Abstract
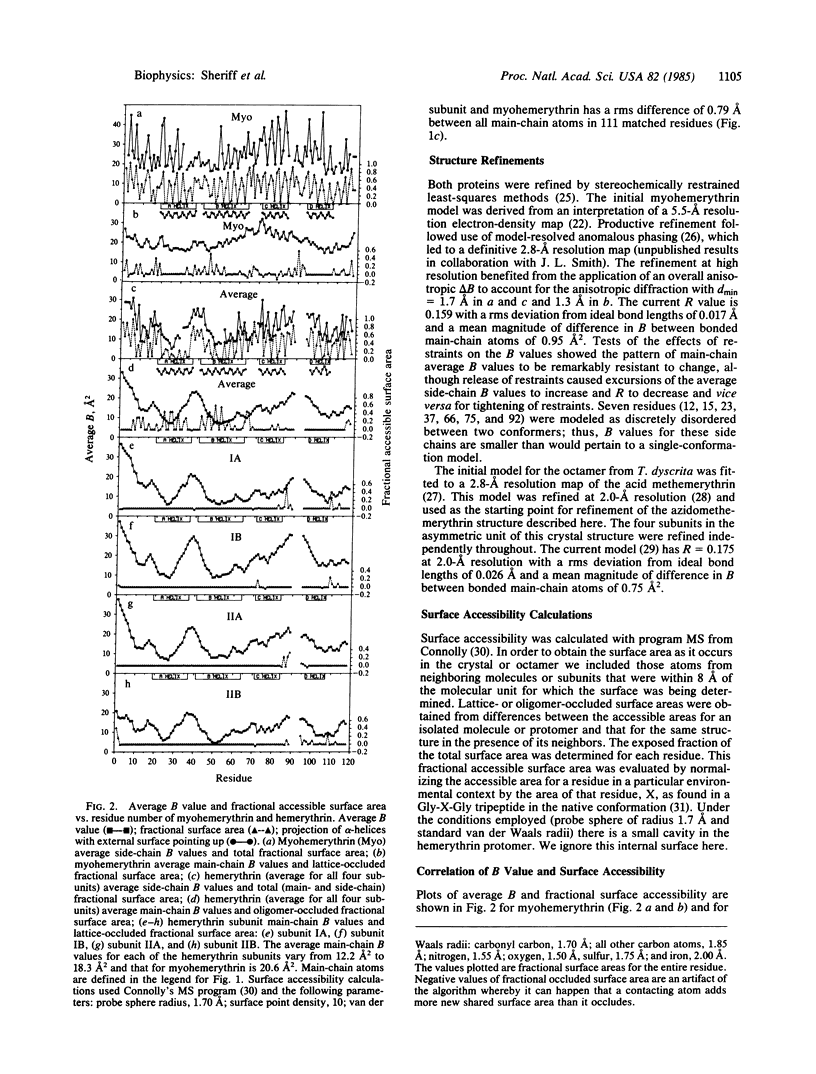
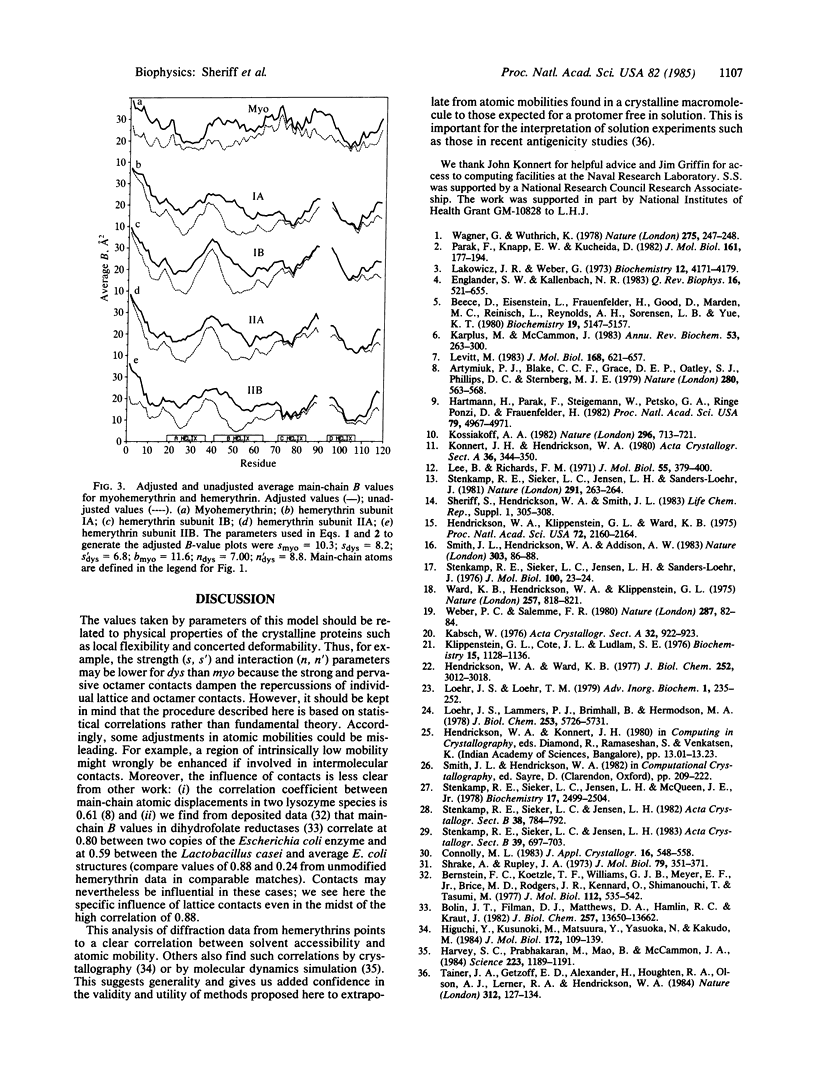
Thermal factor parameters (B values) have been compared from the refined crystal structures of the myohemerythrin from Themiste zostericola and of the octameric hemerythrin from Themiste dyscrita. These B values, which are directly related to atomic mobilities, were found to correlate rather closely with the solvent accessible areas within the respective crystals. Although protomeric units of the two molecules have exceptionally similar three-dimensional structures, there are marked differences between the patterns of relative atomic mobilities along the polypeptide chains. The differences correspond to lattice and oligomer contacts. An adjustment of the B values based on the fraction of accessible area occluded by contacts yields values that correlate well between the independent subunits and that should pertain more closely to those for the protomer free in solution.
Full text
PDF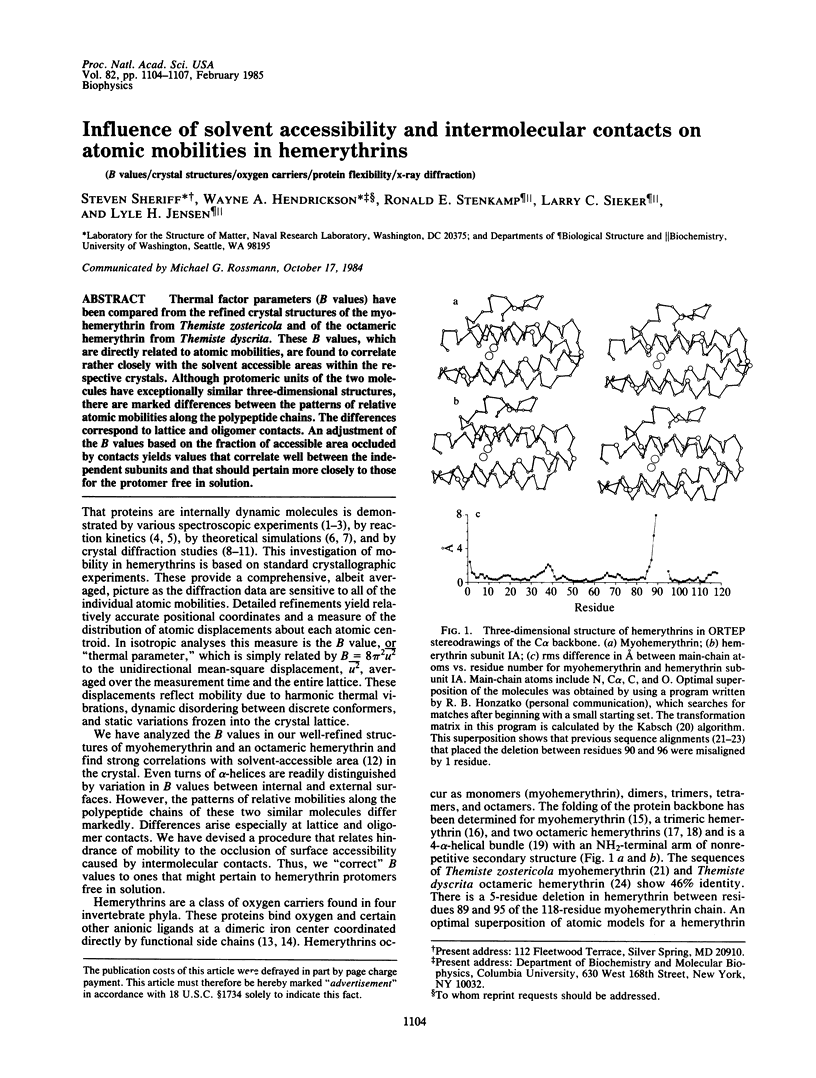

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artymiuk P. J., Blake C. C., Grace D. E., Oatley S. J., Phillips D. C., Sternberg M. J. Crystallographic studies of the dynamic properties of lysozyme. Nature. 1979 Aug 16;280(5723):563–568. doi: 10.1038/280563a0. [DOI] [PubMed] [Google Scholar]
- Beece D., Eisenstein L., Frauenfelder H., Good D., Marden M. C., Reinisch L., Reynolds A. H., Sorensen L. B., Yue K. T. Solvent viscosity and protein dynamics. Biochemistry. 1980 Nov 11;19(23):5147–5157. doi: 10.1021/bi00564a001. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. J Biol Chem. 1982 Nov 25;257(22):13650–13662. [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Hartmann H., Parak F., Steigemann W., Petsko G. A., Ponzi D. R., Frauenfelder H. Conformational substates in a protein: structure and dynamics of metmyoglobin at 80 K. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4967–4971. doi: 10.1073/pnas.79.16.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. C., Prabhakaran M., Mao B., McCammon J. A. Phenylalanine transfer RNA: molecular dynamics simulation. Science. 1984 Mar 16;223(4641):1189–1191. doi: 10.1126/science.6560785. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A., Klippenstein G. L., Ward K. B. Tertiary structure of myohemerythrin at low resolution. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2160–2164. doi: 10.1073/pnas.72.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A., Ward K. B. Pseudosymmetry in the structure of myohemerythrin. J Biol Chem. 1977 May 10;252(9):3012–3018. [PubMed] [Google Scholar]
- Higuchi Y., Kusunoki M., Matsuura Y., Yasuoka N., Kakudo M. Refined structure of cytochrome c3 at 1.8 A resolution. J Mol Biol. 1984 Jan 5;172(1):109–139. doi: 10.1016/0022-2836(84)90417-0. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Klippenstein G. L., Cote J. L., Ludlam S. E. The primary structure of myohemerythrin. Biochemistry. 1976 Mar 9;15(5):1128–1136. doi: 10.1021/bi00650a027. [DOI] [PubMed] [Google Scholar]
- Kossiakoff A. A. Protein dynamics investigated by the neutron diffraction-hydrogen exchange technique. Nature. 1982 Apr 22;296(5859):713–721. doi: 10.1038/296713a0. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Weber G. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry. 1973 Oct 9;12(21):4171–4179. doi: 10.1021/bi00745a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M. Molecular dynamics of native protein. II. Analysis and nature of motion. J Mol Biol. 1983 Aug 15;168(3):621–657. doi: 10.1016/s0022-2836(83)80306-4. [DOI] [PubMed] [Google Scholar]
- Loehr J. S., Lammers P. J., Brimhall B., Hermodson M. A. Amino acid sequence of hemerythrin from Themiste dyscritum. J Biol Chem. 1978 Aug 25;253(16):5726–5731. [PubMed] [Google Scholar]
- Parak F., Knapp E. W., Kucheida D. Protein dynamics. Mössbauer spectroscopy on deoxymyoglobin crystals. J Mol Biol. 1982 Oct 15;161(1):177–194. doi: 10.1016/0022-2836(82)90285-6. [DOI] [PubMed] [Google Scholar]
- Shrake A., Rupley J. A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J Mol Biol. 1973 Sep 15;79(2):351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- Smith J. L., Hendrickson W. A., Addison A. W. Structure of trimeric haemerythrin. Nature. 1983 May 5;303(5912):86–88. doi: 10.1038/303086a0. [DOI] [PubMed] [Google Scholar]
- Stenkamp R. E., Sieker L. C., Jensen L. H., Loehr J. S. Structure of methemerythrin at 5 A resolution. J Mol Biol. 1976 Jan 5;100(1):23–34. doi: 10.1016/s0022-2836(76)80031-9. [DOI] [PubMed] [Google Scholar]
- Stenkamp R. E., Sieker L. C., Jensen L. H., McQueen J. E., Jr Structure of methemerythrin at 2.8-Angstrom resolution: computer graphics fit of an averaged electron density map. Biochemistry. 1978 Jun 27;17(13):2499–2504. doi: 10.1021/bi00606a007. [DOI] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Alexander H., Houghten R. A., Olson A. J., Lerner R. A., Hendrickson W. A. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984 Nov 8;312(5990):127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Dynamic model of globular protein conformations based on NMR studies in solution. Nature. 1978 Sep 21;275(5677):247–248. doi: 10.1038/275247a0. [DOI] [PubMed] [Google Scholar]
- Ward K. B., Hendrickson W. A., Klippenstein G. L. Quaternary and tertiary structure of haemerythrin. Nature. 1975 Oct 30;257(5529):818–821. doi: 10.1038/257818a0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Salemme F. R. Structural and functional diversity in 4-alpha-helical proteins. Nature. 1980 Sep 4;287(5777):82–84. doi: 10.1038/287082a0. [DOI] [PubMed] [Google Scholar]



