates with arrow; references to the specific proteins are reported in parenthesis [76, 78, 90, 92, 97, 98, 100, 101, 148–150]. PTB polypyrymidine tract, PSF protein-associated splicing factor, IRS-4 insulin receptor substrate 4, BKS/STAP2 breast tumor kinase substrate
Like other protein tyrosine kinases, Brk mediates a range of cellular processes related to the development or maintenance of malignancy [106]. Brk expression sensitizes mammary epithelial cells to the mitogenic effects of EGF [87] and enhances PI3K signaling through increased ErbB3 phosphorylation [88], therefore increasing the strength of pro-survival (potentially oncogenic) signaling events [102, 103]. Notably, Brk promotes anchorage independent growth when expressed in non-transformed mammary epithelial cells [87] and prevents detachment-induced autophagic cell death in cancer cells [107, 108], suggesting a potential mechanism for Brk-positive cancer cells to survive the dissemination phase of metastasis. Brk also promotes EGF or heregulin-induced Erk5 and p38 MAPK activation as well as increased cyclin D1 expression [75] and mediates HGF-induced Erk5 activation [78]; these events were required for breast cancer cell migration (further discussed below) [44, 75, 78]. Recently, Lofgren et al. showed that WAP-driven Brk expression in mammary epithelium of transgenic mice results in delayed post-weaning mammary involution and early formation of mammary tumors, most likely via promoting activation of p38 MAPK. These events may prolong survival of mammary epithelium normally programmed to undergo apoptosis and alter the rate or progress of tissue remodeling. Surviving cells may thus accumulate enough genetic damage to form tumors [108].
Sam68 is a Tumor- and Brk-Associated RNA-Binding Protein
Src-associated substrate during mitosis of 68 kDa (Sam68) is an RNA-binding protein that was the first identified substrate for Brk phosphorylation in vivo [91]. Brk colocalizes with Sam68 in dynamic spherical nuclear structures called Sam68/SLM nuclear bodies (SNBs) [109]. SNBs measure less than 1 μM and are located in proximity to the nucleoli. SNBs disassemble during mitosis and upon treatment with transcriptional inhibitors [109]. Interestingly, SNBs were observed in immortalized and transformed cells but absent in normal cells; in addition, they correlate with differentiation status and tumorigenicity in multiple breast cancer cell lines [109]. Thus, SNBs could potentially be used as a marker for cancer cells.
Sam68 Domain Structure
Sam68 belongs to the heteronuclear ribonucleoprotein particle K (hnRNP K) homology (KH) domain family of RNA-binding proteins. Sam68 is also a member of the signal transduction and activation of RNA (STAR) family of proteins [110], as it contains a single extended KH domain, embedded in a larger GSG domain, and motifs recognized by downstream members of the signaling pathways (Fig. 3). These domains and motifs allow Sam68 to process RNA in response to extracellular signals. The GSG domain (GRP33, Sam68, and GLD-1) is required for Sam68 RNA binding and homodimerization [4, 111]. While the mechanisms of action are unclear, this domain is also essential for androgen-dependent proliferation and survival of prostrate cancer cells [112]. Sam68 recognizes bipartite RNA sequences and binds to RNA with high affinity for poly (U) [113] and poly (A) [111] ribonucleotide homopolymers [114]. Recently, the N-terminus of the KH domain, referred to as Qua1, was identified as sufficient for Sam68 homodimerization in vitro [115]. Tyrosine phosphorylation on Sam68 by soluble kinases, such as Brk, causes a disruption of these dimers, reducing the ability of Sam68 to bind RNA as compared to its dephosphorylated state [91, 111, 116, 117].
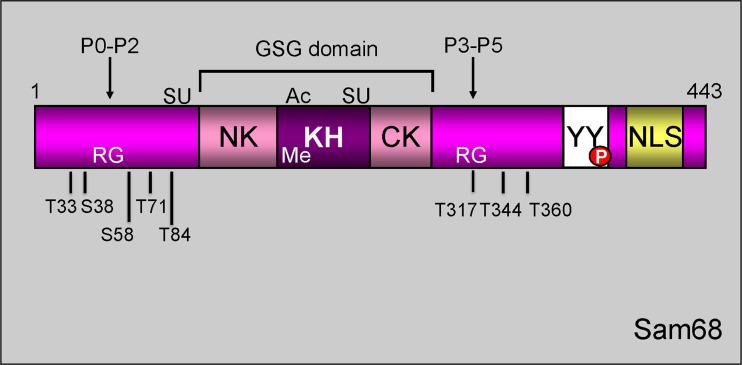
Fig. 3.
Schematic representation of Sam68 structural/functional domains. Sam68 is composed of a GSG domain, consisting of a single RNA-binding KH domain flanked by NK (N-terminal of KH) and CK (C-terminal of KH) segments, six consensus proline-rich motifs (P0–P5), arginine-rich boxes (RG), C-terminal tyrosine-rich domain (YY), and an NLS. Sam68 undergoes post-translational modification such as SUMOylation (SU), acetylation (Ac), methylation (Me), and phosphorylation (P)
In addition to the GSG domain, Sam68 contains six proline-rich sequences (denoted P0 through P5) that are responsible for Sam68 interaction with SH3 (Src homology 3) and WW domain (a short conserved sequence with two signature Trp (W) residues)-containing proteins [118] (Fig. 3). Sam68 was originally identified as a substrate for Src kinase phosphorylation during mitosis [113, 119, 120]; the interaction between Src and Sam68 has been mapped to the SH2 and SH3 domains of Src [113, 120]. In addition to c-Src, the SH3 domain of BRK/Sik (the mouse orthologue of Brk) [91], p85 subunit of PI3K [121], PLC γ-1 [122], and Grb2 [123] interact with proline-rich sequences of Sam68; in particular, P0, P3, P4, and P5 mediate the interaction with Src family tyrosine kinases, leading to phosphorylation of Sam68 [122].
Upon phosphorylation, the tyrosine residues in the C-terminal domain of Sam68 are important for binding to proteins containing SH2 domains [91, 113, 119, 120, 122] including BRK/Sik [116]. Association of Sam68 with SH3 domain-containing proteins decreases its RNA-binding activity and induces its redistribution within the nucleus [116]. Similar to other proteins involved in RNA metabolism, Sam68 contains arginine-glycine (RG)-rich regions and RGG boxes, flanked by proline-rich motifs P0, P3, and P4 (Fig. 3), which are potential sites for arginine methylation [124]. Methylation of Sam68 induces localization to the nuclear compartment [125] [126]. Sam68 exhibits a non-conventional nuclear localization signal (NLS) that is embedded in the last 24 amino acids (from 420 to 443) of the C-terminal region [127] (Fig. 3). Finally, Sam68 also contains two additional nuclear targeting motifs: PPXXR, which is conserved in certain RNA-binding proteins [127], and RXHPYQ/GR, which harbors arginine residues vital for nuclear localization [128].
Sam68 Function and Cell Biology
Divergent roles for Sam68 in cancer have been defined and are suggestive of diverse (context/cell type dependent) activities that determine cell fate. Sam68 was originally defined as a tumor suppressor protein [129, 130]; Sam68 knockout fibroblast cells exhibit anchorage-independent growth, defective contact inhibition, and form metastatic tumors in nude mice [129]. However, other studies suggest that Sam68 contributes to cell growth via regulation of alternative mRNA splicing: in addition to CD44v5 [131], Sam68 is known to regulate mRNA splicing of Bcl-xS [132] and cyclin D1 [133]. Additionally, Sam68 regulates cell cycle progression by promoting S-phase entry through either its RNA-binding or oligomerization abilities. Sam68 expression is elevated in primary breast cancer compared to adjacent nontumorigenic tissues, and its expression and cytoplasmic localization correlate with clinical stage and ER protein levels [134]. Overexpression of wild-type Sam68 in mouse fibroblasts leads to cell cycle arrest (in G1 phase) by downregulating cyclin D1 expression, whereas overexpression of a natural isoform of Sam68 (Sam68 ΔKH) with a deletion of amino acid residues 170 to 208 (within the KH domain) induced cell transformation (as measured by increased soft-agar colony formation) [17, 135]. Downregulation of Sam68 expression reduces proliferation and anchorage-independent growth of breast cancer cells [134]. Additionally, Sam68−/− mice have defects in breast and uterine development, while Sam68 haploinsufficiency correlates with delayed mammary tumor onset [136]. In breast cancer cells, Sam68 colocalizes with the transcriptional coactivator, CBP, and thus may act as an indirect transcriptional repressor, independently of its RNA binding ability [130]. Alternatively, in prostate, Sam68 interacts with androgen receptors and acts as a transcriptional coactivator [137].
Phosphorylation of Sam68 Occurs on Both Tyr and Ser/Thr Residues
Phosphorylation of Sam68 appears to be an important input to its splicing and pro-metastatic activities. Interestingly, phosphorylation of Sam68 is elevated in specimens from breast cancer patients [116]. Phosphorylation of the Sam68 C-terminal domain at Tyr440 by Brk has been reported in MDA-MB-231 breast cancer cells; upon EGF treatment, Brk phosphorylates this residue leading to decreased Sam68 RNA-binding, Sam68 nuclear localization, and cell cycle progression [116]. Notably, in this same model system, Sam68 was also phosphorylated on Ser/Thr residues in a MAP kinase-dependent manner following HGF/Met stimulation [44]. Thus, Sam68 is a target of Brk and other soluble tyrosine kinases and MAPKs (via Ser/Thr phosphorylation). Sam68 contains eight potential proline-directed MAPK phosphorylation sites [138] which are essential for splicing activity and selectivity (namely, inclusion of exon v5 in CD44 mature mRNA) in T-lymphocytes [131]. Moreover, Sam68 phosphorylation promotes EMT of colon adenocarcinoma cells via an ERK1/2-dependent mechanism and alternative splicing of SF2/ASF transcripts [139]. We found that HGF-induced Ser/Thr phosphorylation of Sam68 occurs via either ERK1/2 or ERK5-dependent pathways; these events were required for HGF-induced migration of keratinocyte (HaCat) cells and highly motile breast cancer (MDA-MB-231 and MDA-MB-435) cells [44].
Thus, Sam68 appears to act as a convergence point for multiple signaling pathways. Interestingly, phosphorylation of Ser/Thr residues located outside of the Sam68 KH domain mediated mRNA splicing (potentially of multiple mRNA targets, including CD44v5) and ultimately increased cell migration [44]. These data suggest a critical role for Sam68 in breast tumor progression and potentially in the process of metastasis. In sum, differential phosphorylation events control Sam68 activity: tyrosine phosphorylation of Sam68 by Brk (or c-Src and Src-like kinases) may primarily induce Sam68 nuclear localization and RNA release required for proliferative responses [116], whereas multisite Ser/Thr phosphorylation downstream of ERK1/2 or Brk-driven ERK5 signaling may control breast cancer cell motility required for invasion and metastasis [44], possibly via regulation of the specificity of Sam68-dependent mRNA splicing (Fig. 4).
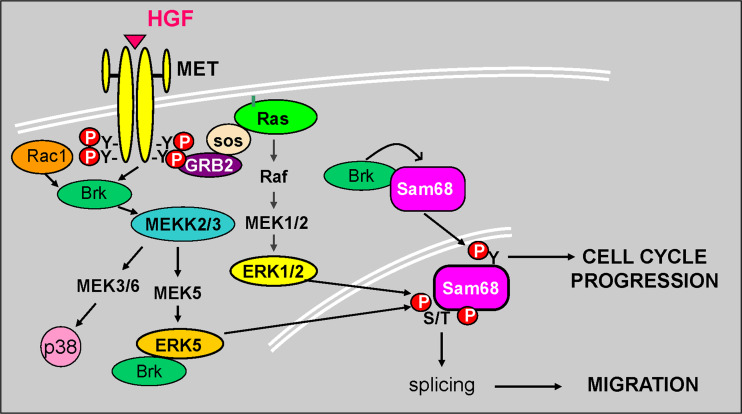
Fig. 4.
Mechanism of action of Brk and Sam68 downstream of activated Met receptors. Brk and Sam68 are essential effectors of HGF-induced cell cycle progression (Brk-mediated Tyr phosphorylation of Sam68) and migration (MAPK-mediated Ser/Thr phosphorylation of Sam68) via an ERK5-dependent mechanism. Growth factor activation facilitates Brk complex formation with ERK5 activating both tyrosine and serine/threonine phosphorylation events which lead to downstream biological outcomes. Brk also associates with and phosphorylates Sam68 to alter its RNA binding activity and thus cell cycle progression. HGF binding to Met receptors induces Brk, Erk5, and Sam68 dependent cellular migration
Novel Met Effectors (Brk, Erk5, and Sam68) are Implicated in Breast Tumor Progression
In HGF-induced breast cancer and keratinocyte cell migration, Brk acts downstream of Met receptors but upstream of Erk5 [78]. We hypothesize that the HGF/Met pathway functions to promote invasion and metastasis in breast cancer (discussed above), in part via activation of Brk and subsequent ERK5-dependent phosphorylation of Sam68 on Ser/Thr residues. Brk likely mediates Met receptor signaling to Erk5 via Brk/Erk5 complex formation. Although the exact details of how Brk signals to Erk5 are unknown, the Brk SH2 or SH3 domain(s) may function to coordinate signaling complexes that recruit one or more upstream kinases in the Erk5 MAPK module (MEKK2/3 and MEK5) and/or their activators, perhaps via linkage to adaptor molecules that commonly recognize these abundant signaling domains (discussed above). Remarkably, both kinase-inactive and wild-type Brk activated endogenous Erk5 and enhanced HGF-driven breast cancer cell motility (Fig. 4). Sam68 was also required for HGF-induced cell migration, and Sam68 phospho-mutants lacking multiple ERK kinase consensus sites blocked migration in response to HGF [78]. These studies indicate that scaffolding actions are an important mediator of Brk function in breast cancer cells. siRNA depletion of Brk, Erk5, or Sam68 abolished HGF-induced cell migration in breast cancer cells [44, 78]. Notably, cellular migration and invasion are mediated by Brk-induced Rac1 activation and phosphorylation of paxillin in response to EGF treatment [76]. Additionally, downstream of Rac-1, Brk-dependent activation of p38 MAPK and increased expression of cyclin D1 appear to contribute to EGF-induced cell migration [75].
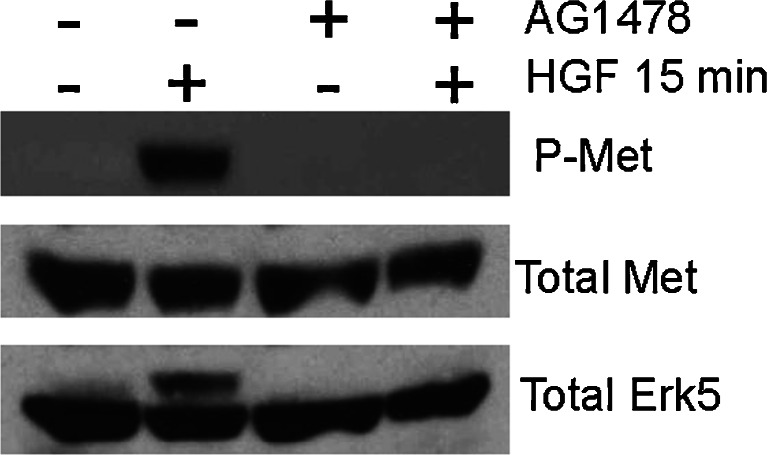
Related to the above findings with EGF, Met can also interact with the epidermal growth factor receptor (EGFR) at multiple signaling levels, signifying an integration of Brk/Erk5/Sam68 upstream activators. In hepatoma and epidermoid carcinoma cell lines, ligand-independent phosphorylation of Met regulates TGFα-activated EGFR, as TGFα and/or EGFR neutralizing antibodies abolished Met phosphorylation [140]. In addition, EGFR activation in thyroid carcinoma cells leads to Met overexpression and constitutive activation [141]; more recently, Met has been shown to collaborate with Her2 and Her3 [142]: Met/ErbB2 cooperation supports invasive growth by promoting breakdown of cell–cell junctions and enhancing cell invasion [143]. Met also contributes to gefitinib resistance in EGFR-activated cells through Met-driven ErbB3 (Her3) activation [144]. Breast cancer cells insensitive to EGFR tyrosine kinase inhibitors rely on Met and c-Src (and possibly Brk) for growth signals [145, 146]. In hepatocellular and pancreatic carcinoma, Met can be trans-activated not only by EGFR but also by G-protein coupled receptors (GPCRs) which increase reactive oxygen species (ROS), resulting in tyrosine phosphatase inhibition and consequent Met receptor activation [147]. Notably, in Brk+ MDA-MB-231 breast cancer cells, HGF-induced activation of Met and Erk5 also requires EGFR; blockade of EGFR kinase activation using AG1478 abolished HGF-induced Met and Erk5 phosphorylation (Fig. 5). Brk activation downstream of EGFR family members is well documented ([106] and discussed above). Thus, EGFR-dependent transactivation of Met receptors provides a potential avenue for amplified Brk signaling as a direct input to selected downstream effectors important for cell migration, including Erk5 and Sam68 [44, 75, 78].
Fig. 5.
EGFR activity is required for HGF-induced phosphorylation of Met receptors and activation of Erk5. MDA-MB-231 breast cancer cells were pretreated with AG1478 (EGFR tyrosine kinase inhibitor) for 30 min prior to HGF stimulation for 15 min. Cells were harvested and Western blotted with antibodies against phospho-Met receptor, total Met receptor, and total Erk5. The up-shifted higher migrating band in the total Erk5 blot represents phosphorylated (i.e., on multiple sites) and activated Erk5
Summary
HGF/Met signaling to Brk, Erk5, and Sam68 is emerging as an important pathway in breast tumorigenesis and breast tumor progression (Fig. 4). Recent work from our group has defined a pathway whereby activation of the Met receptor (and ErbB receptors) induces a Brk/Erk5/Sam68 complex (Lange, unpublished data) which functions to reprogram cell mRNA splicing and thus protein expression to favor breast cancer cell motility [44, 75, 78]. HGF-induced cell migration occurs independently of Brk kinase activity [78], indicating that the ability of Brk to act as a scaffold to increase MAPK (Erk1/2 and Erk5) activity (leading to Sam68 Ser/Thr phosphorylation) is critical to the oncogenic potential of Brk. However, the molecular details of these events remain largely undefined. Perhaps in cancer cells, Brk scaffolding actions ultimately become dominant because overexpressed Brk-SH2 and -SH3 domains signal promiscuously; Brk is absent or expressed at very low levels in normal mammary epithelial cells [77]. Clearly, a better understanding of the complexity and the role(s) played by Brk, various ERKs, and other Sam68-binding proteins functioning downstream of activated Met receptors in breast cancer cells is badly needed; Met, Brk, Erk5, and Sam68 may be considered as part of potential targeted therapies for treatment of metastatic breast cancer or serve as markers for tumors predicted to respond to MAPK (MEK) inhibitors.
Acknowledgements
Work in Carol A. Lange Laboratory was supported by American Cancer Society RSG TBE-107800 (to CAL) and NIH/NCI grant CA107547 (to CAL). We thank Dr. Gwen Dressing for helpful comments.
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- 1.Nakamura T, et al. Myocardial protection from ischemia/reperfusion injury by endogenous and exogenous HGF. J Clin Invest. 2000;106(12):1511–1519. doi: 10.1172/JCI10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huh CG, et al. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edakuni G, et al. Expression of the hepatocyte growth factor/c-Met pathway is increased at the cancer front in breast carcinoma. Pathol Int. 2001;51(3):172–178. doi: 10.1046/j.1440-1827.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- 4.Jin L, et al. Expression of scatter factor and c-met receptor in benign and malignant breast tissue. Cancer. 1997;79(4):749–760. doi: 10.1002/(sici)1097-0142(19970215)79:4<749::aid-cncr12>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Tuck AB, et al. Coexpression of hepatocyte growth factor and receptor (Met) in human breast carcinoma. Am J Pathol. 1996;148(1):225–232. [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Nawa K, Ichihara A. Partial purification and characterization of hepatocyte growth factor from serum of hepatectomized rats. Biochem Biophys Res Commun. 1984;122(3):1450–1459. doi: 10.1016/0006-291x(84)91253-1. [DOI] [PubMed] [Google Scholar]
- 7.Russell WE, McGowan JA, Bucher NL. Partial characterization of a hepatocyte growth factor from rat platelets. J Cell Physiol. 1984;119(2):183–192. doi: 10.1002/jcp.1041190207. [DOI] [PubMed] [Google Scholar]
- 8.Stoker M, et al. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 1987;327:239–242. doi: 10.1038/327239a0. [DOI] [PubMed] [Google Scholar]
- 9.Weidner KM, et al. Evidence for the identity of human scatter factor and human hepatocyte growth factor. Proc Natl Acad Sci U S A. 1991;88(16):7001–7005. doi: 10.1073/pnas.88.16.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuyama R, et al. Regional localization of the hepatocyte growth factor (HGF) gene to human chromosome 7 band q21.1. Genomics. 1991;11(2):410–415. doi: 10.1016/0888-7543(91)90149-9. [DOI] [PubMed] [Google Scholar]
- 11.Donate LE, et al. Molecular evolution and domain structure of plasminogen-related growth factors (HGF/SF and HGF1/MSP) Protein Sci. 1994;3(12):2378–2394. doi: 10.1002/pro.5560031222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura T. Molecular characterization of hepatocyte growth factor (HGF) Seikagaku. 1989;61(10):1243–1247. [PubMed] [Google Scholar]
- 13.Miyazawa K, Shimomura T, Kitamura N. Activation of hepatocyte growth factor in the injured tissues is mediated by hepatocyte growth factor activator. J Biol Chem. 1996;271(7):3615–3618. doi: 10.1074/jbc.271.7.3615. [DOI] [PubMed] [Google Scholar]
- 14.Shimomura T, et al. Activation of the zymogen of hepatocyte growth factor activator by thrombin. J Biol Chem. 1993;268(30):22927–22932. [PubMed] [Google Scholar]
- 15.Mars WM, Zarnegar R, Michalopoulos GK. Activation of hepatocyte growth factor by the plasminogen activators uPA and tPA. Am J Pathol. 1993;143(3):949–958. [PMC free article] [PubMed] [Google Scholar]
- 16.Naldini L, et al. Biological activation of pro-HGF (hepatocyte growth factor) by urokinase is controlled by a stoichiometric reaction. J Biol Chem. 1995;270(2):603–611. doi: 10.1074/jbc.270.2.603. [DOI] [PubMed] [Google Scholar]
- 17.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem. 2000;275(47):36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 18.Shimomura T, et al. Activation of hepatocyte growth factor by two homologous proteases, blood-coagulation factor XIIa and hepatocyte growth factor activator. Eur J Biochem. 1995;229(1):257–261. doi: 10.1111/j.1432-1033.1995.tb20463.x. [DOI] [PubMed] [Google Scholar]
- 19.Jiang W, et al. Hepatocyte growth factor/scatter factor, its molecular, cellular and clinical implications in cancer. Crit Rev Oncol Hematol. 1999;29(3):209–248. doi: 10.1016/s1040-8428(98)00019-5. [DOI] [PubMed] [Google Scholar]
- 20.Trusolino L, Comoglio PM. Scatter factor and semaphorin receptors: cell signaling for invasive growth. Nat Rev Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- 21.Jiang WG, Hiscox S. Hepatocyte growth factor/scatter factor, a cytokine playing multiple and converse roles. Histol Histopathol. 1997;12(2):537–555. [PubMed] [Google Scholar]
- 22.Bottaro DP, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251(4995):802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 23.Naldini L, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. 1991;6(4):501–504. [PubMed] [Google Scholar]
- 24.Nusrat A, et al. Hepatocyte growth factor/scatter factor effects on epithelia. Regulation of intercellular junctions in transformed and nontransformed cell lines, basolateral polarization of c-met receptor in transformed and natural intestinal epithelial, and induction of wound reapir in a transformed model epithelium. J Clin Invest. 1994;93:2056–2065. doi: 10.1172/JCI117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bussolino F, et al. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J Cell Biol. 1992;119(3):629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beilmann M, et al. Neoexpression of the c-met/hepatocyte growth factor-scatter factor receptor gene in activated monocytes. Blood. 1997;90(11):4450–4458. [PubMed] [Google Scholar]
- 27.Park M, et al. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci U S A. 1987;84(18):6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper CS, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311(5981):29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 29.Park M, et al. Mechanism of met oncogene activation. Cell. 1986;45(6):895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 30.Abella JV, et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol Cell Biol. 2005;25(21):9632–9645. doi: 10.1128/MCB.25.21.9632-9645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peschard P, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8(5):995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 32.Qi J, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71(3):1081–1091. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponzetto C, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77(2):261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 34.Royal I, Fournier TM, Park M. Differential requirement of Grb2 and PI3-kinase in HGF/SF-induced cell motility and tubulogenesis. J Cell Physiol. 1997;173(2):196–201. doi: 10.1002/(SICI)1097-4652(199711)173:2<196::AID-JCP20>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 35.Xiao GH, et al. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A. 2001;98(1):247–252. doi: 10.1073/pnas.011532898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birchmeier C, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4(12):915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 37.Boccaccio C, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391(6664):285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 38.Thomas RM, et al. The RON receptor tyrosine kinase mediates oncogenic phenotypes in pancreatic cancer cells and is increasingly expressed during pancreatic cancer progression. Cancer Res. 2007;67(13):6075–6082. doi: 10.1158/0008-5472.CAN-06-4128. [DOI] [PubMed] [Google Scholar]
- 39.Follenzi A, et al. Cross-talk between the proto-oncogenes Met and Ron. Oncogene. 2000;19(27):3041–3049. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- 40.Conrotto P, et al. Interplay between scatter factor receptors and B plexins controls invasive growth. Oncogene. 2004;23(30):5131–5137. doi: 10.1038/sj.onc.1207650. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, et al. A mechanism of cell survival: sequestration of Fas by the HGF receptor Met. Mol Cell. 2002;9(2):411–421. doi: 10.1016/s1097-2765(02)00439-2. [DOI] [PubMed] [Google Scholar]
- 42.van der Voort R, et al. Heparan sulfate-modified CD44 promotes hepatocyte growth factor/scatter factor-induced signal transduction through the receptor tyrosine kinase c-Met. J Biol Chem. 1999;274(10):6499–6506. doi: 10.1074/jbc.274.10.6499. [DOI] [PubMed] [Google Scholar]
- 43.Orian-Rousseau V, et al. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16(23):3074–3086. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locatelli A, Lange CA. Met receptors induce Sam68-dependent cell migration by activation of alternate extracellular signal-regulated kinase family members. J Biol Chem. 2011;286(24):21062–21072. doi: 10.1074/jbc.M110.211409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu ST, et al. Correlation of CD44v5 expression with invasiveness and prognosis in renal cell carcinoma. J Formos Med Assoc. 2003;102(4):229–233. [PubMed] [Google Scholar]
- 46.Birchmeier C, Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol. 1998;8(10):404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 47.Bladt F, et al. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376(6543):768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 48.Dietrich S, et al. The role of SF/HGF and c-Met in the development of skeletal muscle. Development. 1999;126(8):1621–1629. doi: 10.1242/dev.126.8.1621. [DOI] [PubMed] [Google Scholar]
- 49.Chmielowiec J, et al. c-Met is essential for wound healing in the skin. J Cell Biol. 2007;177:151–162. doi: 10.1083/jcb.200701086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borowiak M, et al. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A. 2004;101(29):10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276(5309):60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 52.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5(10):836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 53.Cowin AJ, et al. Hepatocyte growth factor and macrophage-stimulating protein are upregulated during excisional wound repair in rats. Cell Tissue Res. 2001;306(2):239–250. doi: 10.1007/s004410100443. [DOI] [PubMed] [Google Scholar]
- 54.Kawaida K, et al. Hepatocyte growth factor prevents acute renal failure and accelerates renal regeneration in mice. Proc Natl Acad Sci U S A. 1994;91(10):4357–4361. doi: 10.1073/pnas.91.10.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohmichi H, Matsumoto K, Nakamura T. In vivo mitogenic action of HGF on lung epithelial cells: pulmotrophic role in lung regeneration. Am J Physiol. 1996;270(6 Pt 1):L1031–L1039. doi: 10.1152/ajplung.1996.270.6.L1031. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida S, et al. Neutralization of hepatocyte growth factor leads to retarded cutaneous wound healing associated with decreased neovascularization and granulation tissue formation. J Invest Dermatol. 2003;120(2):335–343. doi: 10.1046/j.1523-1747.2003.12039.x. [DOI] [PubMed] [Google Scholar]
- 57.Nayeri F, et al. Hepatocyte growth factor may accelerate healing in chronic leg ulcers: a pilot study. J Dermatolog Treat. 2002;13(2):81–86. doi: 10.1080/095466302317584449. [DOI] [PubMed] [Google Scholar]
- 58.Nayeri F, et al. Hepatocyte growth factor; expression, concentration and biological activity in chronic leg ulcers. J Dermatol Sci. 2005;37(2):75–85. doi: 10.1016/j.jdermsci.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Bevan D, et al. Diverse and potent activities of HGF/SF in skin wound repair. J Pathol. 2004;203(3):831–838. doi: 10.1002/path.1578. [DOI] [PubMed] [Google Scholar]
- 60.Niranjan B, et al. HGF/SF: a potent cytokine for mammary growth, morphogenesis and development. Development. 1995;121(9):2897–2908. doi: 10.1242/dev.121.9.2897. [DOI] [PubMed] [Google Scholar]
- 61.Montesano R, et al. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell. 1991;67(5):901–908. doi: 10.1016/0092-8674(91)90363-4. [DOI] [PubMed] [Google Scholar]
- 62.Ferracini R, et al. The Met/HGF receptor is over-expressed in human osteosarcomas and is activated by either a paracrine or an autocrine circuit. Oncogene. 1995;10(4):739–749. [PubMed] [Google Scholar]
- 63.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19(10):542–551. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 64.Natali PG, et al. Overexpression of the met/HGF receptor in renal cell carcinomas. Int J Cancer. 1996;69(3):212–217. doi: 10.1002/(SICI)1097-0215(19960621)69:3<212::AID-IJC11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 65.Di Renzo MF, et al. Overexpression of the Met/HGF receptor in ovarian cancer. Int J Cancer. 1994;58(5):658–662. doi: 10.1002/ijc.2910580507. [DOI] [PubMed] [Google Scholar]
- 66.Takeo S, et al. Examination of oncogene amplification by genomic DNA microarray in hepatocellular carcinomas: comparison with comparative genomic hybridization analysis. Cancer Genet Cytogenet. 2001;130(2):127–132. doi: 10.1016/s0165-4608(01)00479-4. [DOI] [PubMed] [Google Scholar]
- 67.Olivero M, et al. Overexpression and activation of hepatocyte growth factor/scatter factor in human non-small-cell lung carcinomas. Br J Cancer. 1996;74(12):1862–1868. doi: 10.1038/bjc.1996.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Renzo MF, et al. Expression of the Met/hepatocyte growth factor receptor in human pancreatic cancer. Cancer Res. 1995;55(5):1129–1138. [PubMed] [Google Scholar]
- 69.Humphrey PA, et al. Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am J Pathol. 1995;147(2):386–396. [PMC free article] [PubMed] [Google Scholar]
- 70.Ivan M, et al. Activated ras and ret oncogenes induce over-expression of c-met (hepatocyte growth factor receptor) in human thyroid epithelial cells. Oncogene. 1997;14(20):2417–2423. doi: 10.1038/sj.onc.1201083. [DOI] [PubMed] [Google Scholar]
- 71.Gambarotta G, et al. Ets up-regulates MET transcription. Oncogene. 1996;13(9):1911–1917. [PubMed] [Google Scholar]
- 72.Boon EM, et al. Wnt signaling regulates expression of the receptor tyrosine kinase met in colorectal cancer. Cancer Res. 2002;62(18):5126–5128. [PubMed] [Google Scholar]
- 73.Graveel CR, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106(31):12909–12914. doi: 10.1073/pnas.0810403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ponzo MG, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106(31):12903–12908. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ostrander JH, et al. Breast tumor kinase (protein tyrosine kinase 6) regulates heregulin-induced activation of ERK5 and p38 MAP kinases in breast cancer cells. Cancer Res. 2007;67(9):4199–4209. doi: 10.1158/0008-5472.CAN-06-3409. [DOI] [PubMed] [Google Scholar]
- 76.Chen HY, et al. Brk activates rac1 and promotes cell migration and invasion by phosphorylating paxillin. Mol Cell Biol. 2004;24(24):10558–10572. doi: 10.1128/MCB.24.24.10558-10572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Llor X, et al. BRK/Sik expression in the gastrointestinal tract and in colon tumors. Clin Cancer Res. 1999;5(7):1767–1777. [PubMed] [Google Scholar]
- 78.Castro NE, Lange CA. Breast tumor kinase and extracellular signal-regulated kinase 5 mediate Met receptor signaling to cell migration in breast cancer cells. Breast Cancer Res. 2010;12(4):R60. doi: 10.1186/bcr2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitchell PJ, et al. Cloning and characterisation of cDNAs encoding a novel non-receptor tyrosine kinase, brk, expressed in human breast tumours. Oncogene. 1994;9(8):2383–2390. [PubMed] [Google Scholar]
- 80.Lee H, et al. Exon-intron structure of the human PTK6 gene demonstrates that PTK6 constitutes a distinct family of non-receptor tyrosine kinase. Mol Cells. 1998;8(4):401–407. [PubMed] [Google Scholar]
- 81.Derry JJ, et al. Altered localization and activity of the intracellular tyrosine kinase BRK/Sik in prostate tumor cells. Oncogene. 2003;22(27):4212–4220. doi: 10.1038/sj.onc.1206465. [DOI] [PubMed] [Google Scholar]
- 82.Schmandt RE, et al. The BRK tyrosine kinase is expressed in high-grade serous carcinoma of the ovary. Cancer Biol Ther. 2006;5(9):1136–1141. doi: 10.4161/cbt.5.9.2953. [DOI] [PubMed] [Google Scholar]
- 83.Lin HS, et al. Identification of tyrosine kinases overexpressed in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2004;130(3):311–316. doi: 10.1001/archotol.130.3.311. [DOI] [PubMed] [Google Scholar]
- 84.Easty DJ, et al. Loss of expression of receptor tyrosine kinase family genes PTK7 and SEK in metastatic melanoma. Int J Cancer. 1997;71(6):1061–1065. doi: 10.1002/(sici)1097-0215(19970611)71:6<1061::aid-ijc24>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 85.Qiu H, Miller WT. Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition. J Biol Chem. 2002;277(37):34634–34641. doi: 10.1074/jbc.M203877200. [DOI] [PubMed] [Google Scholar]
- 86.Serfas MS, Tyner AL. Brk, Srm, Frk, and Src42A form a distinct family of intracellular Src-like tyrosine kinases. Oncol Res. 2003;13(6–10):409–419. doi: 10.3727/096504003108748438. [DOI] [PubMed] [Google Scholar]
- 87.Kamalati T, et al. Brk, a breast tumor-derived non-receptor protein-tyrosine kinase, sensitizes mammary epithelial cells to epidermal growth factor. J Biol Chem. 1996;271(48):30956–30963. doi: 10.1074/jbc.271.48.30956. [DOI] [PubMed] [Google Scholar]
- 88.Kamalati T, et al. Expression of the BRK tyrosine kinase in mammary epithelial cells enhances the coupling of EGF signalling to PI 3-kinase and Akt, via erbB3 phosphorylation. Oncogene. 2000;19(48):5471–5476. doi: 10.1038/sj.onc.1203931. [DOI] [PubMed] [Google Scholar]
- 89.Xiang B, et al. Brk is coamplified with ErbB2 to promote proliferation in breast cancer. Proc Natl Acad Sci U S A. 2008;105(34):12463–12468. doi: 10.1073/pnas.0805009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vasioukhin V, Tyner AL. A role for the epithelial-cell-specific tyrosine kinase Sik during keratinocyte differentiation. Proc Natl Acad Sci U S A. 1997;94(26):14477–14482. doi: 10.1073/pnas.94.26.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Derry JJ, et al. Sik (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol Cell Biol. 2000;20(16):6114–6126. doi: 10.1128/mcb.20.16.6114-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mitchell PJ, Sara EA, Crompton MR. A novel adaptor-like protein which is a substrate for the non-receptor tyrosine kinase, BRK. Oncogene. 2000;19(37):4273–4282. doi: 10.1038/sj.onc.1203775. [DOI] [PubMed] [Google Scholar]
- 93.Ikeda O, et al. STAP-2 is phosphorylated at tyrosine-250 by Brk and modulates Brk-mediated STAT3 activation. Biochem Biophys Res Commun. 2009;384(1):71–75. doi: 10.1016/j.bbrc.2009.04.076. [DOI] [PubMed] [Google Scholar]
- 94.Haegebarth A, et al. The nuclear tyrosine kinase BRK/Sik phosphorylates and inhibits the RNA-binding activities of the Sam68-like mammalian proteins SLM-1 and SLM-2. J Biol Chem. 2004;279(52):54398–54404. doi: 10.1074/jbc.M409579200. [DOI] [PubMed] [Google Scholar]
- 95.Lukong KE, Richard S. Breast tumor kinase BRK requires kinesin-2 subunit KAP3A in modulation of cell migration. Cell Signal. 2008;20(2):432–442. doi: 10.1016/j.cellsig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 96.Shen CH, et al. Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res. 2008;68(19):7779–7787. doi: 10.1158/0008-5472.CAN-08-0997. [DOI] [PubMed] [Google Scholar]
- 97.Lukong KE, Huot ME, Richard S. BRK phosphorylates PSF promoting its cytoplasmic localization and cell cycle arrest. Cell Signal. 2009;21(9):1415–1422. doi: 10.1016/j.cellsig.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 98.Palka-Hamblin HL, et al. Identification of beta-catenin as a target of the intracellular tyrosine kinase PTK6. J Cell Sci. 2010;123(Pt 2):236–245. doi: 10.1242/jcs.053264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng Y, et al. Protein tyrosine kinase 6 directly phosphorylates AKT and promotes AKT activation in response to epidermal growth factor. Mol Cell Biol. 2010;30(17):4280–4292. doi: 10.1128/MCB.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu L, et al. Identification of STAT3 as a specific substrate of breast tumor kinase. Oncogene. 2006;25(35):4904–4912. doi: 10.1038/sj.onc.1209501. [DOI] [PubMed] [Google Scholar]
- 101.Weaver AM, Silva CM. Signal transducer and activator of transcription 5b: a new target of breast tumor kinase/protein tyrosine kinase 6. Breast Cancer Res. 2007;9(6):R79. doi: 10.1186/bcr1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu X, Robinson GW, Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol Endocrinol. 1996;10(12):1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 103.Philp JA, Burdon TG, Watson CJ. Differential activation of STATs 3 and 5 during mammary gland development. FEBS Lett. 1996;396(1):77–80. doi: 10.1016/0014-5793(96)01069-1. [DOI] [PubMed] [Google Scholar]
- 104.Chapman RS, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13(19):2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walker NI, Bennett RE, Kerr JF. Cell death by apoptosis during involution of the lactating breast in mice and rats. Am J Anat. 1989;185(1):19–32. doi: 10.1002/aja.1001850104. [DOI] [PubMed] [Google Scholar]
- 106.Ostrander JH, Daniel AR, Lange CA. Brk/PTK6 signaling in normal and cancer cell models. Curr Opin Pharmacol. 2010;10(6):662–669. doi: 10.1016/j.coph.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harvey AJ, et al. Brk protects breast cancer cells from autophagic cell death induced by loss of anchorage. Am J Pathol. 2009;175(3):1226–1234. doi: 10.2353/ajpath.2009.080811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lofgren KA, et al. Mammary gland specific expression of Brk/PTK6 promotes delated involution and tumor formation associated with activation of p38 MAPK. Breast Cancer Res. 2011;13(5):R89. doi: 10.1186/bcr2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen T, et al. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Cell Biol. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13(12):479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 111.Chen T, et al. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17(10):5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Busa R, et al. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 2007;26(30):4372–4382. doi: 10.1038/sj.onc.1210224. [DOI] [PubMed] [Google Scholar]
- 113.Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368(6474):867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 114.Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272(43):27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 115.Meyer NH, et al. Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain. J Biol Chem. 2010;285(37):28893–28901. doi: 10.1074/jbc.M110.126185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lukong KE, et al. Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J Biol Chem. 2005;280(46):38639–38647. doi: 10.1074/jbc.M505802200. [DOI] [PubMed] [Google Scholar]
- 117.Wang LL, Richard S, Shaw AS. P62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270(5):2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- 118.Wong G, et al. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69(3):551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- 119.Weng Z, et al. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol Cell Biol. 1994;14(7):4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fumagalli S, et al. A target for Src in mitosis. Nature. 1994;368(6474):871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 121.Taylor SJ, et al. Functional interaction between c-Src and its mitotic target, Sam 68. J Biol Chem. 1995;270(17):10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- 122.Richard S, et al. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family tyrosine kinases, Grb2, and phospholipase C gamma-1. Mol Cell Biol. 1995;15(1):186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Trub T, et al. The role of a lymphoid-restricted, Grb2-like SH3-SH2-SH3 protein in T cell receptor signaling. J Biol Chem. 1997;272(2):894–902. doi: 10.1074/jbc.272.2.894. [DOI] [PubMed] [Google Scholar]
- 124.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 125.Cote J, et al. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol Biol Cell. 2003;14(1):274–287. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bedford MT, et al. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275(21):16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 127.Ishidate T, et al. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 1997;409(2):237–241. doi: 10.1016/s0014-5793(97)00455-9. [DOI] [PubMed] [Google Scholar]
- 128.Wu J, et al. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J Biol Chem. 1999;274(41):29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- 129.Liu K, et al. Neoplastic transformation and tumorigenesis associated with sam68 protein deficiency in cultured murine fibroblasts. J Biol Chem. 2000;275(51):40195–40201. doi: 10.1074/jbc.M006194200. [DOI] [PubMed] [Google Scholar]
- 130.Hong W, et al. Physical and functional interaction between the transcriptional cofactor cbp and the kh domain protein Sam68. Mol Cancer Res. 2002;1(1):48–55. [PubMed] [Google Scholar]
- 131.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420(6916):691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 132.Paronetto MP, et al. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176(7):929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Paronetto MP, et al. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70(1):229–239. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Song L, et al. Sam68 up-regulation correlates with, and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. J Pathol. 2010;222(3):227–237. doi: 10.1002/path.2751. [DOI] [PubMed] [Google Scholar]
- 135.Barlat I, et al. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J Biol Chem. 1997;272(6):3129–3132. doi: 10.1074/jbc.272.6.3129. [DOI] [PubMed] [Google Scholar]
- 136.Richard S, et al. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene. 2008;27(4):548–556. doi: 10.1038/sj.onc.1210652. [DOI] [PubMed] [Google Scholar]
- 137.Rajan P, et al. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J Pathol. 2008;215(1):67–77. doi: 10.1002/path.2324. [DOI] [PubMed] [Google Scholar]
- 138.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 139.Valacca C, et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol. 2010;191(1):87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jo M, et al. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem. 2000;275(12):8806–8811. doi: 10.1074/jbc.275.12.8806. [DOI] [PubMed] [Google Scholar]
- 141.Bergstrom JD, Westermark B, Heldin NE. Epidermal growth factor receptor signaling activates met in human anaplastic thyroid carcinoma cells. Exp Cell Res. 2000;259(1):293–299. doi: 10.1006/excr.2000.4967. [DOI] [PubMed] [Google Scholar]
- 142.Agarwal S, et al. Association of constitutively activated hepatocyte growth factor receptor (Met) with resistance to a dual EGFR/Her2 inhibitor in non-small-cell lung cancer cells. Br J Cancer. 2009;100(6):941–949. doi: 10.1038/sj.bjc.6604937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khoury H, et al. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol Biol Cell. 2005;16(2):550–561. doi: 10.1091/mbc.E04-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 145.Mueller KL, et al. EGFR/Met association regulates EGFR TKI resistance in breast cancer. J Mol Signal. 2010;5:8. doi: 10.1186/1750-2187-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mueller KL, et al. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68(9):3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fischer OM, et al. Reactive oxygen species mediate Met receptor transactivation by G protein-coupled receptors and the epidermal growth factor receptor in human carcinoma cells. J Biol Chem. 2004;279(28):28970–28978. doi: 10.1074/jbc.M402508200. [DOI] [PubMed] [Google Scholar]
- 148.Qiu H, Miller WT. Role of the Brk SH3 domain in substrate recognition. Oncogene. 2004;23(12):2216–2223. doi: 10.1038/sj.onc.1207339. [DOI] [PubMed] [Google Scholar]
- 149.Qiu H, et al. Interaction between Brk kinase and insulin receptor substrate-4. Oncogene. 2005;24(36):5656–5664. doi: 10.1038/sj.onc.1208721. [DOI] [PubMed] [Google Scholar]
- 150.Zhang P, et al. Regulated association of protein kinase B/Akt with breast tumor kinase. J Biol Chem. 2005;280(3):1982–1991. doi: 10.1074/jbc.M412038200. [DOI] [PubMed] [Google Scholar]