Abstract
RHM1 (1) and RHM2 (2) are highly N-methylated linear octapeptides produced by an atypical strain of Acremonium sp., cultured from a marine sponge collected in Papua New Guinea. The known peptaibiotic efrapeptin G (3) was also isolated from this fungus. The planar structures of 1 and 2 were assigned based on 1D- and 2D-NMR experiments and fragmentation patterns from ESIMS. The absolute configuration of 1 was determined via Marfey's method; the absolute configuration of 2 is proposed to be identical. Efrapeptin G (3) displayed potent cytotoxicity against murine cancer cell lines, while RHM1 (1) and RHM2 (2) showed weak cytotoxicity against murine cancer cell lines; efrapeptin G (3) and RHM1 (1) also demonstrated antibacterial activity.
It is not currently possible to predict the chemical signatures of filamentous fungi isolated from marine sources.1 Low molecular weight (<500 amu) bioactive polyketides often dominate the secondary metabolites isolated from terrestrial fungal cultures. Two important therapeutically active compounds produced by well-studied fungal genera Penicillium and Aspergillus illustrate this chemistry. Mycophenolic acid (C17H20O6), discovered over 70 years ago from Penicillium sp.,2 is an immunosuppressive agent (1995: mycophenolate mofetil, first marketed in the U.S.) that has also shown promising antiviral properties.3 The members of the well-defined structural class headed by mevinolin (C24H36O5), originally isolated from Aspergillus, are potent HMG-CoA inhibitors of cholesterol biosynthesis (1987: first FDA approval of lovastatin).4
Exploration of structural diversity of marine-derived fungal natural products has been in progress for more than a decade. Early discoveries of the chloriolins5 and the fumiquinazolines6 validated the hypothesis that marine-derived strains of fungi could serve as useful sources of unprecedented molecular frameworks. Compounds such as asperazine7 from the sponge-derived, chemically rich Aspergillus niger, or the aigilomycins8 from a mangrove isolate of Aigialus parVus, provided further validation that new chemotypes could be discovered from genera previously isolated from terrestrial sources. Similarly, the outstanding cytotoxic properties of the gymnastatins from Gymnascella9 as well as the trichoverroids and their related macrolides, especially isororidin A from Myrothecium Verrucaria,10 are excellent examples of structures with therapeutic potential obtained from sponge-derived fungi.
This work describes the constituents of a marine sponge-derived Acremonium strain grown in saltwater culture. At the outset we recognized numerous previous studies on terrestrial members of this genus, with the antibiotic cephalosporin C11 as the most notable metabolite produced. Only five previous studies had examined the chemistry of marine-derived strains of Acremonium, resulting in different biosynthetic products isolated from distinct sources. This interesting pattern included reports of (a) polyketides12 and hydroquinones13 from algal-derived cultures, (b) ketide-terpenes from a strain isolated from submerged wood,14 (c) two families of alkaloids from a tunicate-derived fungus,15 and (d) diterpene glycosides from a holothurin source.16 In contrast to these outcomes, our initial NMR spectra of the crude extracts from the sponge-derived Acremonium culture displayed signals characteristic of polypeptides. These findings are described below with the structure elucidation of two new octapeptides, RHM 1 (1) and RHM2 (2), and efrapeptin G (3),17 a mitochondrial ATPase inhibitor previously isolated only from the genus Tolypocladium.
Results and Discussion
A crude extract from a small-scale (100 mL) culture of an Acremonium sp. (UCSC coll. no. 021172 cKZ) cultured from a Teichaxinella sp. marine sponge (coll. no. 02172; collection locale: Milne Bay, Papua New Guinea) displayed potent cytotoxicity in a primary screen using leukemia and solid tumor murine and human cancer cell lines (Table 1).18 This prompted the growth of a larger-scale culture (20 L) to facilitate the purification of metabolites from dichloromethane-soluble broth (EFD) and mycelial extracts (TFD) (Table 1, Figure S1). Bioassay against a murine lymphocytic cell line (L1210) and Staphylococcus epidermidis and NMR data were used to guide the fractionation, resulting in two new related linear octapeptides, RHM1 (1) and RHM2 (2), and the known peptaibiotic efrapeptin G (3).
Table 1.
Bioactive Properties of Extracts and Pure Compounds from Strain 021172c
| sample | relative potencya | MICb (μg/mL) |
|---|---|---|
| EtOAc extract (100 mL culture) | 92 | nt |
| CH2Cl2 extract of broth (20 L culture) | 580 | 125 |
| efrapeptin G (3) | 3300 | 80 |
| RHM1 (1) | 16 | 50 |
| CH2Cl2 extract of mycelia (20 L culture) | 34 | 250 |
| RHM2 (2) | 1.0 | >400 |
| RHM1 (1) | 4.0 | 25 |
| vancomycin | nt | 0.63 |
Potency against murine L1210 (lymphocytic leukemia) is calculated by multiplying the reciprocal of the dilution required for potent inhibition (zone of 500 units) × 100/mg/mL; larger values correspond to greater potency; potency is relative to the value for 3.
MIC is the minimum inhibitory concentration against Staphylococcus epidermidis; nt = not tested.
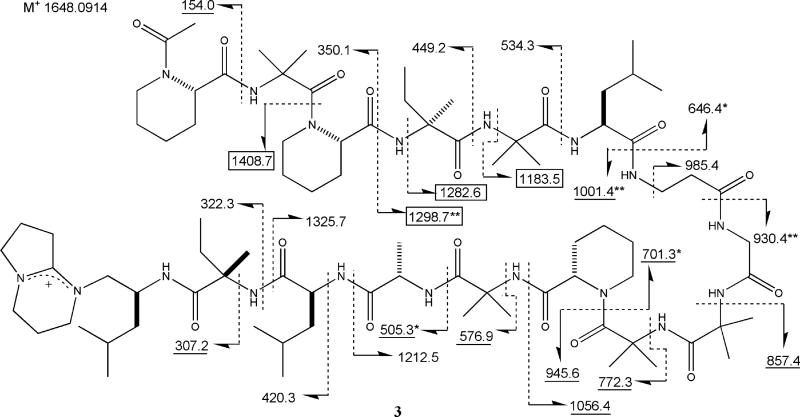
A dereplication approach provided the identity of 3, which displayed an ESIMS peak at m/z 1649.4. Natural product data bases were searched for compounds with masses between 1647 and 1651, which resulted in 11 hits. Refining the search to metabolites of fungal origin revealed efrapeptin G as the single remaining match. A comparison of analytical data for 3 and efrapeptin G indicated similar profiles; the FTMS data, m/z 1648.0914, required the [M]+ molecular formula of C83H143N18O16 (Δ 1.0 mmu of calcd) and confirmed that it was a salt. The presence of 15 amino acid units and a carboxy terminal imminium ion moiety was also evident from diagnostic resonances in the NMR spectrum as discussed below and consistent with prominent ESIMS fragment ions (m/z 703.5 and 945.6) observed via fragmentation at the amide bond between the 10th and 11th amino acids of efrapeptin G.
The next challenge was to unequivocally define the sequence of the 15 amino acids present in 3. The arguments advanced to establish the structure of efrapeptin G17a were based on amino acid analysis, a comparison of four FABMS fragment ions which highlighted the difference between efrapeptins F and G, and the results of directed biosynthetic experiments.19 To proceed, we undertook a detailed analysis of the literature spectra of efrapeptin G and data we collected for 3 (Figures S2a, S2b, S3, and S4). The FAB mass spectrum for efrapeptin G was included in the supplementary literature,17a and these data are overlaid in Figure 1 with the FABMS we obtained on 3. Three types of fragment ions20 identified in efrapeptin G were used to set the proposed amino acid sequence, and these consisted of (a) type B, N-terminal acylium ions, (b) type Y′, C-terminal amino fragments, and (c) type Z, C-terminal alkyl fragments. There were only four fragment ions discussed in the structure elucidation of efrapeptin G (boxed values, Figure 1), described as m/z 1185 (observed as 1184, Z11), 1284 (barely visible in spectrum, Z12), 1299 (Y′12), and 1410 (observed as 1408,Y′13), and each of these agreed with our experimental data. We observed an additional 20 fragment ions, which were used to verify the sequence of amino acids (Figure 1). The eight underlined values in Figure 1 were present in the FABMS spectrum of efrapeptin G but were not discussed in the literature. There are an additional eight FABMS peaks observed for both efrapeptin G and 3, which are related to side-chain fragmentation including m/z 250.1, 292.2, 377.2, 618.4, 729.3, 772.3, 814.3, and 857.6. This analysis confirmed that the two samples were identical and bolstered confidence that the original structure of efrapeptin G was correct.
Figure 1.
FABMS fragmentation (m/z) for efrapeptin G (3). Codes: underlined values this work and ref 17a; boxed values discussed in ref 17a; values ± 1 amu (+*/–**).32
The lack of published NMR data for efrapeptin G (3) stimulated us to assemble a complete data set as shown in Table 2. All NMR resonances for the 83 carbons, the 130 protons attached to carbon, and the 13 protons attached to nitrogen could be located and identified with the aid of a DEPT spectrum. Some assignments could be readily made (Table 2) on the basis of literature values for a hydrolysis product of efrapeptin G,17a which was composed of the imminium capping group and the last two amino acids (Iva and Leu). The individual resonances of the remaining sets of closely related functional groups were only provisionally made and were assisted by published NMR data for the nonstandard amino acids Pip, Iva, and β-Ala21 as well as calculated NMR data (ACD v 8.0). Finally, the NMR analysis added the penultimate evidence to verify the structure of 3, and the configuration shown is identical to that of efrapeptin G.
Table 2.
Provisional Assignments of 13C/1H NMR (125/500 MHz, MeOH-d4) Resonances for Efrapeptin G (3)
| type | 13C δ | lit. 13C δa | 1H δ; mult. (J Hz) no. | lit. 1H δa mult. (J Hz) no. | |
|---|---|---|---|---|---|
| Ac | |||||
| Me | CH3 | 17.5 | 2.18; s 3H | ||
| C=O | C | 171.7 | |||
| Pip | |||||
| α | CH | 57.5, 56.7, 56.1 | 5.14; bd (6.0) 1H, 5.05; bt (4.0) 1H, 4.86; m 1H | ||
| β | CH2 | 27.5, 26.7, 26.6 | 1.93–1.58; m 6H | ||
| γ | CH2 | 21.7, 21.6, 21.4 | 2.26; m 3H, 1.93–1.58; m 3H | ||
| δ | CH2 | 26.2, 26.0, 25.8 | 1.56–1.41; m 6H | ||
| ε | CH2 | 45.5, 44,9, 42.2 | 4.34; m 2H, 4.22; m 1H | ||
| 3.75–3.69; m 2H, 3.41–3.36; m 1H | |||||
| C=O | C | 175.6, 174.0, 173.5 | |||
| Aib | |||||
| NH | 8.28; s 1H, 8.18; s 1H, 8.09; s 1H | ||||
| 7.91; s 1H, 7.89; s 1H | |||||
| α c | C | 58.7, 58.4, 58.3, 58.3, 58.1 | |||
| β1 | CH3 | 27.8, 27.3, 25.6, 25.2, 24.1 | 1.56–1.41; m 15H | ||
| β2 | CH3 | 27.1, 26.9, 25.3, 24.4, 23.9 | 1.56–1.41; m 15H | ||
| C=O | C | 178.0, 177.6, 177.4, 176.9, 175.6 | |||
| Iva | |||||
| NH | 7.89; s 1H, 7.19; s 1H | ||||
| α | C6 | 1.9, 61.3 | 61.8 | ||
| β1 | CH3 | 23.6, 23.3 | 23.6 | 1.56–1.41; m 6H | 1.37; s 3H |
| β2 | CH2 | 34.2, 28.3 | 27.9 | 1.93–1.58; m 4H | 1.9; m 1H, 2.15; m 1H |
| γ | CH3 | 8.7, 8.6 | 8.3 | 0.95–0.84; m 6H | 0.85; t (7.32) 3H |
| C=O | C | 178.4, 177.0 | 176.8 | ||
| Leu | |||||
| NH | 7.93; d (7.0) 1H, 7.10; d (9.5) 1H | ||||
| α | CH | 54.6, 54.1 | 53.2 | 4.16; ddd (3.0, 6.5, 10.5) 1H | |
| 3.93; ddd (4.5, 6.0, 10.5) 1H | 3.92; m 1H | ||||
| β | CH2 | 41.1, 37.4 | 41.7 | 1.93–1.58; m 2H, 1.56–1.41; m 2H | 1.61–1.77; m 2H |
| γ | CH2 | 6.4, 26.2 | 25.5 | 1.93–1.58; m 2H | 1.61–1.77; m 1H |
| δ1 | CH3 | 21.7, 21.6 | 23.1 | 0.95–0.84; m 6H | 1.00;b d (6.14) 3H |
| δ2 | CH3 | 20.8, 21.1 | 22.1 | 0.95–0.84; m 6H | 1.02;b d (6.14) 3H |
| C=O | C | 176.4, 176.1 | 169.9 | ||
| β-Ala | |||||
| NH | 7.77; m 1H | ||||
| α | CH2 | 34.2 | 3.85; m 1H, 2.92; ddd (6.0, 10.0, 17.0) 1H | ||
| β | CH2 | 37.0 | 2.62; m 1H, 2.44; m 1H | ||
| C=O | C | 173.7 | |||
| Gly | |||||
| NH | 7.87; m 1H | ||||
| α | CH2 | 45.5 | 3.75–3.69; d (17.5) 2H | ||
| C=O | 175.0 | ||||
| Ala | |||||
| NH | 7.74; d (7.5) 1H | ||||
| α | CH | 53.5 | 4.01; dq (3.5, 7.5) 1H | ||
| β | CH3 | 17.5 | 1.56–1.41; m 3H | ||
| C=O | 173.5 | ||||
| Imminium Cap | |||||
| 2′ | CH2 | 46.0 | 45.9 | 3.41–3.36; m 1H, 3.75–3.69; m 1H | 3.43; m 1H, 3.64;c m 1H |
| 3′ | CH2 | 20.2 | 19.9 | 2.06; m 2H | 2.06; m 2H |
| 4′ | CH2 | 43.6 | 43.6 | 3.41–3.36; m 2H | 3.41;c m 2H |
| 6′ | CH2 | 55.7 | 55.6 | 3.81; ddd (6.5, 9.5 10.0) 1H | 3.81; m 1H |
| 3.75–3.65; m 1H | 3.73; m 1H | ||||
| 7′ | CH2 | 19.2 | 19.2 | 2.13; m 2H | 2.15; m 2H |
| 8′ | CH2 | 32.0 | 32.2 | 3.03; m 1H, 3.18; m 1H | 3.03; m 1H, 3.18; m 1H |
| 8a′ | C | 166.9 | 166.1 | ||
| 1″ | NH | 8.36; d (4.5) 1H | |||
| 1 | CH | 46.2 | 46.9 | 4.34; m 1H | 4.37; m 1H |
| 2 | CH2 | 58.0 | 57.9 | 3.64; m 1H, 3.41–3.36; m 1H | 3.58; m 1H, 3.39; m 1H |
| 3 | CH2 | 41.1 | 41.1 | 1.93–1.58; m 1H, 1.20; ddd (3.5, 10.5, 14.0) 1H | 1.69; m 1H, 1.22; m 1H |
| 4 | CH | 25.6 | 25.8 | 1.93–1.58; m 1H | 1.66; m 1H |
| 5 | CH3 | 21.8 | 21.8 | 0.95–0.84; m 3H | 0.94; d (6.34) 3H |
| 6 | CH3 | 23.7 | 23.8 | 0.95–0.84; m 3H | 0.94; d (6.34) 3H |
The structures of 1 and 2 were assembled in an iterative process. This began with identifying their constitutive amino acid groups via a combination of degradation and NMR studies. Next, the subunits were assembled on the basis of gHMBC correlations considered in parallel with the fragmentation patterns observed by ESIMS. The characterization of RHM1 (1), isolated as a crystalline white solid, proceeded with establishing the molecular formula C53H97N9O11 based on FTMS data [m/z 1036.7386 (MH)+, Δ 0.5 mmu of calcd]. The ESIMS data included fragment ions at m/z 891.6, 764.5, 637.4, 626.5, and 411.3, as shown in Figure 2.
Figure 2.
ESIMS fragmentation pattern:1m/z for RHM1 (1),2m/z for RHM2 (2).
Shifts from the 13C NMR, and carbon types derived from a DEPT experiment, revealed the peptide backbone features of 1 (Table 3). Ten carbonyls (between δ 174 and 169), eight α carbons (between δ 59 and 52), five N-methyl carbons (between δ 31 and 29), and NMR resonances characteristic of side-chain methyl carbons for five isoleucines (Iles), one valine (Val), and one leucine (Leu) were observed. Features of the 1H NMR (DMSO-d6, Figure S4) supported conclusions drawn from the 13C data including three NH protons (δ 8.31, 8.14, and 7.93), eight α protons (δ 5.20, 5.17, 5.10, 5.07, 4.78, 4.52, 4.46, and 4.31), and five N-methyl groups (δ 3.08, 3.02, two at 2.95, and 2.94). A sharp methyl singlet located at δ 1.80 (overlapping with five other aliphatic protons) suggested an N-acetyl terminus.
Table 3.
13C/1H NMR Data (500 MHz/125 MHz, DMSO-d6) for RHM 1 (1)
| position | 1H δ; mult. (J Hz) | 13C δ; type | gHMBC | gCOSY | 15N δ | |
|---|---|---|---|---|---|---|
| OAc-Gln1 | 1 | 1.80; s | 22.5; CH3 | C-2 | ||
| 2 | 169.0; C | |||||
| NH | 7.93; d (8.5) | C-2, C-7 | H-3 | 124.3 | ||
| 3 | 4.31; dt (5.0, 8.5) | 52.1; CH | C-7 | Gln NH, H-4b | ||
| 4 | 1.80; m, 1.66; ddt (13, 8.5, 5.0) | 28.5; CH2 | H-3 | |||
| 5 | 2.03; m | 31.6; CH2 | ||||
| 6 | 173.7; C | |||||
| NH2 | 7.26; s, 6.74; s | C-5, C-6 | 115.4 | |||
| 7 | 171.4; C | |||||
| Ile2 | NH | 8.14; d(8.5) | C-7, C-13 | H-10 | 124.3 | |
| 8 | 4.52; t (9.0) | 52.5; CH | Ile NH, H-9 | |||
| 9a | 1.80; m | 36.0; CH | H-8 | |||
| 10b | 1.44; m, 1.06; m | 24.0; CH2 | ||||
| 11c | 0.77; m | 10.6; CH3 | ||||
| 12d | 0.77; m | 14.9; CH3 | ||||
| 13 | 172.6; C | |||||
| MeLeu3 | 14e | 2.94; s | 31.1; CH3 | C-15 | 115.4 | |
| 15 | 5.07; dd (4.5, 12.0) | 53.8; CH | C-13, C-16, C-20 | H-16a | ||
| 16 | 1.78; m, 1.55; ddd (4.5, 11.0, 14.5) | 36.4; CH2 | H-15 | |||
| 17 | 1.80; m | 24.5; CH | ||||
| 18f | 0.77; m | 21.2; CH3 | ||||
| 19f | 0.77; m | 20.8; CH3 | ||||
| 20 | 169.9; C | |||||
| Ile4 | NH | 8.31; d (8.0) | C-20 | H-21 | 115.4 | |
| 21 | 4.46; dd (8.5, 9.5) | 52.7; CH | C-20, C-22 | Ile NH, H-22 | ||
| 22 | 1.80; m | 35.5; CH | H-21 | |||
| 23b | 1.44; m, 1.06 m | 24.0; CH2 | ||||
| 24c | 0.77; m | 10.5; CH3 | ||||
| 25d | 0.77; m | 14.9; CH3 | ||||
| 26g | 170.3; C | |||||
| MeVal5 | 27e | 3.02; s | 30.7; CH3 | C-28 | 116.2 | |
| 28 | 5.10; d (11.0) | 57.2; CH | C-29, C-32 | H-29 | ||
| 29 | 2.19; dq (6.5, 11.0) | 26.7; CH | H-28 | |||
| 30h | 0.77; m | 18.8; CH3 | ||||
| 31h | 0.77; m | 17.9; CH3 | ||||
| 32 | 172.3; C | |||||
| MeIle6 | 33e | 2.94; s | 30.0; CH3 | C-32 | 117.8 | |
| 34 | 5.20; d (11.0) | 55.5; CH | H-35 | |||
| 35a | 2.03; m | 32.6; CH | H-34 | |||
| 36b | 1.39; m, 1.30; m | 23.6; CH2 | ||||
| 37c | 0.77; m | 10.4; CH3 | ||||
| 38d | 0.77; m | 14.8; CH3 | ||||
| 39 | 172.1; C | |||||
| MeIle7 | 40e | 3.08; s | 29.9; CH3 | C-39, C-41 | 116.0 | |
| 41 | 4.78; d (11.0) | 58.8; CH | C-39, C-42, C-46 | H-42 | ||
| 42 | 1.80; m | 32.2; CH | H-41 | |||
| 43b | 1.39; m, 1.13; m | 23.4; CH2 | ||||
| 44c | 0.77; m | 10.4; CH3 | ||||
| 45d | 0.77; m | 14.6; CH3 | ||||
| 46 | 169.9; C | |||||
| MeIle8OH | 47e | 2.94; s | 29.9; CH3 | C-46, C-48 | 117.4 | |
| 48 | 5.17; d (11.0) | 55.2; CH | C-46 | H-49 | ||
| 49a | 2.03; m | 32.6; CH | H-48 | |||
| 50b | 1.39; m, 1.30; m | 23.2; CH2 | ||||
| 51c | 0.77; m | 10.2; CH3 | ||||
| 52d | 0.77; m | 14.6; CH3 | ||||
| 53g | 170.1; C | |||||
| OH | n.o. |
Interchangeable carbons
Interchangeable carbons
Interchangeable carbons
Interchangeable carbons
Interchangeable carbons
Interchangeable carbons
Interchangeable carbons
Interchangeable carbons
n.o.: not observed; low-field diastereotopic protons are labeled “a”, high-field diastereotopic protons are labeled “b”.
Amino acid (AA) analysis was not straightforward; it indicated the presence of glutamic acid (Glu), Ile, and Leu, while the 1H NMR indicated a –CH–CH–(CH3)2 backbone of a Val group. Consideration of the AA analysis and 1H and 13C NMR data side-by-side revealed the initial assessment of Leu and Glu to be incorrect. A glutamine (Gln) group was identified from signature diastereotopic NH2 protons (δ 7.26 and 6.74) upfield of the backbone NH protons and individually coupled to C-5 and C-6 (gHMBC data) and to each other (1H–1H COSY). The observation of glutamic acid by AA analysis can be attributed to hydrolysis of glutamine during the workup step. The presence of N-MeLeu is illustrated by both a gHMBC correlation from an N-Me group, H3-14 (δ 2.94) to C-15, and the multiplicity of H-15, a dd (J = 4.5 and 12.0 Hz), rather than a ddd, which would be expected for a nonmethylated Leu. One of the other nonstandard amino acids that composes 1 (N-MeVal or N-MeIle) may have a retention time similar to Leu, resulting in a misidentified peak in the AA analysis.
The substructural features responsible for the partial formula C10N9O11, based on the 10 carbonyl carbons observed by 13C NMR and heteroatoms established by MS, were revisited to identify the terminal carboxylic acid. This partial formula consists of C9N9O9 from (a) the single primary amide, (b) the three secondary amides, and (c) the five tertiary amides, resulting in a residual atom count of CO2. This observation along with the lack of resonances in the δ 60–80 region could only be rationalized by attaching a carboxylic acid group to C-48. Finally, all 10 degrees of unsaturation were accounted for by these aforementioned structural elements.
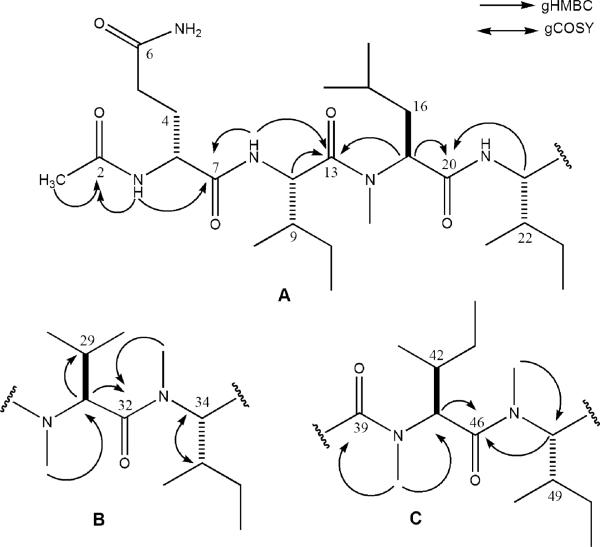
The three substructures A–C (Figure 3) were assembled after obtaining 2D-NMR data including gHMBC, gHSQC, gHMQC, gCOSY, and TOCSY. A natural abundance 15N gHMBC NMR experiment was also conducted in order to clarify overlapping proton shifts due to the large number of aliphatic amino acids. While the data did not provide new correlations for the construction of substructures, the shifts included in Table 3 verified the presence of the nine amide groups discussed above.
Figure 3.
Substructures A–C for RHM1 (1) illustrating key gHMBC and gCOSY correlations.
The justification for the substructures proposed in Figure 3 for 1 hinged primarily on the 1H–1H COSY and gHMBC data, while the biggest challenge involved the unambiguous placement of the seven isopropyl, isobutyl, and sec-butyl moieties. The terminus of substructure A was established through correlations from the acetyl methyl group (δ 1.80) to the acetyl carbonyl, C-2 (δ 169.0), and from the secondary NH of Gln1 to C-2 and the amide carbonyl, C-7 (δ 171.4). The additional backbone portion was deduced on the basis of the correlations from the Ile2 NH proton (δ 8.14) to C-7 and its amide carbonyl, C-13 (δ 172.6). The MeLeu3 α proton (δ 5.07) also correlated to C-13 and the MeLeu3 amide carbonyl, C-20 (δ 169.9), and the final key correlation was from the Ile4 α proton, H-21 (δ 4.46), to C-20. The side-chain assignments of the α and β protons were provided through COSY (Table 3), while TOCSY data confirmed that the spin systems were properly assigned for the δ and γ carbons. Other highlights of the logic to assign side-chain protons included the multiplicity and shifts at CH2-4 to locate the propianomide group as well as the dt for H-3, and the COSY correlation to H-4b (δ 1.66 ddt) indicated it was flanked by one geminal and three vicinal protons. The 13C shift at C-4 (δ 28.5) was also diagnostic of this environment, as it matched the calculated shift (δ 27.5) for this substructure. Similarly, the side-chain protons attached to C-8 and C-15 could be deciphered on the basis of the multiplicities of the β protons (H-9, multiplet via coupling to a CH3; H-16a ddd) and the reasonable agreement in the observed shift at C-16 (δ 36.4) versus the calculated shift (δ 41).
A dipeptide fragment (substructure B) consisting of two N-Me amino acids was identified in the interior backbone. Initially, the necessary 2D correlations were not observed to immediately unravel its exact placement, but this issue was resolved from the MS data. The N-MeVal was defined on the basis of gHMBC correlations from the H3-27 (δ 3.02) to α carbon C-28 (δ 57.2) and from H-28 (δ 5.10) to the amide carbonyl C-32 (δ 172.3) and to C-29 (δ 26.7), with the latter correlation securing the position of the sole isopropyl group. The N-MeIle was characterized by observing the clean doublet for H-34 (δ 5.20, Figure 3) coupling only to H-35 (δ 2.03). The pattern for these two protons indicated that a sec-butyl must be attached to C-34. The gHMBC correlation from H3-33 (δ 2.94) to C-32 defined the peptide bond between these two amino acids.
The final substructure (C) also contained a dipeptide fragment made up of the two remaining N-MeIle groups. The N-Me groups were located on the basis of gHMBC correlations from H3-40 (δ 3.08) to α carbon C-41 (δ 58.8) and C-39 (δ 172.1) and from H3-47 (δ 2.94) to C-48 (δ 55.2). Both N-MeIle7 and N-MeIle8 α protons H-41 and H-48 (δ 4.78 and δ 5.17) were clean doublets requiring, as noted above, the attachment of sec-butyl groups to C-41 and C-48. Finally, both N-MeIle7 and N-MeIle8 α (δ 4.78 and 5.17) protons displayed gHMBC correlations to the amide carbonyl of N-MeIle7, C-46 (δ 169.9).
At this point there were two possible ways to embed the carboxylic acid group to complete the structure of 1. Alternative I consisted of joining substructures A–B–C with C-53 as the carboxylate position, whereas possibility II had an A–C–B sequence with C-39 as the carboxylate location. The MS data proved essential for making an unambiguous choice between the two possible connectivities. Four intense ESIMS fragment ions were observed along with the [M + H]+ ion for 1. These acylium ions were generated through cleavage (type B) at the amide bond (Figure 2). The smallest fragment ion, m/z 411.3, was consistent with the NMR-derived sequence of the first three amino acids of substructure A: Ac-Gln1-Ile2-N-MeLeu3. According to the 2D-NMR data, the fourth amino acid must be Ile4. The next ion in the series was at m/z 637.4, reflecting the addition of 226.1 amu, equivalent to the combined masses of N-MeVal and Ile, and required their connection in this sequence (Figure 2), validating proposed structure I. The subsequent mass fragment ions, m/z 764.5 and 891.6, required the successive addition of two N-MeIle groups (Figure 2), consistent with the sequence established by NMR.
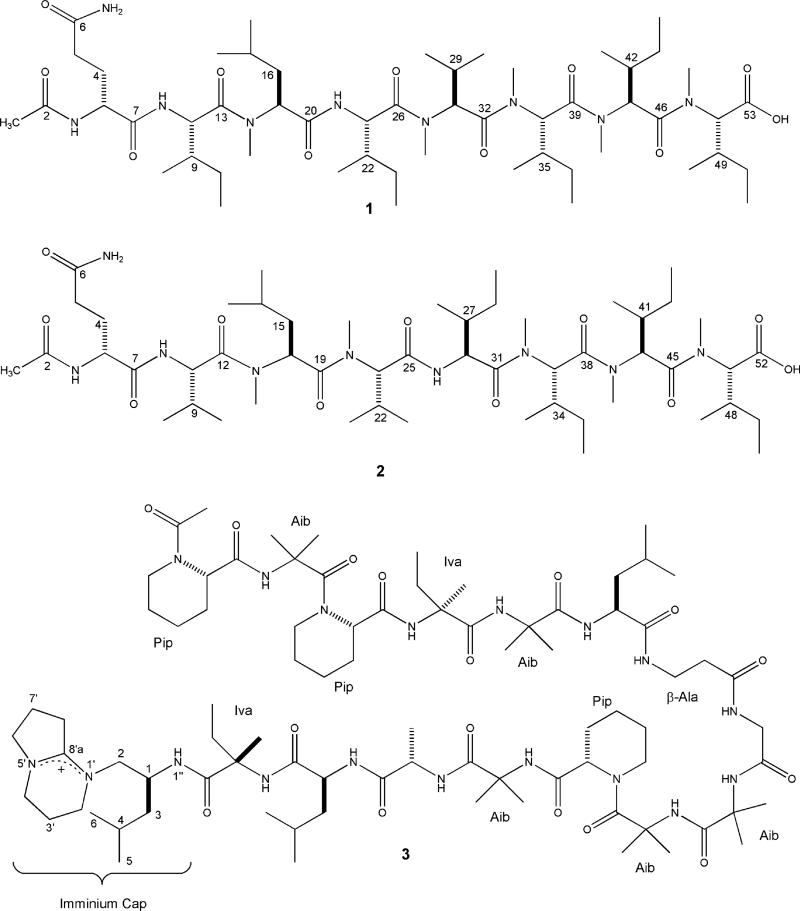
The configuration of each amino acid was determined by Marfey’s method, and the derivatives were analyzed with LCMS. The hydrolysis step converted the glutamine to glutamic acid, as was the case with the amino acid analysis, so R and S standards for glutamic acid were also examined. Retention times and masses of standards matched the R isomer of glutamic acid and the S stereoisomer for the N-Me-Leu and N-Me-Val and the 2S, 3S stereoisomer for both Ile and N-Me-Ile. Thus the stereostructure of 1 was determined to be Ac-(R)-Gln-(2S,3S)-Ile-(S)-N-Me-Leu-(2S,3S)-Ile-(S)-N-Me-Val-(2S,3S)-N-Me-Ile-(2S,3S)-N-Me-Ile-(2S,3S)-N-Me-Ile-OH.
A similar approach was employed to characterize RHM2 (2). It was isolated as an amorphous white solid with the molecular formula C52H95N9O11 obtained through FTMS for 2 [m/z 1022.7442 (M + H)+, Δ 0.2 mmu]. Four fragment ions similar to those seen in 1 were also observed for 2 at m/z 877.6, 750.5, 623.4, and 397.3 (Figure 2). The molecular formula difference between compounds 1 and 2 amounted to 14 amu, signifying one less CH2. Consistent with this minor difference, the 13C NMR and DEPT spectra for compound 2 were similar to compound 1 with 10 carbonyl carbons, eight α carbons, and five N-Mes. However, the 13C shifts for the side-chain methyl carbons were diagnostic of four Ile, two Val, and one Leu residue (Table 4), indicating one of the Ile groups of 1 was replaced as Val in 2. The 1H NMR spectra for 2 also contained evidence for a substantial population of rotational isomers at the Gln1 NH, the Val2 NH, the Ile5 NH, and protons 21, 33, and 40 (Figure S19).
Table 4.
13C/1H NMR Data (600 MHz/150 MHz, DMSO-d6) for RHM2 (2)
| position | 1H δ; mult. (J Hz) | 13C δ; type | gHMBC | gCOSY | |
|---|---|---|---|---|---|
| OAc-Gln1 | 1 | 1.81; m | 22.3; CH3 | C-2 | |
| 2 | 168.8; C | ||||
| NHa | 7.94; d (8.5) | C-2, C-3 | |||
| 3 | 4.31; dt (4.0, 9.0) | 51.9; CH | C-2, C-4, C-5, C-7 | ||
| 4 | 1.81; m, 1.66; ddt (13.0, 9.0, 5.0) | 28.4; CH2 | C-3, C-5, C-6 | ||
| 5 | 2.03; m | 31.5; CH2 | C-6 | ||
| 6 | 173.5; C | ||||
| NH2 | 7.20; m, 6.80; m | C-6 | |||
| 7 | 171.2; C | ||||
| Val2 | NHa | 8.13; d (8.5) | C-7, C-8 | H-8 | |
| 8 | 4.47; t (8.7) | 52.3; CH | C-9, C-10, C-12 | Val NH, H-9 | |
| 9 | 1.43; m | 35.4; CH | H-8 | ||
| 10b | 0.77; m | 17.7; CH3 | |||
| 11b | 0.77; m | 18.6; CH3 | |||
| 12 | 172.1; C | ||||
| MeLeu3 | 13c | 3.02; s | 31.0; CH3 | C-12, C-14 | |
| 14 | 5.05; dd (4.5, 12.0) | 57.1; CH | C-15, C-16, C-19 | H-15a, H-16 | |
| 15 | 1.78; m, 1.55; ddd (4.0, 10.5, 14.5) | 36.3; CH2 | H-14 | ||
| 16 | 1.81;m | 24.3; CH | H-14 | ||
| 17d | 0.77; m | 23.0; CH3 | |||
| 18d | 0.77; m | 20.7; CH3 | |||
| 19 | 172.5; C | ||||
| MeVal4 | 20c | 2.94; s | 29.9; CH3 | C-21 | |
| 21a | 5.09; d (11.0) | 55.3; CH | C-22, C-23, C-24 | H-22 | |
| 22 | 2.20; m | 26.6; CH | H-21 | ||
| 23b | 0.77; m | 17.8; CH3 | |||
| 24b | 0.77; m | 18.6; CH3 | |||
| 25e | 169.7; C | ||||
| Ile5 | NHa | 8.29; d (8.5) | C-25, C-26 | ||
| 26 | 4.52; t (9.3) | 52.6; CH | C-25, C-27, C-30, C-31 | ||
| 27 | 1.81; m | 35.8; CH | |||
| 28f | 1.43; m, 1.14; m | 23.9; CH2 | H-28a to C-29, C-30 | ||
| 29g | 0.77; m | 10.5; CH3 | |||
| 30h | 0.77; m | 14.8; CH3 | |||
| 31 | 172.0; C | ||||
| MeIle6 | 32 | 3.08; s | 30.5; CH3 | C-31, C-33 | |
| 4.79; d (11.0) | 58.7; CH | C-32, C-34 | H-34 | ||
| 34 | 1.80; m | 32.1; CH | H-33 | ||
| 35f | 1.43; m, 1.14; m | 23.9; CH2 | |||
| 36g | 0.77; m | 10.6; CH3 | |||
| 37h | 0.77; m | 14.8; CH3 | |||
| 38e | 169.6; C | ||||
| MeIle7 | 39c | 2.94; s | 29.8; CH3 | C-38 | |
| 40a | 5.12; d (11.0) | 53.8; CH | C-41, C-45 | H-41 | |
| 41 | 2.03; m | 32.5; CH | H-40 | ||
| 42f | 1.23; m, 1.07; m | 23.4; CH2 | |||
| 43g | 0.77; m | 10.6; CH3 | |||
| 44h | 0.77; m | 14.6; CH3 | |||
| 45 | 170.1; C | ||||
| MeIle8OH | 46c | 2.94; s | 29.8; CH3 | C-45 | |
| 47 | 5.16; d (11.0) | 55.1; CH | C-48, C-52 | H-48 | |
| 48 | 2.03; m | 32.5; CH | H-47 | ||
| 49f | 1.23; m, 1.07; m | 23.3; CH2 | |||
| 50g | 0.77; m | 10.1; CH3 | |||
| 51h | 0.77; m | 14.4; CH3 | |||
| 52 | 170.0; C | ||||
| OHi | 10.78; bs |
Multiple proton signals appear due to rotation about amide bonds.
Interchangeable carbons.
Interchangeable carbons.
Interchangeable carbons.
Interchangeable carbons.
Interchangeable carbons.
Interchangeable carbons.
Interchangeable carbons.
In dioxane-d8; low-field diastereotopic protons are labeled “a”, high-field diastereotopic protons are labeled “b”.
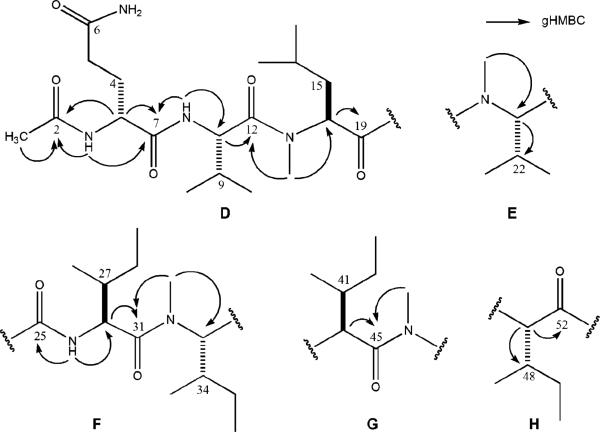
Exhaustive analysis of 2D-NMR for 2 including gHSQC, gHMQC, gHMBC, gCOSY, and TOCSY aided the construction of five substructures, D–H (Figure 4). Substructure D possessed the same terminal group as 1. This consisted of an N-Ac group attached to the Gln, established through gHMBC correlations from the acetyl methyl group (δ 1.81) to the acetyl carbonyl C-2 (δ 168.8) and from the secondary NH of Gln1 to C-2 and the amide carbonyl, C-7 (δ 171.2). Extending this unit into an Ac-Gln1-Val2-MeIle3 (D) array was based on the additional 2D-NMR correlations (Table 4). The first set (Figure 4) was from the Gln1 α proton, H-3 (δ 4.31) to C-2 and C-7 (δ 171.2) and from the Val2 NH (δ 8.13) to C-7, along with an additional correlation between the Val2 NH and the Val2 α carbon (δ 52.3). The connection between the second and third amino acids to complete substructure M was made through gHMBC correlations from H-8 to C-12 (δ 172.1), from N-MeLeu3 N-Me (δ 3.02) to C-12 and C-14 (δ 57.1), and from H-14 (δ 5.05) to C-19 (δ 172.5). The side-chain α and β proton assignments of 2 were also established through COSY (Table 4), while TOCSY data confirmed that the spin systems had been properly assigned for the δ and γ protons.
Figure 4.
Substructures D–H for RHM2 (2) illustrating key gHMBC correlations.
Three additional partial structures of N-Me amino acid subunits and one dipeptide subunit were identified in a straightforward manner on the basis of the data of Table 4. The methylated amino acids consisted of E as N-MeVal, and G and H as N-MeIles were each identified in a straightforward fashion. The remaining dipeptide substructure F, composed of Ile5-N-MeIle6, was established through gHMBC correlations from N-H (δ 8.29) to C-26 (δ 52.6) and C-25 (δ 169.7) and from Ile5 α proton H-26 (δ 4.52) to C-31 (δ 172.0), along with correlations from H3-32 (δ 3.08) to C-31 and C-33 (δ 58.7). At this point there were several ways to connect the five substructures of 2 (D–H). When no correlations were present (carbons 35, 42, and 49), protons were assigned on the basis of the shifts for parallel amino acids from compound 1.
Making the final unique choice among the several possible final structures for 2 was based primarily on ion fragmentation observed in ESIMS (Figure 2). The connectivities deduced for D by NMR matched the tripeptide required by the mass of the smallest fragment ion, m/z 397.3. Similar to the case for 1, the next highest fragment ion, m/z 632.4, required the addition of an N-MeVal and Ile to rationalize the mass increment of 226.1 amu. To adhere to the mass requirements, substructure E must be followed by G. Substructures D and F could not be consecutive due to the correlations of H-14 and NH of Ile5 to distinct carbonyls (C-19 and C-25, respectively). This allows the construction of the sequence D–E–F with only G and H undefined. The correlation from H3-46 (δ 2.94) to C-45 meant substructure G could not be the terminal amino acid in the molecule; therefore the sequence of the substructures for 2 was D–E–F–G–H with C-52 assigned as the carboxylic acid group. The configuration of each amino acid in 2 is postulated to be the same as 1 on the basis of the congruent origin of biosynthetic materials for 1 and 2, resulting in the sequence Ac-(R)-Glu-(S)-Val-(S)-N-MeLeu-(S)-N-Me-Val-(2S,3S)-Ile-(2S,3S)-N-Me-Ile-(2S,3S)-N-Me-Ile-(2S,3S)-N-Me-Ile-OH.
The remaining issue deserving comment for 2 was the 1H NMR differences between 1 and 2 in the region of NH and α proton shifts. Unexpectedly, a minimum set of four amide bond rotamers could be observed for 2, while two (one major, one minor) rotational isomers could be observed for 1. The differing circumstances can be seen by comparing the intense and small doublets in the ratio 85:15 observed for H-41 (δ 4.78) in 1 versus the cluster for 2 of at least four distinct H-33 doublets (δ 4.70–4.80) in the ratio of 38: 33:20:9 (A:B:C:D), as shown in Figure S19. Other distinct rotational isomers visible for 2 include three for the Ile5 NH (26:35:39) and Gln1 NH (32:47:21), two at the Val2 NH (55:45) and α proton H-40 (58:42), and four at H-21 (31:24:23:20). The presence of rotational isomers in 1 and 2 can also be observed in the 13C NMR (Figure S20). For example, each of the α and γ carbons of N-MeVal5 (C-28, C-30, C-31) of 1 demonstrated the minor rotamer, which was also observed in the 1H NMR. Similarly, at least three rotamers are visible in the 13C NMR of 2, which are most apparent at the N-MeVal4 and Val2 γ carbons (C-10, C-11 and C-23, C-24 in Figure S20), the N-MeLeu3 α carbon (C-14), and the N-Me of N-MeIle6 (C-32).
Data from a variable-temperature 1H NMR (VT NMR) experiment was sought to further define the presence of the rotamers discussed above for 1 and 2. The latter was chosen for this analysis because it contained several well-resolved proton signals. As expected, the apparent doubled doublet for H-40 (Figure S19) coalesced to a single doublet (J = 10.5 Hz, Figure S19) at 160 °C. The coalescence temperature of protons H-21, H-14, and H-33 was not reached due to limitations of the solvent (DMSO-d6). Another interesting feature of the VT NMR experiment was the eventual collapse and upfield shift of the NH protons (Ile5, Val2, and Gln1, Figure S19) due to a faster exchange rate at the increased temperature. The results of the VT NMR experiment suggest the four stable isomers we observed can be attributed to the arrangement of R groups in multiple sets of trans–trans, trans–cis, cis–trans, or cis–cis conformations.
The final step was to explore the bioactivity of the three compounds, each of which was isolated in sufficient amounts to facilitate further studies. The antimicrobial assay against S. epidermidis directed fractionation to RHM1 (1), which possesses antibiotic properties (MIC 25-50 μg/mL, Table 1). Efrapeptin G (3) was slightly less active against S. epidermidis (MIC 80 μg/mL, Table 1), while RHM2 (2) did not exhibit antimicrobial activity (MIC > 400 μg/mL, Table 1). Both 1 and 2 exhibited mild cytotoxicity against murine L1210 cells in a disk diffusion soft agar colony-forming assay18 (Table 1). Alternatively, 3 exhibited potent cytotoxicity against murine L1210 cells and against HCT-116 with an IC50 of 3.5 × 10–3 μg/mL. Developmental therapeutics work has been initiated on this compound including clonogenic studies, and these results will be reported elsewhere.
Previous studies suggest that the C-terminal blocking group may play a role in the biological activity of the efrapeptins,17 possibly accounting for the differential bioactivity against murine cell lines between efrapeptin G (3) and the RHMs (1 and 2). Efrapeptin G (3) is largely composed of unusual amino acids such as isovaline, aminoisobutyric acid, β-alanine, and pipecolic acid, which may also contribute to the differences in bioactivity. Efrapeptins are most likely produced by a nonribosomal peptide synthetase (NRPS), due to the presence of a high proportion of unusual amino acids and modified C- and N-termini. RHMs may also be the product of an NRPS, as indicated by the large number of methylated amino acids and the presence of (R)-glutamine.
Previously reported N-methylated peptides from marine-derived fungi include the cyclin-dependent kinase inhibitors dictyonamides22 from an unidentified fungus, cytotoxic scytalidmides23 from Scytalidium sp., and the aspergillamides24 produced by an Aspergillus. Compounds 1 and 2 are most similar to the dictyonamides, which are linear and contain a large proportion of N-methylated and aliphatic amino acids. However, 1 and 2 are markedly different from other peptides of this group by the presence of the (R)-glutamine. The production of chlorofusin by the fungi genera Fusarium,25 the trikoningins by Trichoderma,26 and integramides by Dendrodochium27 have all preceded the RHMs as natural products from fungi containing at least one (R)-amino acid. However, none of these strains were marine-derived, nor are the structures methylated at the amide nitrogens, making 1 and 2 the first N-methylated peptides with an (R)-amino acid from a marine-derived fungus.
Experimental Section
General Experimental Procedures
The NMR spectra for 1 were recorded at 500 MHz (1H) and 125 MHz (13C). NMR spectra for 2 were recorded at 600 MHz (1H) and 150 MHz (13C). The natural abundance 15N gHMBC experiment used the same pulse sequence as standard gHMBC experiments. High-resolution mass measurements were obtained from a FT-ICR hybrid mass spectrometer, which combines ion trap and Fourier transform cyclotron resonance technology into a single instrument. Additional mass spectra were acquired with a benchtop Mariner ESITOF mass spectrometer. Preparative HPLC was performed using columns of 6 μm ODS; semipreparative HPLC was performed using columns of 5 μm ODS.
Biological Materials
Strain 021172cKZ was cultured via previously reported methods5 from a Teichaxinella sp. marine sponge collected with scuba in 2002, during an expedition to Papua New Guinea (sponge collection no. 02172, voucher maintained at UCSC). Strain 021172cKZ was identified as Leucosphaerina indica (3% different) via molecular methods10 and as an atypical Acremonium sp. through morphology-based identification. When grown on potato dextrose agar media, the strain demonstrated large, creamy, white moist areas that were considered sporodochia. The white color was unusual, as most Acremonium spp. are buff to some shade of salmon. Microscopically, the conidiophores were branched, which is also unusual for Acremonium, and the conidia were pointed at both ends and contained two guttules (oil droplets).28 The strain is hereto referred to as a notably atypical Acremonium sp. due to the large genetic difference presented by the molecular method and its unusual morphological characteristics. The fungus is maintained as a cryopreserved glycerol stock at UCSC.
Culture Conditions
The large-scale culture (20 L) was grown in Czapek-Dox media made with filtered Monterey Bay seawater-based media adjusted to pH 7.3 with shaking (150 rpm) for 21 days at room temperature (28 °C).
Biological Assays
The disk diffusion soft agar colony-forming assay is used to identify extracts and pure compounds with potent cytotoxicity and solid tumor selectivity. Activity is expressed in zone unit differentials between solid tumor cells (murine Colon38 and human ColonH116 and LungH125) and leukemia (murine L1210, human CEM) or normal cells (CFU-GM). Selectivity is defined as a zone unit differential greater than or equal to 250.18 The antimicrobial assay is carried out in microtiter format as previously described.29
Extraction and Isolation
The broth and mycelia were separated through vacuum filtration and extracted independently: the broth with a column of XAD-16 resin, and the mycelia by soaking in 1 L of methanol three times, each for 24 h. The XAD-16 column was washed with water followed by methanol, and the eluent was concentrated in vacuo. The resulting crude oil was partitioned between 90% aqueous methanol and hexanes, followed by a 50% aqueous methanol–dichloromethane partition. Both the soft agar disk diffusion and antimicrobial assay of the crude extracts from 021172cKZ indicated that the broth- and mycelial-derived dichloromethane fractions (EFD and TFD, respectively) contained the active components. The EFD (1.44 g) was fractionated with preparatory HPLC (25 to 75% MeCN in H2O with 0.1% formic acid over 50 min) followed by two consecutive rounds of semipreparative HPLC (50 to 100% MeCN in H2O with 0.1% formic acid over 30 min, and 42 to 48% MeCN in H2O with 0.1% formic acid over 20 min) to afford 21.1 mg of 3 and 13.3 mg of 1. The TFD (4.76 g) was fractionated with preparatory HPLC (25 to 75% MeCN in H2O with 0.1% formic acid over 30 min), followed by semipreparative HPLC (50 to 88% MeCN in H2O with 0.1% formic acid) to afford 345 mg of 1 and 33.5 mg of 2.
RHM1 (1)
fine white crystals; [α]25D –125.0 (c 0.08, MeOH); UV (MeOH) λmax (log ε) 205 nm (4.33); 1H NMR (500 MHz, DMSO-d6) and 13C NMR (125 MHz, DMSO-d6), see Table 3; FTMS m/z [M + H]+ 1036.7386 (calcd for C53H98N9O11 1036.7391).
RHM2 (2)
fine white crystals; [α]25D –137.5 (c 0.08, MeOH); UV (MeOH) λmax (log ε) 207 nm (4.47); 1H NMR (600 MHz, DMSO-d6) and 13C NMR (150 MHz, DMSO-d6), see Table 4; FTMS m/z [M + H]+ 1022.7242 (calcd for C52H96N9O11 1022.7244).
Efrapeptin G (3)
white solid; 1H NMR (500 MHz, MeOH-d4) and 13C NMR (125 MHz, MeOH-d4), see Table 2; FTMS m/z [M]+ 1648.0914 (calcd for C83H143N18O16 1648.0924).
Preparation of (2S,3R)-N-Me-Ile and (2R,3S)-N-Me-Ile.30
The procedure consisted of combining N-Boc-(2R,3S)-Ile (155.6 mg) dissolved in THF under an N2 atmosphere with anhydrous CH3I (0.335 mL) at 0 °C followed by the addition of 60% oil suspension of NaH (70 mg) while gently stirring under a flow of N2. The resulting mixture was stirred for 16 h at room temperature under N2, then diluted with saturated aqueous NH4Cl (5 mL) and extracted with diethyl ether (5 mL). The organic layer was extracted with saturated aqueous NaHCO3–water (1:1) three times (2.5 mL). The aqueous layers were combined and adjusted to pH 3 with 10% citric acid and extracted with EtOAc (2.5 mL) two times. The organic layers were combined and concentrated through rotary evaporation, yielding 80.4 mg of pale yellow oil. The material was deprotected by dissolving 18.0 mg of the crude oil in 1 mL of CH2Cl2 and adding 1 mL of TFA while stirring under an N2 atmosphere at 0 °C for 1.5 h. The same procedure was carried out with N-Boc-(2S,3R)-Ile (131.4 mg), resulting in 70.8 mg of a pale yellow oil. The identity of the products was confirmed with 1H NMR and LCMS; the purities were found to be sufficient for use as standards for the Marfey's analysis.
Determination of R/S Configuration Using Marfey's Method.31
Approximately 1 mg of 1 was hydrolyzed with 1 mL of 6 N HCl at 110 °C for 20 h. The hydrolysate was evaporated to dryness and treated with the following: 0.100 mL of 1 M NaHCO3 and 0.050 mL of a 10 mg/mL solution of 1-fluoro-2,4-dinitrophenyl-5-l-alanine amide (l-FDAA). The mixture was heated at 80 °C for 30 min, cooled to room temperature, and quenched with 0.050 mL of 2 N HCl. Then 0.300 mL of MeCN was added and samples were evaluated with LCMS. A linear gradient with aqueous MeCN with 0.01 M TFA was run on an analytical C-18 RP column over 40 min (25 to 100%), and retention times of standards were compared with the derivatized hydrolysate of 1. Retention times (min) of standards: (S)-Gln (13.2), (R)-Gln (13.4), (2S,3S)-N-Me-Ile (24.2), (2R,3R)-N-Me-Ile (25.4), (2S,3R)-N-Me-Ile (24.3), (2R,3S)-N-Me-Ile (25.6), (2S,3S)-Ile (22.6), (2R,3R)-Ile (24.9), (2S,3R)-Ile (22.5), (2R,3S)-Ile (24.8), (S)-N-Me-Val (22.3), (R)-N-Me-Val (23.5), (S)-N-Me-Leu (23.6), (R)-N-Me-Leu (24.5), (S)-Glu (15.4), (R)-Glu (15.9). Retention times (min) for the hydrolysate of 1: (R)-Glu (15.9), (S)-N-Me-Val (22.4), (2S,3S)-Ile (22.6), (S)-N-Me-Leu (23.6), (2S,3S)-N-Me-Ile (24.1).
Supplementary Material
Chart 1.
Acknowledgment
This work was supported by the National Institutes of Health (RO1 CA 47135). We thank B. Gerstenberger for assistance with synthesis of N-Me amino acids, J. Cunniff and D. Kage at Thermo Electron for providing FTMS data, and N. Sevova at Notre Dame for providing FABMS data. We would also like to thank C. Bui for AA analysis, A. Amagata for assistance with fungal cultures, and S. Pomponi for sponge identification. Mr. M. Waddington, Accugenix, a division of Acculabs, INS, is acknowledged for molecular identification, and Dr. D. A. Sutton, Department of Pathology, University of Texas Health Science Center at San Antonio, is acknowledged for taxonomic identification of the fungal strain.
Footnotes
Supporting Information Available: 1H and 13C NMR and FABMS for compound 3, and 1H, 13C NMR, gHMBC, gCOSY, and TOCSY for compounds 1 and 2 along with VT NMR figures are available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.For a recent review of the entire chemistry of marine-derived fungi (273 compounds discovered to 2002), see: Bugni TS, Ireland CM. Nat. Prod. Rep. 2004;21:143–163. doi: 10.1039/b301926h.
- 2.a Clutterbuck PW, Oxford AE, Raistrick H, Smith G, Biochem J. 1932;26:1441–1458. doi: 10.1042/bj0261441. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Campbell IM, Calzadilla CH, McCorkindale NJ. Tetrahedron Lett. 1966;7:5107–5111. [Google Scholar]
- 3.a Cole RJ, Cox RH. Handbook of Toxic Fungal Metabolites. Academic Press; New York: 1981. [Google Scholar]; b Seebacher G, Mallinger R, Laufer G, Grimm M, Griesmacher A, Weigel G, Wolner E, Muller MM. AdV. Exp. Med. Biol. 1998;431:801–803. doi: 10.1007/978-1-4615-5381-6_154. [DOI] [PubMed] [Google Scholar]; c Covarrubias-Zúniga A, Gonzalez-Lucan A, Dominguez MM. Tetrahedron. 2003;59:1989–1994. [Google Scholar]
- 4.a Endo A, Hasumi K, Sakai K, Kanabe T. J. Antibiot. 1985;38:920–925. doi: 10.7164/antibiotics.38.920. [DOI] [PubMed] [Google Scholar]; b Rosen T, Heathcock C. Tetrahedron. 1986;42:4909–4951. [Google Scholar]; c McKenney JM. Clin. Pharm. 1988;7:21–36. [PubMed] [Google Scholar]
- 5.Cheng XC, Varoglu M, Abrell L, Crews P, Lobkovaky E, Clardy J. J. Org. Chem. 1994;59:6344–6348. [Google Scholar]
- 6.Numata A, Takahashi C, Matsushita T, Miyamoto T, Kawai K, Usami Y, Matsumura E, Inoue M, Ohishi H, Shingu T. Tetrahedron Lett. 1992;33:1621–1624. [Google Scholar]
- 7.Varoglu M, Corbett TH, Valeriote FA, Crews P. J. Org. Chem. 1997;62:7078–7079. doi: 10.1021/jo970568z. [DOI] [PubMed] [Google Scholar]
- 8.Isaka M, Suyarnsestakorn C, Tanticharoen M, Kongsaeree P, Thebtaranonth Y. J. Org. Chem. 2002;67:1561–1566. doi: 10.1021/jo010930g. [DOI] [PubMed] [Google Scholar]
- 9.a Numata A, Amagata T, Minoura K, Ito T. Tetrahedron Lett. 1997;38:5675–5678. [Google Scholar]; b Amagata T, Bio M, Ohta T, Minoura K, Numata A. J. Chem. Soc., Perkin Trans 1. 1998:3585–3599. [Google Scholar]
- 10.a Amagata T, Rath C, Rigo FF, Tarlov N, Tenney K, Valeriote FA, Crews P. J. Med. Chem. 2003;46:4342–4350. doi: 10.1021/jm030090t. [DOI] [PubMed] [Google Scholar]; b Laurent D, Guella G, Roquebert MF, Farinole F, Mancini I, Pietra F. Planta Med. 2000;66:63–66. doi: 10.1055/s-2000-11110. [DOI] [PubMed] [Google Scholar]
- 11.Abraham E. In: Cephalosporins and Penicillins: Chemistry and Biology. Flynn EH, editor. Academic Press; New York: 1972. [Google Scholar]
- 12.Chen CY, Imamura N, Nishijima M, Adachi K, Sakai M, Sano H. J. Antibiot. 1996;49:998–1005. doi: 10.7164/antibiotics.49.998. [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Lateff A, Konig GM, Fisch KM, Holler U, Jones PG, Wright AD. J. Nat. Prod. 2002;65:1605–1611. doi: 10.1021/np020128p. [DOI] [PubMed] [Google Scholar]
- 14.Verracarin A, isororidin A. and verrol-4-acetate; see ref 10b.
- 15.The oxepinamides and fumiquinazones: Belofsky GN, Anguera M, Jensen PR, Fenical WM. Chem. Eur. J. 2000;6:1355–1360. doi: 10.1002/(sici)1521-3765(20000417)6:8<1355::aid-chem1355>3.0.co;2-s.
- 16.The virescenosides come from both marine fungi: Afiyatullov SS, Kuznetsova TA, Isakov VV, Pivkin MV, Prokof'eva NG, Elyakov GB. J. Nat. Prod. 2000;63:848–850. doi: 10.1021/np9904004.Afiyatullov SS, Kalinovsky AI, Kuznetsova TA, Isakov VV, Pivkin MV, Dmitrenok PS, Elyakov GB. J. Nat. Prod. 2002;65:641–644. doi: 10.1021/np010503y.Afiyatullov SS, Kalinovsky AI, Kuznetsova TA, Pivkin MV, Pfrkof'eva NG, Dmitrenok PS, Elyakov GB. J. Nat. Prod. 2004;67:1047–1051. doi: 10.1021/np0305324.; and terrestrial fungi: Cagnoli-Bellavita N, Ceccherelli P, Mariani R, Polonsky J, Baskevitch Z. Eur. J. Biochem. 1970;15:356–559. doi: 10.1111/j.1432-1033.1970.tb01015.x.
- 17.Gupta S, Krasnoff SB, Roberts DW, Renvich JAA, Brinen LS, Clardy J. J. Org. Chem. 1992;57:2306–2313. 13C and 1H NMR comparisons in Table 2 are made to compound 11 in this publication. Drasnoff SB, Gupta S, St. Leger RJ, Renwick JAA, Roberts DW. J. InVertebr. Pathol. 1991;58:180–188.
- 18.Valeriote F, Grieshaber CK, Media J, Pietraszkewicz H, Hoffmann J, Pan M, McLaughline S. J. Exp. Ther. Oncol. 2002;2:228–236. doi: 10.1046/j.1359-4117.2002.01038.x. [DOI] [PubMed] [Google Scholar]
- 19.Krasnoff SB, Gupta S. J. Chem. Ecol. 1991;17:1953–1962. doi: 10.1007/BF00992580. [DOI] [PubMed] [Google Scholar]
- 20.For nomenclature of the ions, see: Roepstorff P, Fohlman J. Biomed. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109.
- 21.Pip can be found in the apicidins: Singh SB, Zink DL, Liesch JM, Mosely RT, Dombrowski AW, Bills GF, Darkin-Rattray JS, Schmatz DM, Goetz MA. J. Org. Chem. 2002;67:815–825. doi: 10.1021/jo016088w. and Sch 217048: Hegde VR, Puar MS, Chan TM, Dai P, Das PR, Patel M. 1998;63:9584–9586. β-Ala is found in the theonellapeptolides: Roy MC, Ohtani II, Ichiba T, Tanaka J, Satari R, Higa T. Tetrahedron. 2000;56:9079–9092.Pip Iva , and β-Ala are all present in adenopeptin: Hayakawa Y, Adachi M, Kim JW, Shin-Ya K, Seto H. Tetrahedron. 1998;54:15871–15878.
- 22.Komatsu K, Shigemori H, Kobayashi J. J. Org. Chem. 2001;66:6189–6192. doi: 10.1021/jo0156767. [DOI] [PubMed] [Google Scholar]
- 23.Tan LT, Cheng SC, Jensen PR, Fenical W. J. Org. Chem. 2003;68:8767–8773. doi: 10.1021/jo030191z. [DOI] [PubMed] [Google Scholar]
- 24.Toske SG, Jensen PR, Kauffman CA, Fenical W. Tetrahedron. 1998;54:13459–13466. [Google Scholar]
- 25.Duncan SJ, Gruschow S, Williams DH, McNicholas C, Purewal R, Hajek M, Gerliz M, Martin S, Wrigley SK, Morre M. J. Am. Chem. Soc. 2001;123:554–560. doi: 10.1021/ja002940p. [DOI] [PubMed] [Google Scholar]
- 26.Auvin-Guette C, Rebuffat S, Vuidepot I, Massias M, Bodo B. J. Chem. Soc., Perkin Trans. 1. 1993;2:249–255. [Google Scholar]
- 27.Singh SB, Herath K, Guan ZQ, Zink DL, Dombrowski AW, Polishook JD, Silverman DC, Lingham RB, Felock PJ, Hazuda DJ. Org. Lett. 2002;4:249–255. doi: 10.1021/ol025540a. [DOI] [PubMed] [Google Scholar]
- 28.Sutton DA., Dr. Personal communication. Department of Pathology, University of Texas Health Science Center; [Google Scholar]
- 29.Ralifo P, Crews P. J. Org. Chem. 2004;69:9025–9029. doi: 10.1021/jo048789+. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki Y, Ojika M, Sakagami Y, Kaida K, Fudou R, Kameyama T. J. Antibiot. 2001;54:22–28. doi: 10.7164/antibiotics.54.22. [DOI] [PubMed] [Google Scholar]
- 31.Marfey P. Carlsberg Res. Commun. 1984;49:591–596. [Google Scholar]
- 32.Within the B and Y′ groups certain fragment ions appear 1 amu lower than expected (specified by * in Figure 1) due to loss of a proton via a molecular rearrangement. One possibility is through double-bond formation within the imminium group as previously noted in ref 17a. Fragment ions also appear 1 amu higher than expected in select Y[.minute] fragments (as indicated by ** in Figure 1).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.